Overexpression of β-Ketoacyl CoA Synthase 2B.1 from Chenopodium quinoa Promotes Suberin Monomers’ Production and Salt Tolerance in Arabidopsis thaliana
Abstract
1. Introduction
2. Results
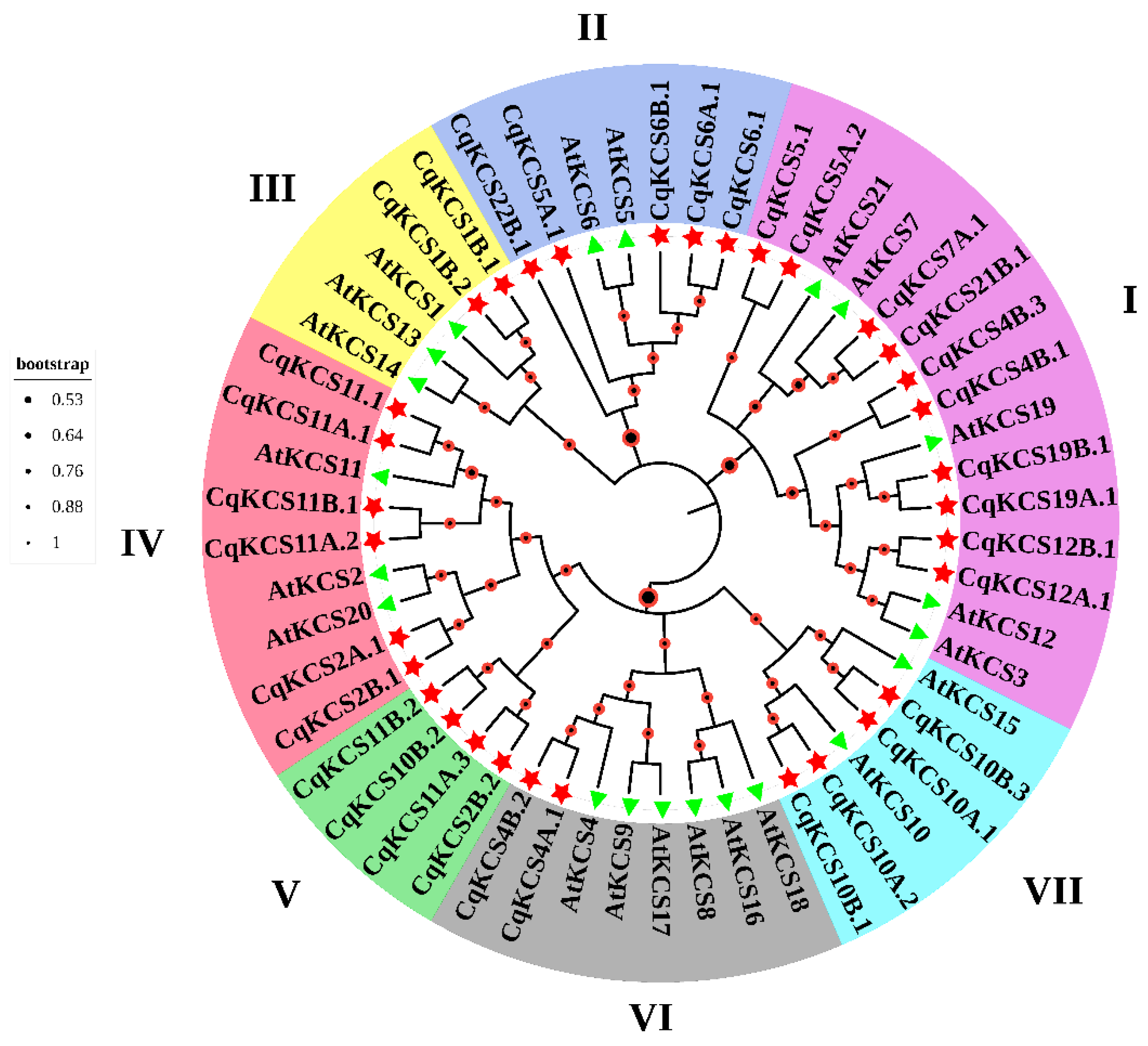
2.1. Phylogenetic Analysis of Quinoa CqKCS
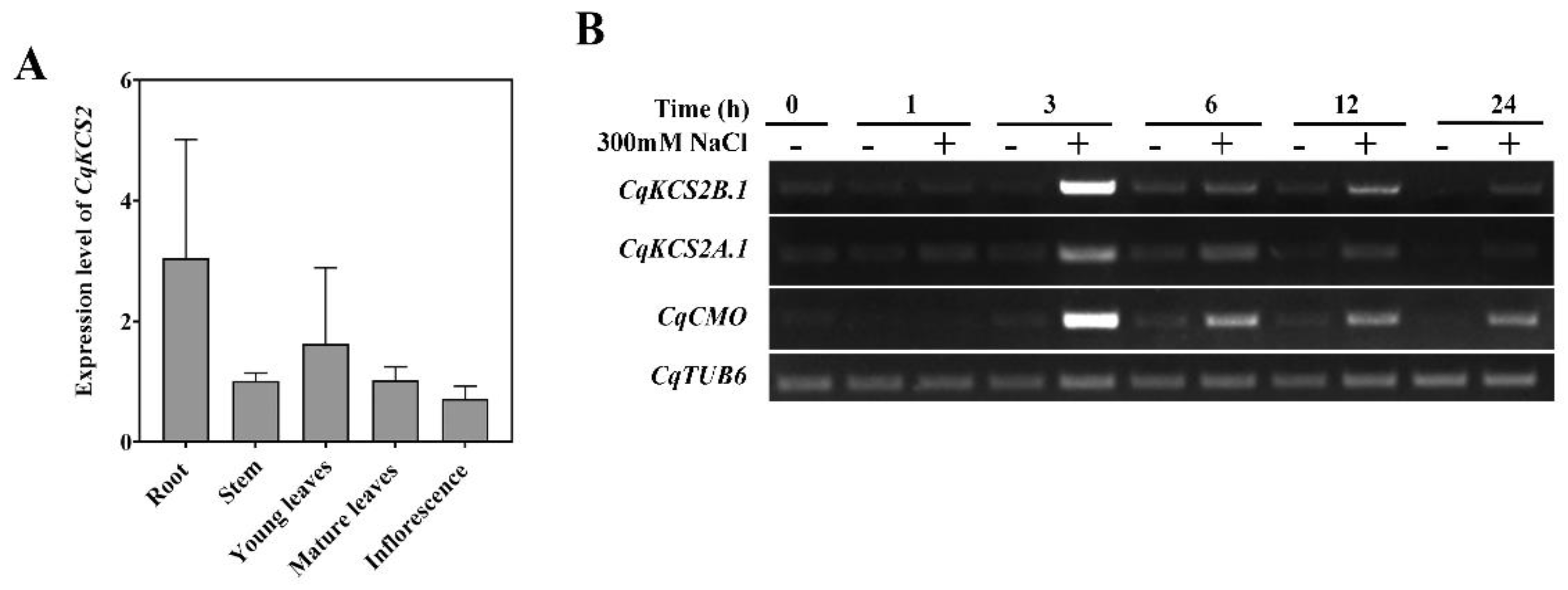
2.2. Expression Pattern of CqKCS2 in C. Quinoa
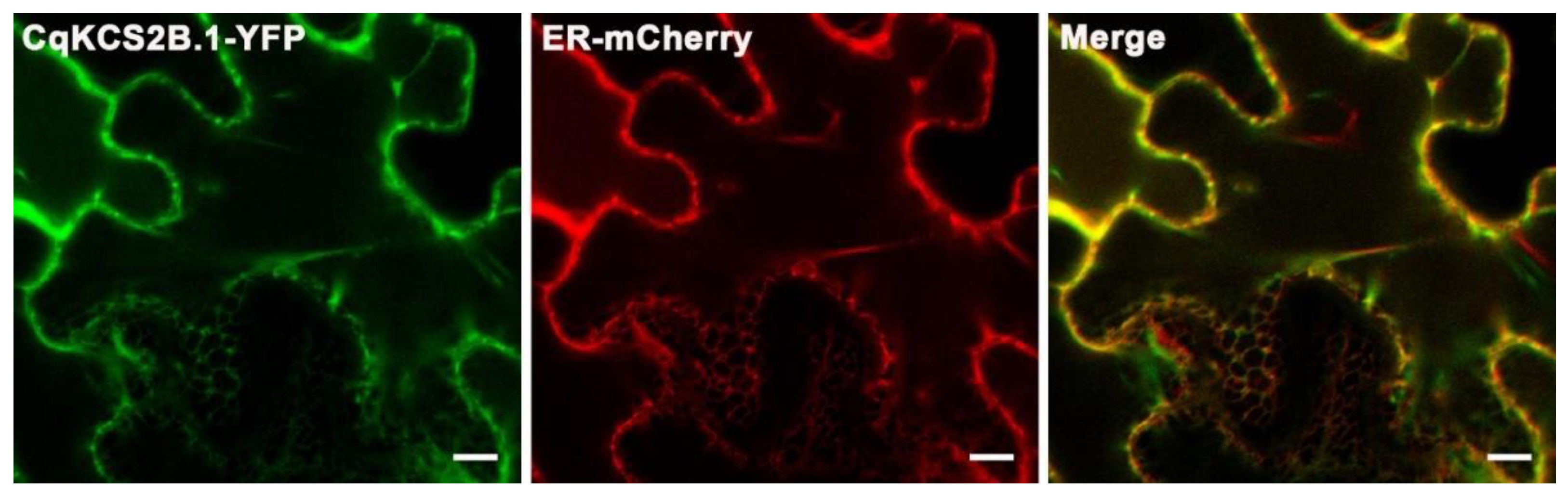
2.3. Localization of CqKCS2B.1 in the Endoplasmic Reticulum
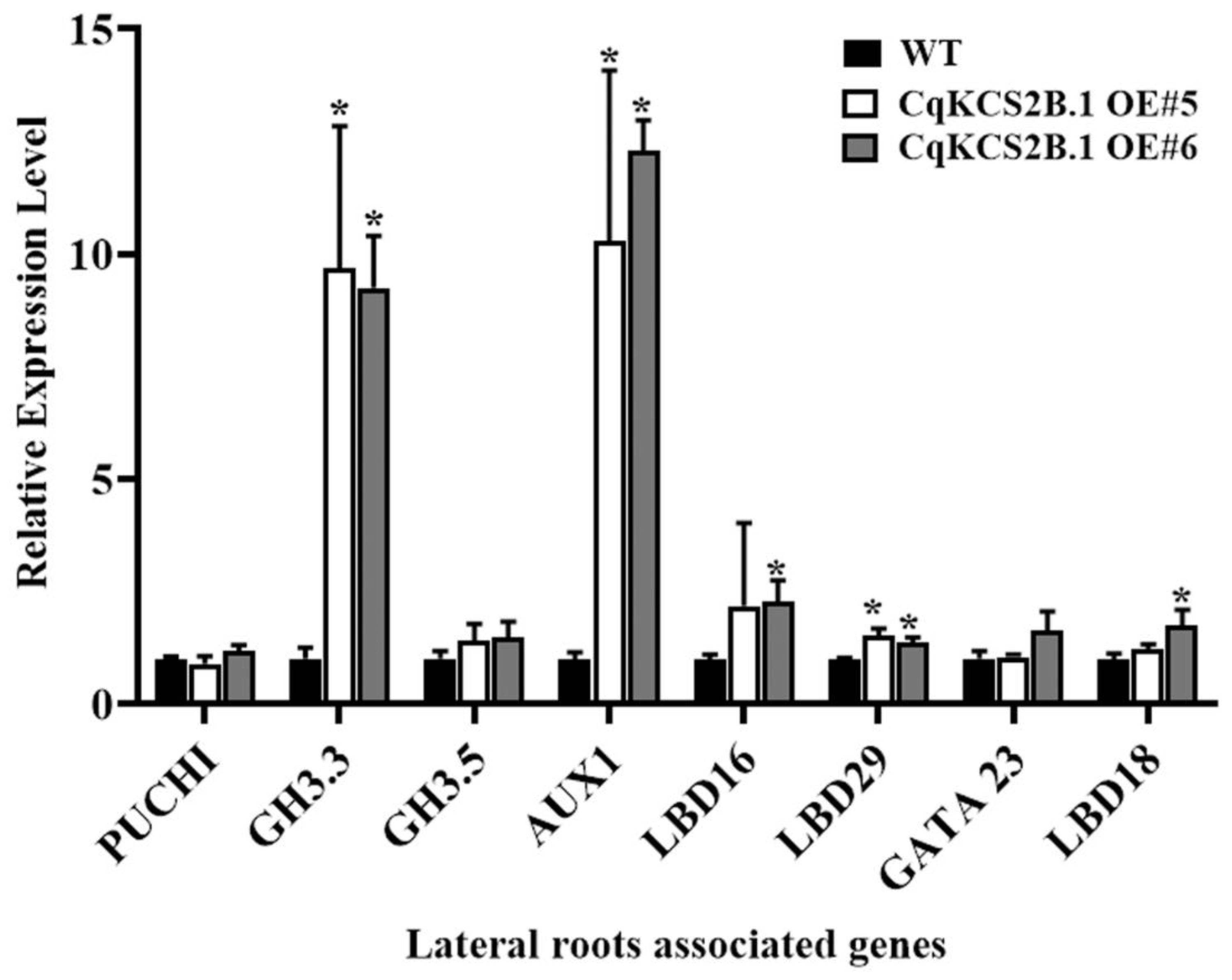
2.4. Ectopic Expression of CqKCS2B.1 Promotes Lateral Root Development in Arabidopsis
2.5. CqKCSB.1 Overexpression Alters Composition of Suberin Monomers in Arabidopsis Roots
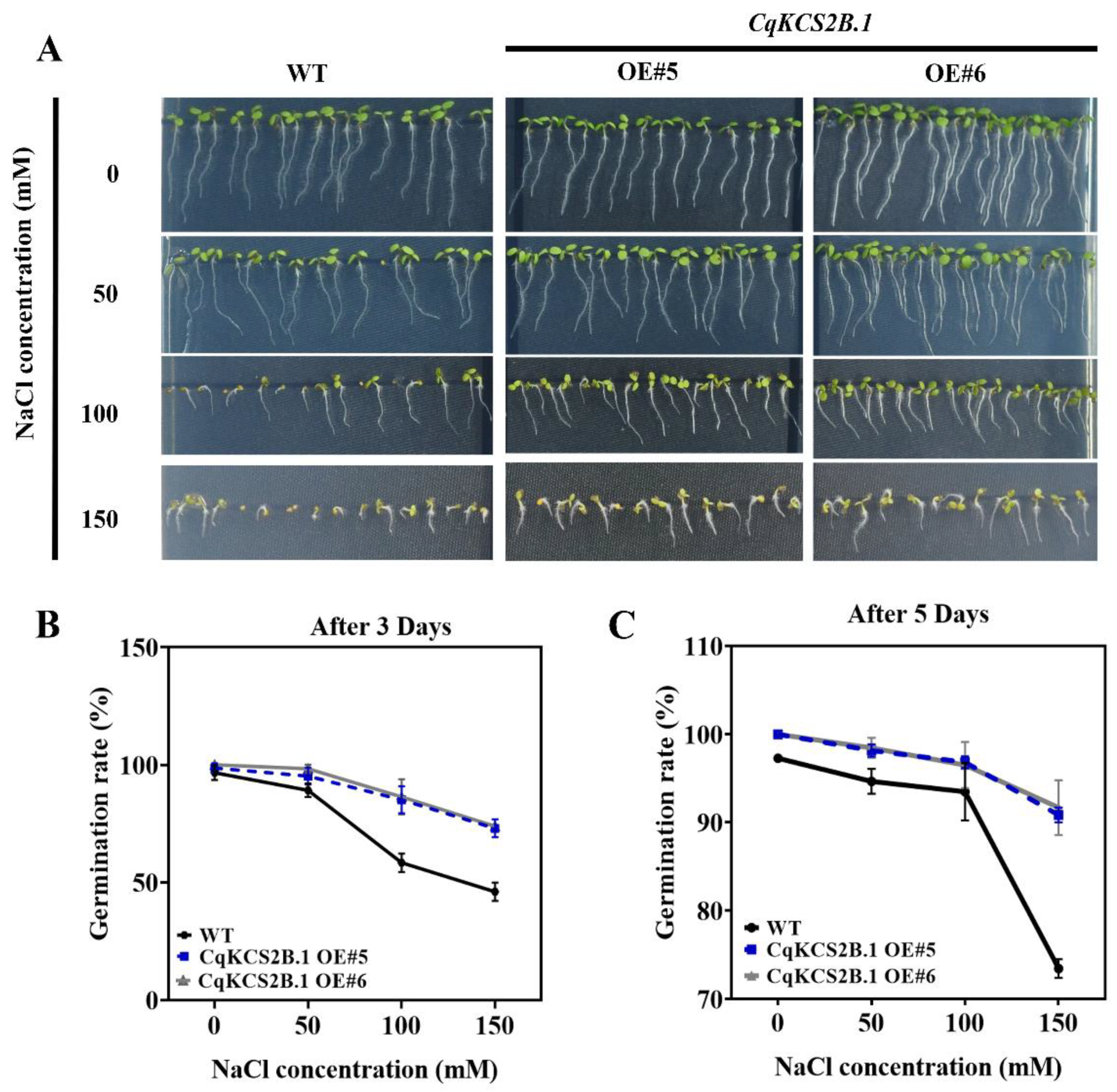
2.6. Ectopic Expression of CqKCS2B.1 Increases the Salt Stress Tolerance of Arabidopsis
3. Discussion
4. Materials and Methods
4.1. Identification of CqKCS Genes
4.2. Phylogenetic Analysis
4.3. Gene Duplication Analysis
4.4. Plant Materials and Growth Conditions
4.5. RNA Extraction, RT-PCR and RT-qPCR Analysis
4.6. Subcellular Localization of CqKCS2B.1
4.7. Generation of CqKCS2B.1 Overexpression Lines in Arabidopsis
4.8. Determination of Salt Tolerance of WT and CqKCS2B.1 OE Lines
4.9. Suberin Content Analysis in Plant Root
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Eigenbrode, S.D.; Espelie, K.E. Effects of plant epicuticular lipids on insect herbivores. Annu. Rev. Entomol. 1995, 40, 171–194. [Google Scholar] [CrossRef]
- Franke, R.; Schreiber, L. Suberin—A biopolyester forming apoplastic plant interfaces. Curr. Opin. Plant Biol. 2007, 10, 252–259. [Google Scholar] [CrossRef] [PubMed]
- Xiao, G.H.; Wang, K.; Huang, G.; Zhu, Y.X. Genome-scale analysis of the cotton KCS gene family revealed a binary mode of action for gibberellin A regulated fiber growth. J. Integr. Plant Biol. 2016, 58, 577–589. [Google Scholar] [CrossRef]
- Nobusawa, T.; Okushima, Y.; Nagata, N.; Kojima, M.; Sakakibara, H.; Umeda, M. Synthesis of very-long-chain fatty acids in the epidermis controls plant organ growth by restricting cell proliferation. PLoS Biol. 2013, 11, e1001531. [Google Scholar] [CrossRef]
- Bach, L.; Gissot, L.; Marion, J.; Tellier, F.; Moreau, P.; Satiat-Jeunemaitre, B.; Palauqui, J.C.; Napier, J.A.; Faure, J.D. Very-long-chain fatty acids are required for cell plate formation during cytokinesis in Arabidopsis thaliana. J. Cell Sci. 2011, 124, 3223–3234. [Google Scholar] [CrossRef] [PubMed]
- Raffaele, S.; Vailleau, F.; Leger, A.; Joubes, J.; Miersch, O.; Huard, C.; Blee, E.; Mongrand, S.; Domergue, F.; Roby, D. A MYB transcription factor regulates very-long-chain fatty acid biosynthesis for activation of the hypersensitive cell death response in Arabidopsis. Plant Cell 2008, 20, 752–767. [Google Scholar] [CrossRef]
