Characterization of Glossy Spike Mutants and Identification of Candidate Genes Regulating Cuticular Wax Synthesis in Barley (Hordeum vulgare L.)
Abstract
1. Introduction
2. Results
2.1. Phenotypic and Physiological Analysis of the Spike Cuticular Wax-Deficient Cer-GN1 Mutant
2.2. Genetic Analysis of the Spike Cuticular Wax-Deficient Cer-GN1 Mutant
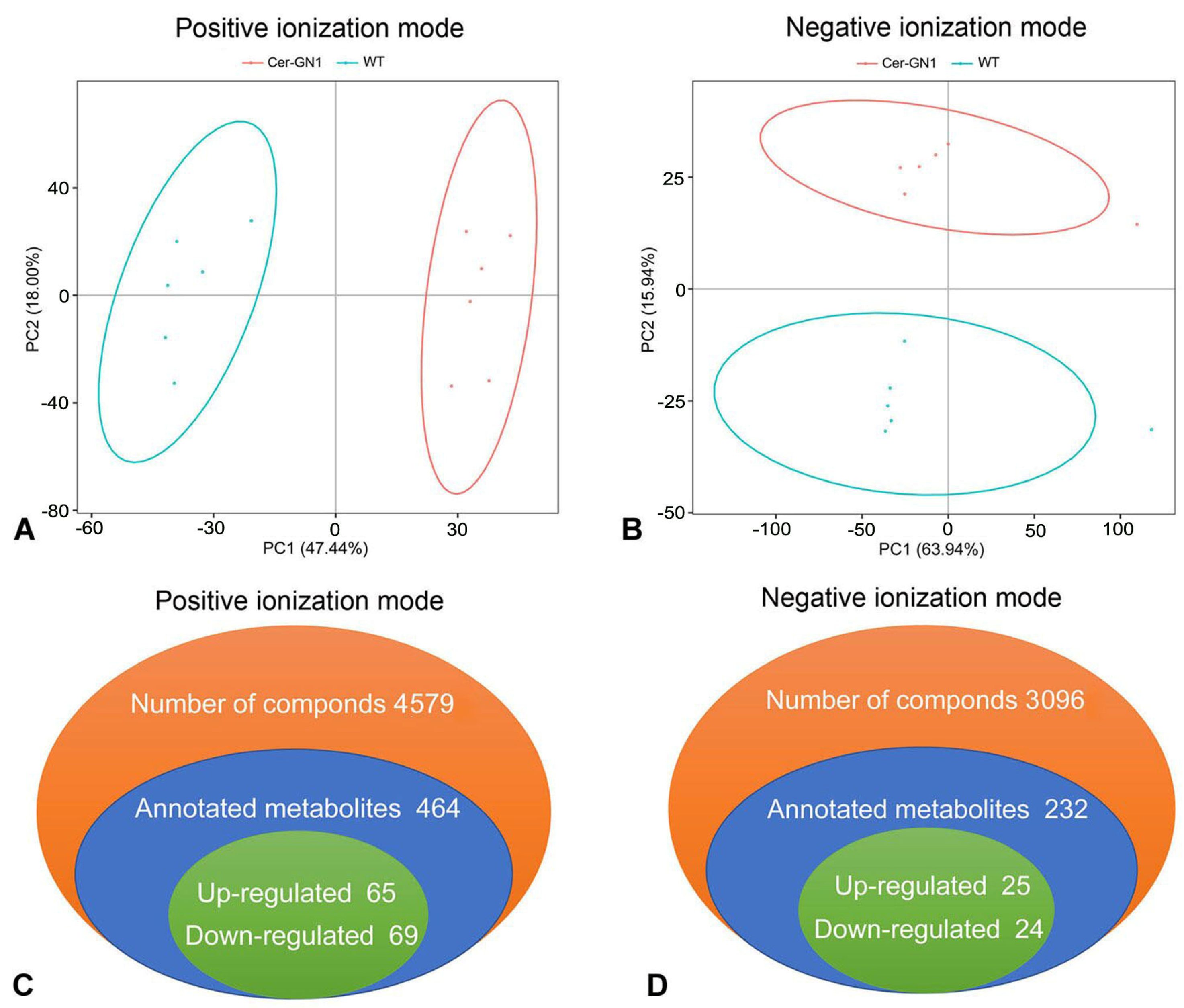
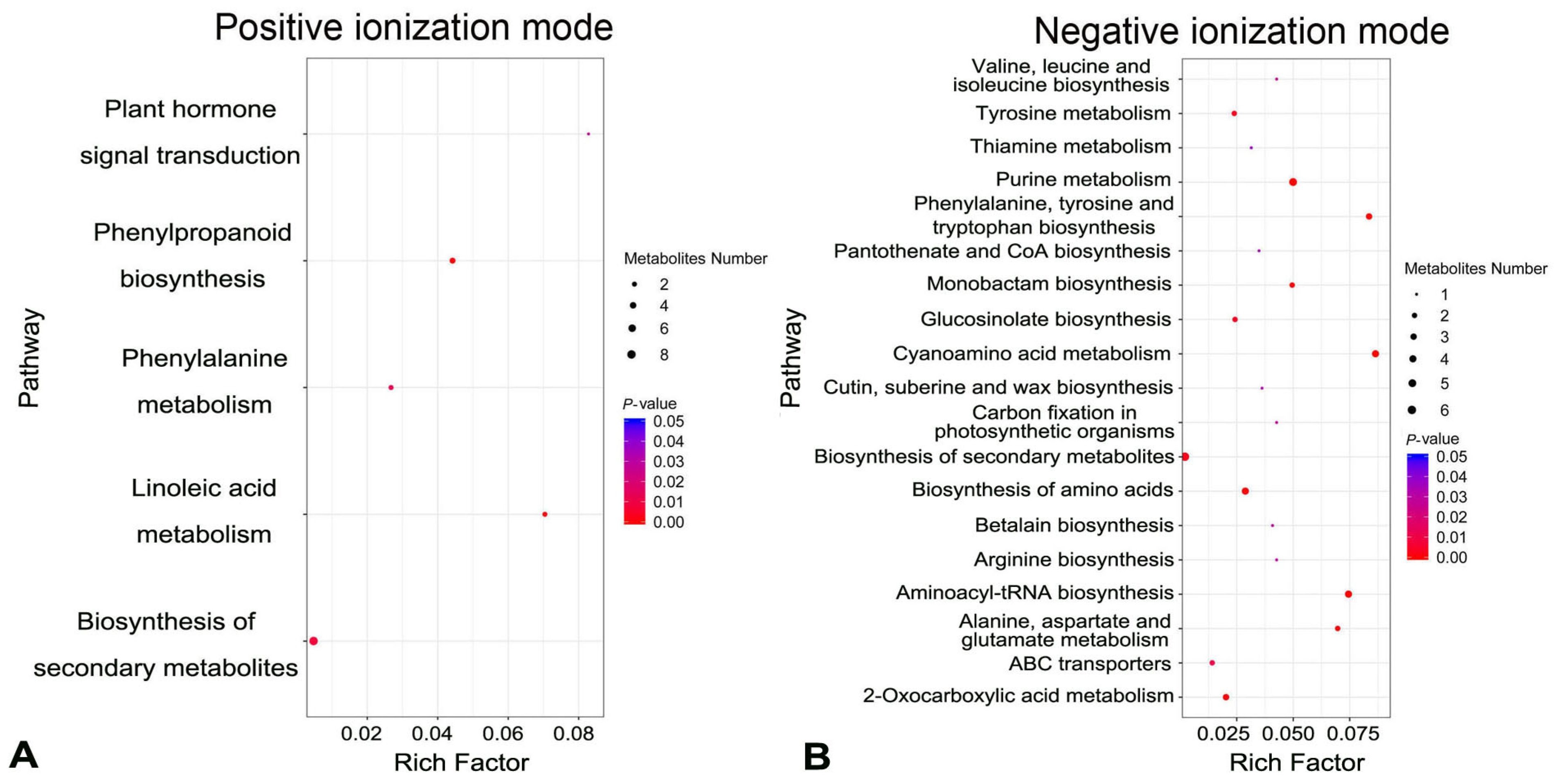
2.3. Identification of Differential Metabolites in Spike Glumes between Cer-GN1 and WT
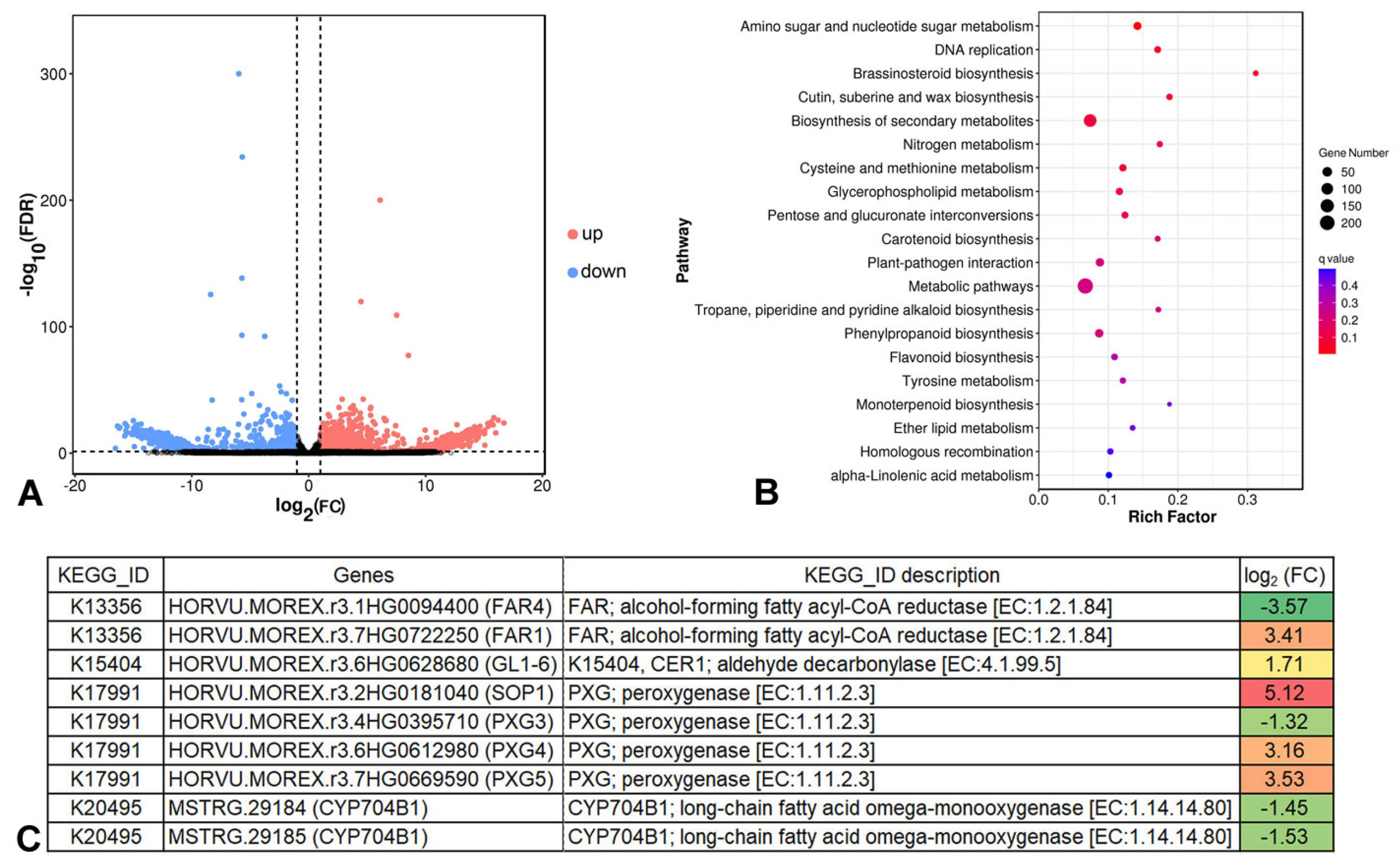
2.4. Global Analysis of DEGs in Cer-GN1 and WT Spike Glumes
2.5. Cer-GN1 DEGs Are Associated with Wax Biosynthesis
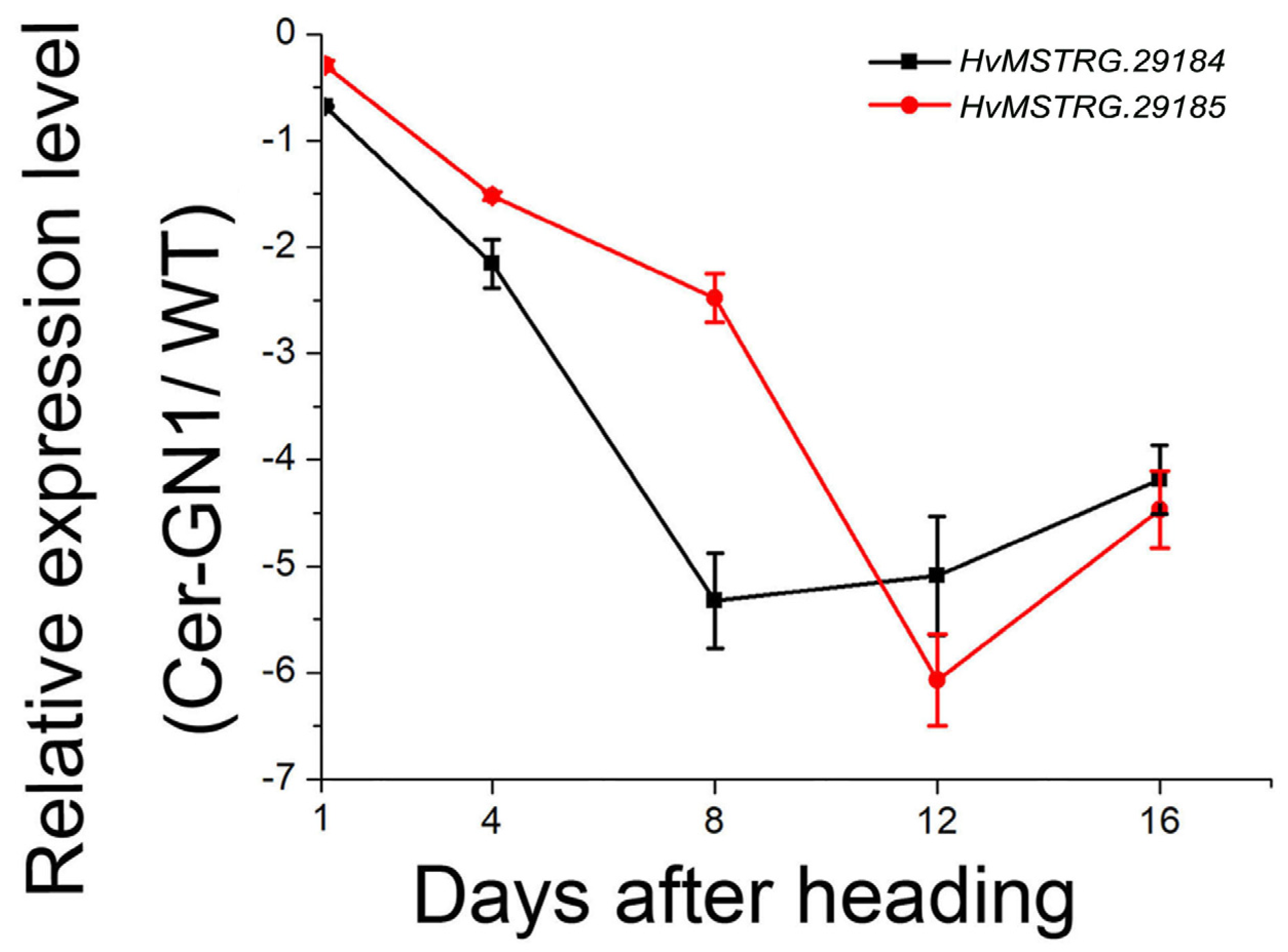
2.6. Verification of HvMSTRG.29184 and HvMSTRG.29185 Gene Expression by qRT-PCR
3. Discussion
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Spike Sampling and Analysis
4.3. Metabolite Extraction, Detection, and Analysis
4.4. RNA Extraction, Library Construction, and Sequencing
4.5. Bioinformatics Analysis
4.6. Quantitative RT-PCR Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Suzuki, N.; Rivero, R.M.; Shulaev, V.; Blumwald, E.; Mittler, R. Abiotic and biotic stress combinations. New Phytol. 2014, 203, 32–43. [Google Scholar] [CrossRef] [PubMed]
- Lewandowska, M.; Keyl, A.; Feussner, I. Wax biosynthesis in response to danger: Its regulation upon abiotic and biotic stress. New Phytol. 2020, 227, 698–713. [Google Scholar] [CrossRef] [PubMed]
