Fumonisin B1 as a Tool to Explore Sphingolipid Roles in Arabidopsis Primary Root Development
Abstract
1. Introduction
2. Results
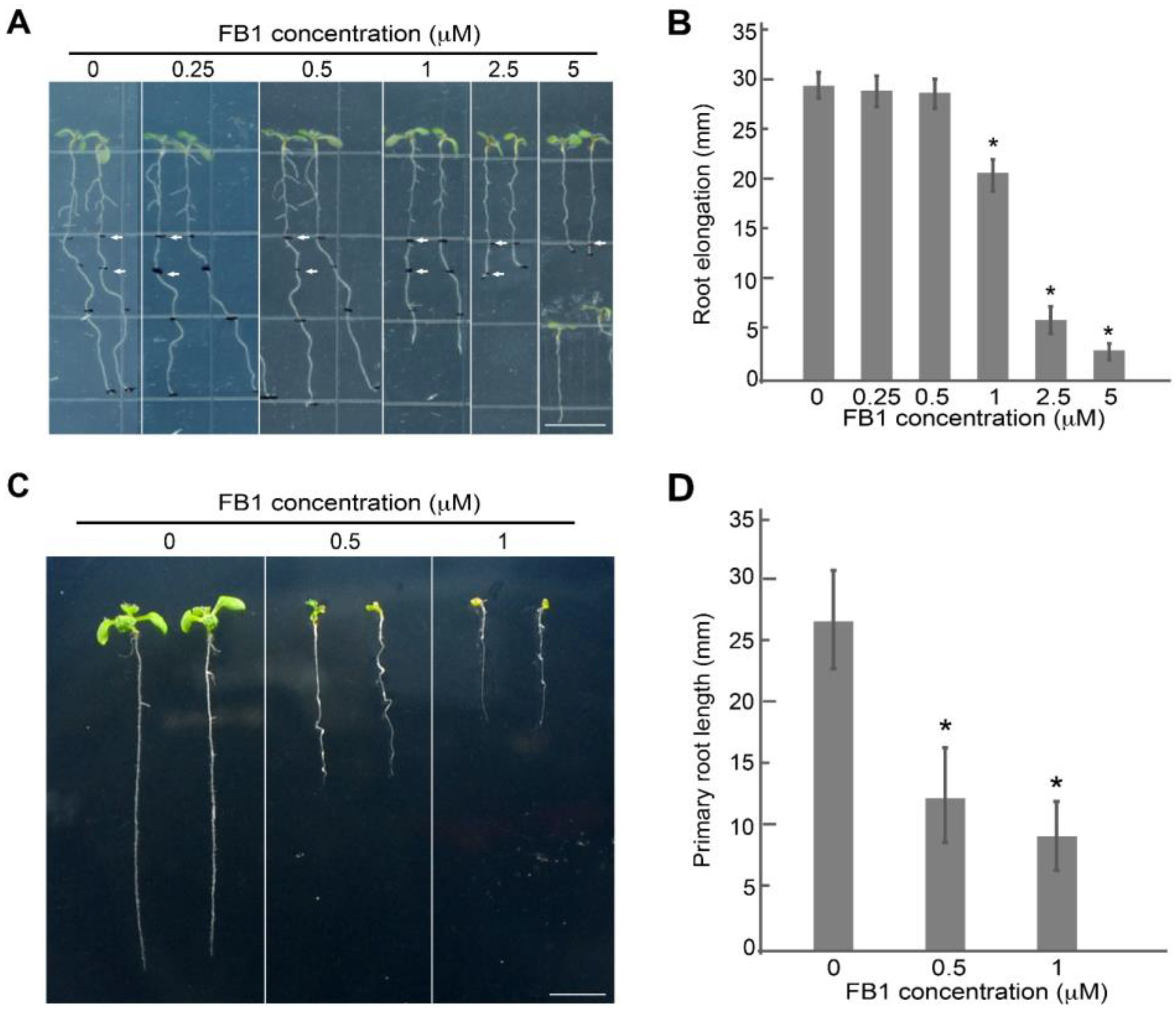