- De Bigault Du Granrut, A.; Cacas, J.L. How very-long-chain fatty acids could signal stressful conditions in plants? Front. Plant Sci. 2016, 7, 1490. [Google Scholar] [CrossRef]
- Ivanova, T.V.; Maiorova, O.V.; Orlova, Y.V.; Kuznetsova, E.I.; Khalilova, L.A.; Myasoedov, N.A.; Balnokin, Y.V.; Tsydendambaev, V.D. Cell ultrastructure and fatty acid composition of lipids in vegetative organs of Chenopodium album L. under salt stress conditions. Russ. J. Plant Physiol. 2016, 63, 763–775. [Google Scholar] [CrossRef]
- Guo, W.; Wu, Q.; Yang, L.; Hu, W.; Liu, D.; Liu, Y. Ectopic Expression of CsKCS6 from Navel Orange Promotes the Production of Very-Long-Chain Fatty Acids (VLCFAs) and Increases the Abiotic Stress Tolerance of Arabidopsis thaliana. Front. Plant Sci. 2020, 11, 564656. [Google Scholar] [CrossRef]
- Sui, N.; Wang, Y.; Liu, S.; Yang, Z.; Wang, F.; Wan, S. Transcriptomic and physiological evidence for the relationship between unsaturated fatty acid and salt stress in peanut. Front. Plant Sci. 2018, 9, 7. [Google Scholar] [CrossRef]
- Beaudoin, F.; Wu, X.; Li, F.; Haslam, R.P.; Markham, J.E.; Zheng, H.; Napier, J.A.; Kunst, L. Functional characterization of the Arabidopsis beta-ketoacyl-coenzyme A reductase candidates of the fatty acid elongase. Plant Physiol. 2009, 150, 1174–1191. [Google Scholar] [CrossRef] [PubMed]
- Bach, L.; Michaelson, L.V.; Haslam, R.; Bellec, Y.; Gissot, L.; Marion, J.; Da Costa, M.; Boutin, J.P.; Miquel, M.; Tellier, F.; et al. The very-long-chain hydroxy fatty acyl-CoA dehydratase PASTICCINO2 is essential and limiting for plant development. Proc. Natl. Acad. Sci. USA 2008, 105, 14727–14731. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Rowland, O.; Kunst, L. Disruptions of the Arabidopsis Enoyl-CoA reductase gene reveal an essential role for very-long-chain fatty acid synthesis in cell expansion during plant morphogenesis. Plant Cell 2005, 17, 1467–1481. [Google Scholar] [CrossRef] [PubMed]
- Denic, V.; Weissman, J.S. A molecular caliper mechanism for determining very long-chain fatty acid length. Cell 2007, 130, 663–677. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-B.; Jung, S.-J.; Go, Y.-S.; Kim, H.-U.; Kim, J.-K.; Cho, H.-J.; Park, O.K.; Suh, M.-C. Two Arabidopsis 3-ketoacyl-CoA synthase genes, KCS20 and KCS2/DAISY, are functionally redundant in cuticular wax and root suberin biosynthesis, but differentially controlled by osmotic stress. Plant J. 2009, 60, 462–475. [Google Scholar] [CrossRef]
- Hooker, T.S.; Millar, A.A.; Kunst, L. Significance of the expression of the CER6 condensing enzyme for cuticular wax production in Arabidopsis. Plant Physiol. 2002, 129, 1568–1580. [Google Scholar] [CrossRef]