- Riederer, M.; Schreiber, L. Protecting against water loss: Analysis of the barrier properties of plant cuticles. J. Exp. Bot. 2001, 52, 2023–2032. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Haslam, T.M.; Krüger, A.; Schneider, L.M.; Mishina, K.; Samuels, L.; Yang, H.; Kunst, L.; Schaffrath, U.; Nawrath, C. The β-ketoacyl-CoA synthase Hv KCS1, encoded by Cer-zh, plays a key role in synthesis of barley leaf wax and germination of barley powdery mildew. Plant Cell Physiol. 2018, 59, 811–827. [Google Scholar] [CrossRef] [PubMed]
- Kwadha, C.A.; Ong Amo, G.O.; Ndegwa, P.N.; Raina, S.K.; Fombong, A.T. The biology and control of the greater wax moth, Galleria mellonella. Insects 2017, 8, 61. [Google Scholar] [CrossRef]
- Bruhn, D.; Mikkelsen, T.N.; Rolsted, M.M.M.; Egsgaard, H.; Ambus, P. Leaf surface wax is a source of plant methane formation under UV radiation and in the presence of oxygen. Plant Biol. 2014, 16, 512–516. [Google Scholar] [CrossRef]
- Busta, L.; Budke, J.M.; Jetter, R. The moss Funaria hygrometrica has cuticular wax similar to vascular plants, with distinct composition on leafy gametophyte, calyptra and sporophyte capsule surfaces. Ann. Bot. 2016, 118, 511–522. [Google Scholar] [CrossRef]
- Kunst, L.; Samuels, A.L. Biosynthesis and secretion of plant cuticular wax. Prog. Lipid Res. 2003, 42, 51–80. [Google Scholar] [CrossRef]
- Yeats, T.H.; Rose, J.K. The formation and function of plant cuticles. Plant Physiol. 2013, 163, 5–20. [Google Scholar] [CrossRef]