2.1. FB1 Inhibits Arabidopsis Primary Root Development
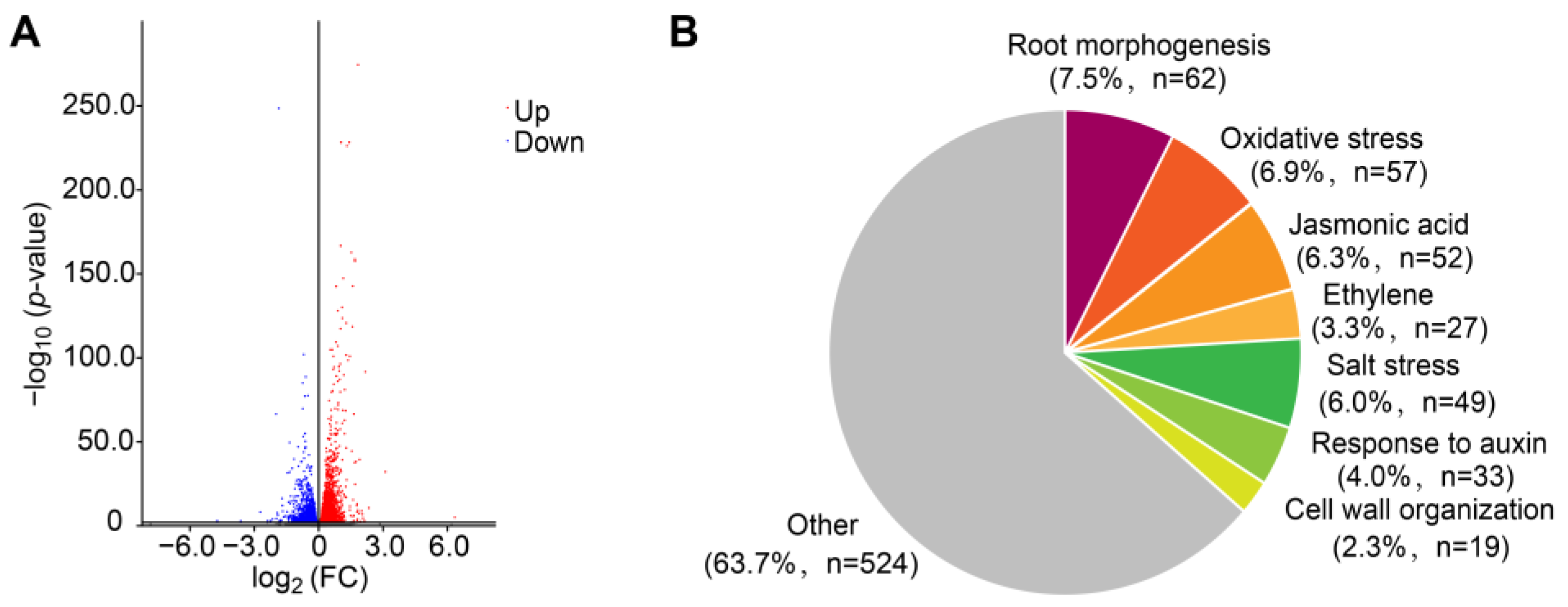
2.2. Transcriptome Analysis in Response to FB1 Treatment