- Pruitt, R.E.; Vielle-Calzada, J.P.; Ploense, S.E.; Grossniklaus, U.; Lolle, S.J. Fiddlehead, a gene required to suppress epidermal cell interactions in Arabidopsis, encodes a putative lipid biosynthetic enzyme. Proc. Natl. Acad. Sci. USA 2000, 97, 1311–1316. [Google Scholar] [CrossRef]
- Kajikawa, M.; Yamaoka, S.; Yamato, K.T.; Kanamaru, H.; Sakuradani, E.; Shimizu, S.; Fukuzawa, H.; Ohyama, K. Functional analysis of a β-Ketoacyl-CoA synthase gene, MpFAE2, by gene silencing in the Liverwort Marchantia polymorpha L. Biosci. Biotechnol. Biochem. 2003, 67, 605–612. [Google Scholar] [CrossRef]
- Huai, D.; Xue, X.; Li, Y.; Wang, P.; Li, J.; Yan, L.; Chen, Y.; Wang, X.; Liu, N.; Kang, Y.; et al. Genome-wide identification of peanut KCS genes reveals that AhKCS1 and AhKCS28 are involved in regulating VLCFA contents in seeds. Front. Plant Sci. 2020, 11, 406. [Google Scholar] [CrossRef]
- Yang, H.; Mei, W.; Wan, H.; Xu, R.; Cheng, Y. Comprehensive analysis of KCS gene family in Citrinae reveals the involvement of CsKCS2 and CsKCS11 in fruit cuticular wax synthesis at ripening. Plant Sci. 2021, 310, 110972. [Google Scholar] [CrossRef]
- Wang, X.; Guan, Y.; Zhang, D.; Dong, X.; Tian, L.; Qu, L.Q. A β-Ketoacyl-CoA Synthase Is Involved in Rice Leaf Cuticular Wax Synthesis and Requires a CER2-LIKE Protein as a Cofactor. Plant Physiol. 2017, 173, 944–955. [Google Scholar] [CrossRef] [PubMed]
- Zurita-Silva, A.; Fuentes, F.; Zamora, P.; Jacobsen, S.-E.; Schwember, A.R. Breeding quinoa (Chenopodium quinoa Willd.): Potential and perspectives. Mol. Breed. 2014, 34, 13–30. [Google Scholar] [CrossRef]
- Adolf, V.I.; Jacobsen, S.-E.; Shabala, S. Salt tolerance mechanisms in quinoa (Chenopodium quinoa Willd.). Environ. Exp. Bot. 2013, 92, 43–54. [Google Scholar] [CrossRef]
- Ruiz, K.B.; Biondi, S.; Martínez, E.A.; Orsini, F.; Antognoni, F.; Jacobsen, S.E. Quinoa–a Model Crop for Understanding Salt-tolerance Mechanisms in Halophytes. Plant Biosyst. 2015, 150, 357–371. [Google Scholar] [CrossRef]
- Mujica, A.; Jacobsen, S.-E. La quinua (Chenopodium quinoa Willd.) y sus parientes silvestres. J. Bót. eco. los Andes Centrales 2006, 32, 449–457. [Google Scholar]
- Jarvis, D.E.; Ho, Y.S.; Lightfoot, D.J.; Schmockel, S.M.; Li, B.; Borm, T.J.; Ohyanagi, H.; Mineta, K.; Michell, C.T.; Saber, N.; et al. The genome of Chenopodium quinoa. Nature 2017, 542, 307–312. [Google Scholar] [CrossRef]
- Schmockel, S.M.; Lightfoot, D.J.; Razali, R.; Tester, M.; Jarvis, D.E. Identification of putative transmembrane proteins involved in salinity tolerance in Chenopodium quinoa by integrating physiological data, RNAseq, and SNP analyses. Front. Plant Sci. 2017, 8, 1023. [Google Scholar] [CrossRef]