- Von Wettstein-Knowles, P. Plant Waxes; John Wiley & Sons, Ltd.: Chichester, UK, 2016. [Google Scholar] [CrossRef]
- Kunst, L.; Samuels, L. Plant cuticles shine: Advances in wax biosynthesis and export. Curr. Opin. Plant Biol. 2009, 12, 721–727. [Google Scholar] [CrossRef]
- Pulsifer, I.P.; Kluge, S.; Rowland, O. Arabidopsis long-chain acyl-CoA synthetase 1 (LACS1), LACS2, and LACS3 facilitate fatty acid uptake in yeast. Plant Physiol. Bioch. 2012, 51, 31–39. [Google Scholar] [CrossRef]
- Fich, E.A.; Segerson, N.A.; Rose, J.K. The plant polyester cutin: Biosynthesis, structure, and biological roles. Annu. Rev. Plant Biol. 2016, 67, 207–233. [Google Scholar] [CrossRef]
- Philippe, G.; Sørensen, I.; Jiao, C.; Sun, X.; Fei, Z.; Domozych, D.S.; Rose, J.K. Cutin and suberin: Assembly and origins of specialized lipidic cell wall scaffolds. Curr. Opin. Plant Biol. 2020, 55, 11–20. [Google Scholar] [CrossRef]
- Von Wettstein-Knowles, P.M. Waxes, cutin, and suberin. In Lipid Metabolism in Plants; CRC Press: Boca Raton, FL, USA, 2018; pp. 127–166. [Google Scholar] [CrossRef]
- Von Wettstein-Knowles, P. Ecophysiology with barley eceriferum (cer) mutants: The effects of humidity and wax crystal structure on yield and vegetative parameters. Ann. Bot. 2020, 126, 301–313. [Google Scholar] [CrossRef]
- McAllister, T.; Campoli, C.; Eskan, M.; Liu, L.; McKim, S.M. A WAX INDUGer1/SHINE transcription factor controls cuticular wax in barley. bioRxiv 2022. [Google Scholar] [CrossRef]