- Maughan, P.J.; Kolano, B.A.; Maluszynska, J.; Coles, N.D.; Bonifacio, A.; Rojas, J.; Coleman, C.E.; Stevens, M.R.; Fairbanks, D.J.; Parkinson, S.E.; et al. Molecular and cytological characterization of ribosomal RNA genes in Chenopodium quinoa and Chenopodium berlandieri. Genome 2006, 49, 825–839. [Google Scholar] [CrossRef]
- Kolano, B.; Gardunia, B.W.; Michalska, M.; Bonifacio, A.; Fairbanks, D.; Maughan, P.J.; Coleman, C.E.; Stevens, M.R.; Jellen, E.N.; Maluszynska, J. Chromosomal localization of two novel repetitive sequences isolated from the Chenopodium quinoa Willd. genome. Genome 2011, 54, 710–717. [Google Scholar] [CrossRef]
- Singh, R.P.; Jha, P.; Jha, P.N. The plant-growth-promoting bacterium Klebsiella sp. SBP-8 confers induced systemic tolerance in wheat (Triticum aestivum) under salt stress. J. Plant Physiol. 2015, 184, 57–67. [Google Scholar] [CrossRef]
- Hinojosa, L.; Gonzalez, J.A.; Barrios-Masias, F.H.; Fuentes, F.; Murphy, K.M. Quinoa abiotic stress responses: A Review. Plants 2018, 7, 106. [Google Scholar] [CrossRef] [PubMed]
- Orsini, F.; Accorsi, M.; Gianquinto, G.; Dinelli, G.; Antognoni, F.; Carrasco, K.B.R.; Martinez, E.A.; Alnayef, M.; Marotti, I.; Bosi, S.; et al. Beyond the ionic and osmotic response to salinity in Chenopodium quinoa: Functional elements of successful halophytism. Funct. Plant Biol. 2011, 38, 818–831. [Google Scholar] [CrossRef]
- Millar, A.A.; Clemens, S.; Zachgo, S.; Giblin, E.M.; Taylor, D.C.; Kunst, L. CUT1, an Arabidopsis gene required for cuticular wax biosynthesis and pollen fertility, encodes a very-long-chain fatty acid condensing enzyme. Plant Cell 1999, 11, 825–838. [Google Scholar] [CrossRef] [PubMed]
- Fiebig, A.; Mayfield, J.A.; Miley, N.L.; Chau, S.; Fischer, R.L.; Preuss, D. Alterations in CER6, a gene identical to CUT1, differentially affect long-chain lipid content on the surface of pollen and stems. Plant Cell 2000, 12, 2001–2008. [Google Scholar] [CrossRef] [PubMed]
- Todd, J.; Post-Beittenmiller, D.; Jaworski, J.G. KCS1 encodes a fatty acid elongase 3-ketoacyl-CoA synthase affecting wax biosynthesis in Arabidopsis thaliana. Plant J. 1999, 17, 119–130. [Google Scholar] [CrossRef]
- Shang, B.; Xu, C.; Zhang, X.; Cao, H.; Xin, W.; Hu, Y. Very-long-chain fatty acids restrict regeneration capacity by confining pericycle competence for callus formation in Arabidopsis. Proc. Natl. Acad. Sci. USA 2016, 113, 5101–5106. [Google Scholar] [CrossRef]
- Franke, R.; Hofer, R.; Briesen, I.; Emsermann, M.; Efremova, N.; Yephremov, A.; Schreiber, L. The DAISY gene from Arabidopsis encodes a fatty acid elongase condensing enzyme involved in the biosynthesis of aliphatic suberin in roots and the chalaza-micropyle region of seeds. Plant J. 2009, 57, 80–95. [Google Scholar] [CrossRef]
- Kim, J.; Jung, J.H.; Lee, S.B.; Go, Y.S.; Kim, H.J.; Cahoon, R.; Markham, J.E.; Cahoon, E.B.; Suh, M.C. Arabidopsis 3-ketoacyl-coenzyme a synthase9 is involved in the synthesis of tetracosanoic acids as precursors of cuticular waxes, suberins, sphingolipids, and phospholipids. Plant Physiol. 2013, 162, 567–580. [Google Scholar] [CrossRef]
- Reina-Pinto, J.J.; Voisin, D.; Kurdyukov, S.; Faust, A.; Haslam, R.P.; Michaelson, L.V.; Efremova, N.; Franke, B.; Schreiber, L.; Napier, J.A.; et al. Misexpression of Fatty Acid Elongation1 in the Arabidopsis epidermis induces cell death and suggests a critical role for phospholipase A2 in this process. Plant Cell 2009, 21, 1252–1272. [Google Scholar] [CrossRef]
- Rossak, M.; Smith, M.; Kunst, L. Expression of the FAE1 gene and FAE1 promoter activity in developing seeds of Arabidopsis thaliana. Plant Mol. Biol. 2001, 46, 717–725. [Google Scholar] [CrossRef]
- James, D.W., Jr.; Lim, E.; Keller, J.; Plooy, I.; Ralston, E.; Dooner, H.K. Directed tagging of the Arabidopsis Fatty Acid Elongation1 (FAE1) gene with the maize transposon activator. Plant Cell 1995, 7, 309–319. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhan, H.; Lu, J.; Xiong, S.; Yang, N.; Yuan, H.; Yang, Z.N. Tapetal 3-Ketoacyl-Coenzyme A synthases are involved in pollen coat lipid accumulation for pollen-stigma interaction in Arabidopsis. Front. Plant Sci. 2021, 12, 770311. [Google Scholar] [CrossRef] [PubMed]
- Lolle, S.J.; Berlyn, G.P.; Engstrom, E.M.; Krolikowski, K.A.; Reiter, W.D.; Pruitt, R.E. Developmental regulation of cell interactions in the Arabidopsis fiddlehead-1 mutant: A role for the epidermal cell wall and cuticle. Dev. Biol. 1997, 189, 311–321. [Google Scholar] [CrossRef] [PubMed]
- Jia, B.; Wang, Y.; Zhang, D.; Wu, X.; Yin, H.; Chen, S.; Guo, S. Screening of reference genes and analysis of gene expression under salt stress in Chenopodium quinoa. J. Yantai Univ. 2020, 33, 283–288. [Google Scholar] [CrossRef]
- Gutierrez, L.; Mongelard, G.; Flokova, K.; Pacurar, D.I.; Novak, O.; Staswick, P.; Kowalczyk, M.; Pacurar, M.; Demailly, H.; Geiss, G.; et al. Auxin controls Arabidopsis adventitious root initiation by regulating jasmonic acid homeostasis. Plant Cell 2012, 24, 2515–2527. [Google Scholar] [CrossRef]
- Marchant, A.; Bhalerao, R.; Casimiro, I.; Eklöf, J.; Casero, P.J.; Bennett, M.; Sandberg, G. AUX1 promotes lateral root formation by facilitating indole-3-acetic acid distribution between sink and source tissues in the Arabidopsis seedling. Plant Cell 2002, 14, 589–597. [Google Scholar] [CrossRef]
- Batsale, M.; Bahammou, D.; Fouillen, L.; Mongrand, S.; Joubes, J.; Domergue, F. Biosynthesis and functions of very-long-chain fatty acids in the responses of plants to abiotic and biotic Stresses. Cells 2021, 10, 1284. [Google Scholar] [CrossRef]