- Fang, Y.; Zhang, X.; Tong, T.; Zhang, Z.; Zhang, X.; Tian, B.; Cui, J.; Zheng, J.; Xue, D. Physiological characterization and gene mapping of a novel cuticular wax-related mutant in barley (Hordeum vulgare L.). Plant Growth Regul. 2021, 93, 221–230. [Google Scholar] [CrossRef]
- Daszkowska-Golec, A.; Karcz, J.; Plociniczak, T.; Sitko, K.; Szarejko, I. Cuticular waxes-A shield of barley mutant in CBP20 (Cap-Binding Protein 20) gene when struggling with drought stress. Plant Sci. 2020, 300, 110593. [Google Scholar] [CrossRef]
- Lundqvist, U.; Wettstein-Knowles, P.V. Dominant mutations at Cer-yy change barley spike wax into leaf blade wax. Carlsberg Res. Commun. 1982, 47, 29–43. [Google Scholar] [CrossRef]
- Von Wettstein Knowles, P. Analyses of barley spike mutant waxes identify alkenes, cyclopropanes and internally branched alkanes with dominating isomers at carbon 9. Plant J. 2007, 49, 250–264. [Google Scholar] [CrossRef]
- Druka, A.; Franckowiak, J.; Lundqvist, U.; Bonar, N.; Alexander, J.; Houston, K.; Radovic, S.; Shahinnia, F.; Vendramin, V.; Morgante, M.; et al. Genetic dissection of barley morphology and development. Plant Physiol. 2011, 155, 617–627. [Google Scholar] [CrossRef]
- Koornneef, M.; Hanhart, C.J.; Thiel, F. A genetic and phenotypic description of Eceriferum (cer) mutants in Arabidopsis thaliana. J. Hered. 1989, 80, 118–122. [Google Scholar] [CrossRef]
- Sturaro, M.; Hartings, H.; Schmelzer, E.; Velasco, R.; Salamini, F.; Motto, M. Cloning and characterization of GLOSSY1, a maize gene involved in cuticle membrane and wax production. Plant Physiol. 2005, 138, 478–489. [Google Scholar] [CrossRef] [PubMed]
- Lundqvist, U.; Lundqvist, A. Mutagen specificity in barley for 1580 eceriferum mutants localized to 79 loci. Hereditas 1988, 108, 1–12. [Google Scholar] [CrossRef]
- Weidenbach, D.; Jansen, M.; Franke, R.B.; Hensel, G.; Weissgerber, W.; Ulferts, S.; Jansen, I.; Schreiber, L.; Korzun, V.; Pontzen, R.; et al. Evolutionary conserved function of barley and Arabidopsis 3-KETOACYL-CoA SYNTHASES in providing wax signals for germination of powdery mildew fungi. Plant Physiol. 2014, 166, 1621–1633. [Google Scholar] [CrossRef] [PubMed]
- Meusel, I.; Neinhuis, C.; Markstädter, C.; Barthlott, W. Chemical composition and recrystallization of epicuticular waxes: Coiled rodlets and tubules. Plant Biol. 2000, 2, 462–470. [Google Scholar] [CrossRef]
- Hen-Avivi, S.; Savin, O.; Racovita, R.C.; Lee, W.; Adamski, N.M.; Malitsky, S.; Almekias-Siegl, E.; Levy, M.; Vautrin, S.; Bergès, H.; et al. A metabolic gene cluster in the wheat W1 and the barley Cer-cqu loci determines β-diketone biosynthesis and glaucousness. Plant Cell 2016, 28, 1440–1460. [Google Scholar] [CrossRef]
- Zabka, V.; Stangl, M.; Bringmann, G.; Vogg, G.; Riederer, M.; Hildebrandt, U. Host surface properties affect prepenetration processes in the barley powdery mildew fungus. New Phytol. 2008, 177, 251–263. [Google Scholar] [CrossRef]
- Haslam, T.M.; Mañas-Fernández, A.; Zhao, L.; Kunst, L. Arabidopsis EGerIFERUM2 is a component of the fatty acid elongation machinery required for fatty acid extension to exceptional lengths. Plant Physiol. 2012, 160, 1164–1174. [Google Scholar] [CrossRef]