- Tresch, S.; Heilmann, M.; Christiansen, N.; Looser, R.; Grossmann, K. Inhibition of saturated very-long-chain fatty acid biosynthesis by mefluidide and perfluidone, selective inhibitors of 3-ketoacyl-CoA synthases. Phytochem 2012, 76, 162–171. [Google Scholar] [CrossRef]
- Lian, X.Y.; Gao, H.N.; Jiang, H.; Liu, C.; Li, Y.Y. MdKCS2 increased plant drought resistance by regulating wax biosynthesis. Plant Cell Rep. 2021, 40, 2357–2368. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, X.; Chen, Z.; Zhang, J.; Si, K.; Xu, R.; He, Y.; Zhu, F.; Cheng, Y. Function and transcriptional regulation of CsKCS20 in the elongation of very-long-chain fatty acids and wax biosynthesis in Citrus sinensis flavedo. Hortic. Res. 2022, 9, uhab027. [Google Scholar] [CrossRef]
- Bach, L.; Faure, J.-D. Role of very-long-chain fatty acids in plant development, when chain length does matter. C. R. Biol. 2010, 333, 361–370. [Google Scholar] [CrossRef] [PubMed]
- Savchenko, T.; Walley, J.W.; Chehab, E.W.; Xiao, Y.; Kaspi, R.; Pye, M.F.; Mohamed, M.E.; Lazarus, C.M.; Bostock, R.M.; Dehesh, K. Arachidonic acid: An evolutionarily conserved signaling molecule modulates plant stress signaling networks. Plant Cell 2010, 22, 3193–3205. [Google Scholar] [CrossRef] [PubMed]
- Fan, M.; Xu, C.; Xu, K.; Hu, Y. Lateral organ boundaries domain transcription factors direct callus formation in Arabidopsis regeneration. Cell Res. 2012, 22, 1169–1180. [Google Scholar] [CrossRef] [PubMed]
- Lavenus, J.; Goh, T.; Roberts, I.; Guyomarc’h, S.; Lucas, M.; De Smet, I.; Fukaki, H.; Beeckman, T.; Bennett, M.; Laplaze, L. Lateral root development in Arabidopsis: Fifty shades of auxin. Trends Plant Sci. 2013, 18, 450–458. [Google Scholar] [CrossRef]
- Roudier, F.; Gissot, L.; Beaudoin, F.; Haslam, R.; Michaelson, L.; Marion, J.; Molino, D.; Lima, A.; Bach, L.; Morin, H.; et al. Very-long-chain fatty acids are involved in polar auxin transport and developmental patterning in Arabidopsis. Plant Cell 2010, 22, 364–375. [Google Scholar] [CrossRef]
- Lv, B.; Wei, K.; Hu, K.; Tian, T.; Zhang, F.; Yu, Z.; Zhang, D.; Su, Y.; Sang, Y.; Zhang, X.; et al. MPK14-mediated auxin signaling controls lateral root development via ERF13-regulated very-long-chain fatty acid biosynthesis. Mol. Plant 2021, 14, 285–297. [Google Scholar] [CrossRef]
- Qin, Y.-M.; Hu, C.-Y.; Pang, Y.; Kastaniotis, A.J.; Hiltunen, J.K.; Zhu, Y.-X. Saturated Very-Long-Chain Fatty Acids Promote Cotton Fiber and Arabidopsis Cell Elongation by Activating Ethylene Biosynthesis. Plant Cell 2007, 19, 3692–3704. [Google Scholar] [CrossRef]
- Wang, Y.; Jin, S.; Xu, Y.; Li, S.; Zhang, S.; Yuan, Z.; Li, J.; Ni, Y. Overexpression of BnKCS1-1, BnKCS1-2, and BnCER1-2 promotes cuticular wax production and increases drought tolerance in Brassica napus. Crop J. 2020, 8, 26–37. [Google Scholar] [CrossRef]