- Kong, L.; Liu, Y.; Zhi, P.; Wang, X.; Xu, B.; Gong, Z.; Chang, C. Origins and evolution of cuticle biosynthetic machinery in land plants. Plant Physiol. 2020, 184, 1998–2010. [Google Scholar] [CrossRef]
- Richardson, A.; Boscari, A.; Schreiber, L.; Kerstiens, G.; Jarvis, M.; Herzyk, P.; Fricke, W. Cloning and expression analysis of candidate genes involved in wax deposition along the growing barley (Hordeum vulgare) leaf. Planta 2007, 226, 1459–1473. [Google Scholar] [CrossRef]
- Kurokawa, Y.; Nagai, K.; Huan, P.D.; Shimazaki, K.; Qu, H.; Mori, Y.; Toda, Y.; Kuroha, T.; Hayashi, N.; Aiga, S. Rice leaf hydrophobicity and gas films are conferred by a wax synthesis gene (LGF 1) and contribute to flood tolerance. New Phytol. 2018, 218, 1558–1569. [Google Scholar] [CrossRef]
- Liu, N.; Chen, J.; Wang, T.; Li, Q.; Cui, P.; Jia, C.; Hong, Y. Overexpression of WAX INDUGer1/SHINE1 gene enhances wax accumulation under osmotic stress and oil synthesis in Brassica napus. Int. J. Mol. Sci. 2019, 20, 4435. [Google Scholar] [CrossRef]
- Dobritsa, A.A.; Shrestha, J.; Morant, M.; Pinot, F.; Matsuno, M.; Swanson, R.; Møller, B.L.; Preuss, D. CYP704B1 is a long-chain fatty acid ω-hydroxylase essential for sporopollenin synthesis in pollen of Arabidopsis. Plant Physiol. 2009, 151, 574–589. [Google Scholar] [CrossRef]