- Smolko, A.; Bauer, N.; Pavlović, I.; Pěnčík, A.; Novák, O.; Salopek-Sondi, B. Altered Root Growth, Auxin Metabolism and Distribution in Arabidopsis thaliana Exposed to Salt and Osmotic Stress. Int. J. Mol. Sci. 2021, 22, 7993. [Google Scholar] [CrossRef]
- Baxter, I.; Hosmani, P.S.; Rus, A.; Lahner, B.; Borevitz, J.O.; Muthukumar, B.; Mickelbart, M.V.; Schreiber, L.; Franke, R.B.; Salt, D.E. Root suberin forms an extracellular barrier that affects water relations and mineral nutrition in Arabidopsis. PLoS Genet. 2009, 5, e1000492. [Google Scholar] [CrossRef]
- Lokesh, U.; Venkatesh, B.; Kiranmai, K.; Nareshkumar, A.; Amarnathareddy, V.; Rao, G.L.; Anthony Johnson, A.M.; Pandurangaiah, M.; Sudhakar, C. Overexpression of ß-Ketoacyl Co-A Synthase1 Gene Improves Tolerance of Drought Susceptible Groundnut (Arachis hypogaea L.) Cultivar K-6 by Increased Leaf Epicuticular Wax Accumulation. Front. Plant Sci. 2018, 9, 1869. [Google Scholar] [CrossRef]
- Mohamed-Yasseen, Y.; Barringer, S.A.; Splittstoesser, W.E.; Costanza, S. The role of seed coats in seed viability. Bot. Rev. 1994, 60, 426–439. [Google Scholar] [CrossRef]
- Beisson, F.; Li, Y.; Bonaventure, G.; Pollard, M.; Ohlrogge, J.B. The acyltransferase GPAT5 is required for the synthesis of suberin in seed coat and root of Arabidopsis. Plant Cell 2007, 19, 351–368. [Google Scholar] [CrossRef]
- Domergue, F.; Vishwanath, S.J.; Joubes, J.; Ono, J.; Lee, J.A.; Bourdon, M.; Alhattab, R.; Lowe, C.; Pascal, S.; Lessire, R.; et al. Three Arabidopsis fatty acyl-coenzyme A reductases, FAR1, FAR4, and FAR5, generate primary fatty alcohols associated with suberin deposition. Plant Physiol. 2010, 153, 1539–1554. [Google Scholar] [CrossRef] [PubMed]
- Cohen, H.; Dong, Y.; Szymanski, J.; Lashbrooke, J.; Meir, S.; Almekias-Siegl, E.; Zeisler-Diehl, V.V.; Schreiber, L.; Aharoni, A. A multilevel study of melon fruit reticulation provides insight into skin ligno-suberization hallmarks. Plant Physiol. 2019, 179, 1486–1501. [Google Scholar] [CrossRef] [PubMed]
- Legay, S.; Cocco, E.; Andre, C.M.; Guignard, C.; Hausman, J.F.; Guerriero, G. Differential lipid composition and gene expression in the semi-russeted “Cox Orange Pippin” Apple variety. Front. Plant Sci. 2017, 8, 1656. [Google Scholar] [CrossRef] [PubMed]
- Negi, B.; Salvi, P.; Bhatt, D.; Majee, M.; Arora, S. Molecular cloning, in-silico characterization and functional validation of monodehydroascorbate reductase gene in Eleusine coracana. PLoS ONE 2017, 12, e0187793. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Derbyshire, M.K.; Gonzales, N.R.; Lu, S.; Chitsaz, F.; Geer, L.Y.; Geer, R.C.; He, J.; Gwadz, M.; Hurwitz, D.I.; et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015, 43, D222–D226. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Nelson, B.K.; Cai, X.; Nebenfuhr, A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 2007, 51, 1126–1136. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chen, M.; Siemiatkowska, B.; Toleco, M.R.; Jing, Y.; Strotmann, V.; Zhang, J.; Stahl, Y.; Fernie, A.R. A highly efficient agrobacterium-mediated method for transient gene expression and functional studies in multiple plant species. Plant Commun. 2020, 1, 100028. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Alonso, J.M.; Hirayama, T.; Roman, G.; Nourizadeh, S.; Ecker, J.R. EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 1999, 284, 2148–2152. [Google Scholar] [CrossRef]
- Zaidi, M.A.; O’Leary, S.J.B.; Gagnon, C.; Chabot, D.; Wu, S.; Hubbard, K.; Tran, F.; Sprott, D.; Hassan, D.; Vucurevich, T.; et al. A triticale tapetal non-specific lipid transfer protein (nsLTP) is translocated to the pollen cell wall. Plant Cell Rep. 2020, 39, 1185–1197. [Google Scholar] [CrossRef]
- Lichtenthaler, H.K. Chlorophylls and carotenoids: Pigments of photosynthetic biomembranes. In Plant Cell Membranes; Methods in Enzymology; Academic Press: Cambridge, MA, USA, 1987; Volume 148, pp. 350–382. [Google Scholar]
- Jenkin, S.; Molina, I. Isolation and compositional analysis of plant cuticle lipid polyester monomers. J. Vis. Exp. 2015, 105, e53386. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tariq, F.; Zhao, S.; Ahmad, N.; Wang, P.; Shao, Q.; Ma, C.; Yang, X. Overexpression of β-Ketoacyl CoA Synthase 2B.1 from Chenopodium quinoa Promotes Suberin Monomers’ Production and Salt Tolerance in Arabidopsis thaliana. Int. J. Mol. Sci. 2022, 23, 13204. https://doi.org/10.3390/ijms232113204
Tariq F, Zhao S, Ahmad N, Wang P, Shao Q, Ma C, Yang X. Overexpression of β-Ketoacyl CoA Synthase 2B.1 from Chenopodium quinoa Promotes Suberin Monomers’ Production and Salt Tolerance in Arabidopsis thaliana. International Journal of Molecular Sciences. 2022; 23(21):13204. https://doi.org/10.3390/ijms232113204
Chicago/Turabian StyleTariq, Faheem, Shuangshuang Zhao, Naveed Ahmad, Pingping Wang, Qun Shao, Changle Ma, and Xianpeng Yang. 2022. "Overexpression of β-Ketoacyl CoA Synthase 2B.1 from Chenopodium quinoa Promotes Suberin Monomers’ Production and Salt Tolerance in Arabidopsis thaliana" International Journal of Molecular Sciences 23, no. 21: 13204. https://doi.org/10.3390/ijms232113204
APA StyleTariq, F., Zhao, S., Ahmad, N., Wang, P., Shao, Q., Ma, C., & Yang, X. (2022). Overexpression of β-Ketoacyl CoA Synthase 2B.1 from Chenopodium quinoa Promotes Suberin Monomers’ Production and Salt Tolerance in Arabidopsis thaliana. International Journal of Molecular Sciences, 23(21), 13204. https://doi.org/10.3390/ijms232113204