- Silva, J.; Sukweenadhi, J.; Myagmarjav, D.; Mohanan, P.; Yu, J.; Shi, J.; Jung, K.; Zhang, D.; Yang, D.; Kim, Y. Overexpression of a novel cytochrome P450 monooxygenase gene, CYP704B1, from Panax ginseng increase biomass of reproductive tissues in transgenic Arabidopsis. Mol. Biol. Rep. 2020, 47, 4507–4518. [Google Scholar] [CrossRef]
- Lai, Y. Characeration and Analysis of Wax Mutant Induced by EMS in Barley; Gansu Agricultural University: Lanzhou, China, 2014. [Google Scholar]
- Richardson, A.; Fricke, W.; Wojciechowski, T.; Schreiber, L.; Kerstiens, G. Cuticular permeance and its relation to profiles of waxes and cutin along the developing barley leaf. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2006, 143, S48–S49. [Google Scholar]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Mascher, M.; Wicker, T.; Jenkins, J.; Plott, C.; Lux, T.; Koh, C.S.; Ens, J.; Gundlach, H.; Boston, L.B.; Tulpová, Z. Long-read sequence assembly: A technical evaluation in barley. Plant Cell 2021, 33, 1888–1906. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Trevaskis, B.; Hemming, M.N.; Peacock, W.J.; Dennis, E.S. HvVRN2 responds to daylength, whereas HvVRN1 is regulated by vernalization and developmental status. Plant Physiol. 2006, 140, 1397–1405. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]



| Crossing Combination | F1 Plants | F2 Populations | ||||
|---|---|---|---|---|---|---|
| Total Plants | Glaucous Plants | Glossy Plants | χ2 3:1 | χ2 0.05 | ||
| Cer-GN1 × Mei 42 | Glaucous | 488 | 372 | 116 | 0.3307 | 3.84 |
| Cer-GN1 × Ganpi 6 | Glaucous | 552 | 417 | 129 | 0.0604 | 3.84 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bian, X.; Yao, L.; Si, E.; Meng, Y.; Li, B.; Ma, X.; Yang, K.; Lai, Y.; Shang, X.; Li, C.; et al. Characterization of Glossy Spike Mutants and Identification of Candidate Genes Regulating Cuticular Wax Synthesis in Barley (Hordeum vulgare L.). Int. J. Mol. Sci. 2022, 23, 13025. https://doi.org/10.3390/ijms232113025
Bian X, Yao L, Si E, Meng Y, Li B, Ma X, Yang K, Lai Y, Shang X, Li C, et al. Characterization of Glossy Spike Mutants and Identification of Candidate Genes Regulating Cuticular Wax Synthesis in Barley (Hordeum vulgare L.). International Journal of Molecular Sciences. 2022; 23(21):13025. https://doi.org/10.3390/ijms232113025
Chicago/Turabian StyleBian, Xiuxiu, Lirong Yao, Erjing Si, Yaxiong Meng, Baochun Li, Xiaole Ma, Ke Yang, Yong Lai, Xunwu Shang, Chengdao Li, and et al. 2022. "Characterization of Glossy Spike Mutants and Identification of Candidate Genes Regulating Cuticular Wax Synthesis in Barley (Hordeum vulgare L.)" International Journal of Molecular Sciences 23, no. 21: 13025. https://doi.org/10.3390/ijms232113025
APA StyleBian, X., Yao, L., Si, E., Meng, Y., Li, B., Ma, X., Yang, K., Lai, Y., Shang, X., Li, C., Wang, J., & Wang, H. (2022). Characterization of Glossy Spike Mutants and Identification of Candidate Genes Regulating Cuticular Wax Synthesis in Barley (Hordeum vulgare L.). International Journal of Molecular Sciences, 23(21), 13025. https://doi.org/10.3390/ijms232113025