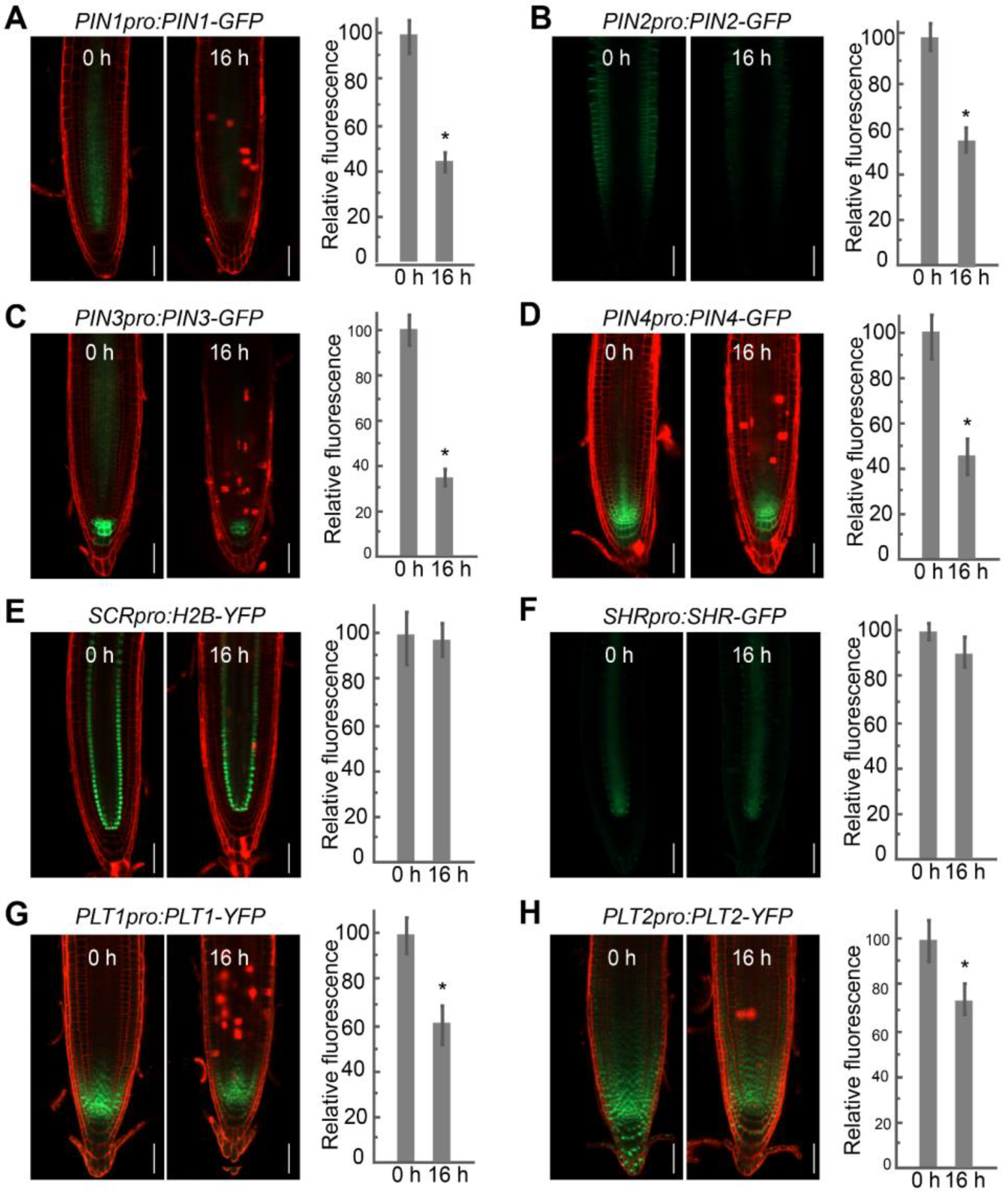
2.3. FB1 Alters PIN and PLT Expression Levels in the Roots
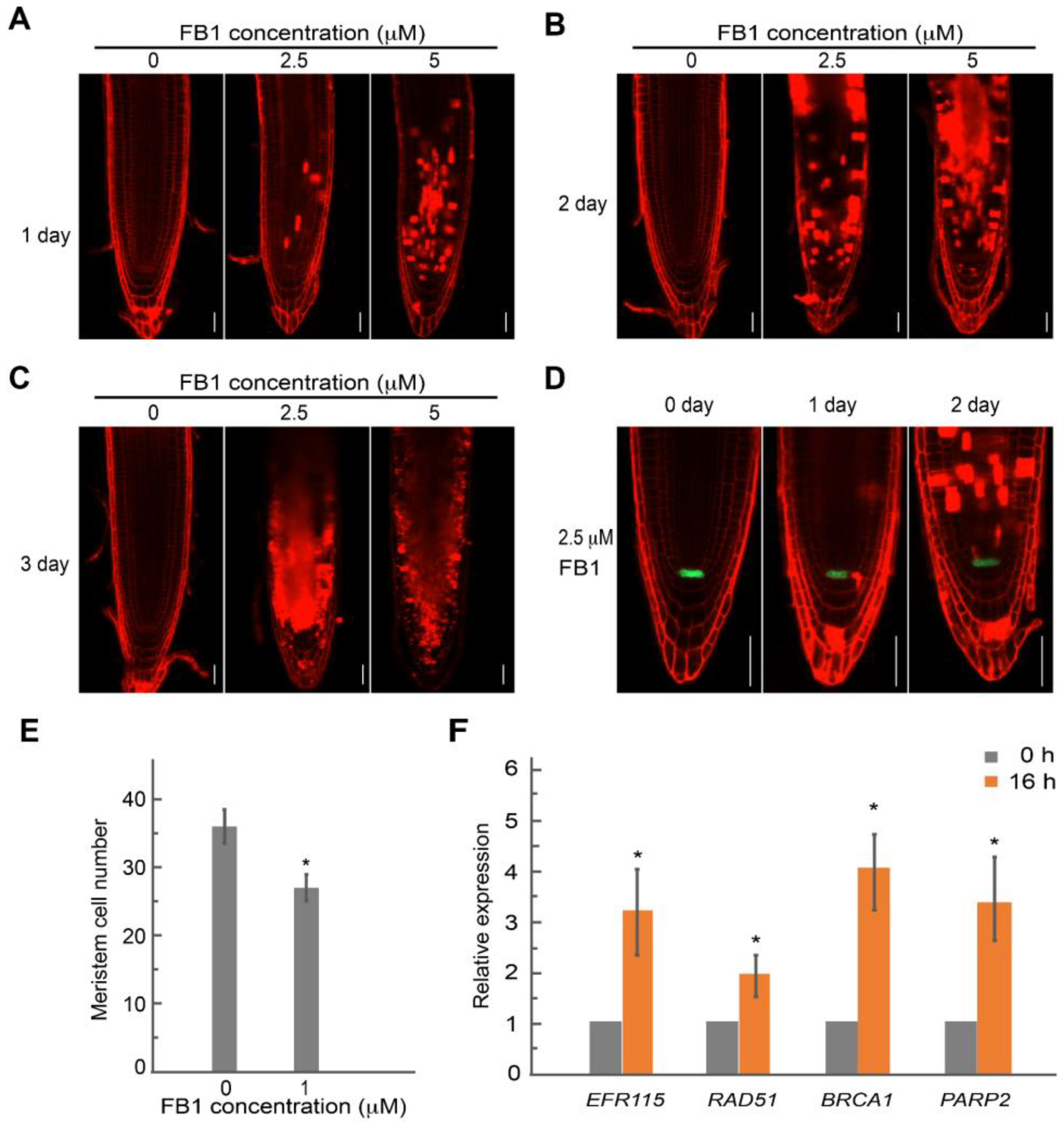
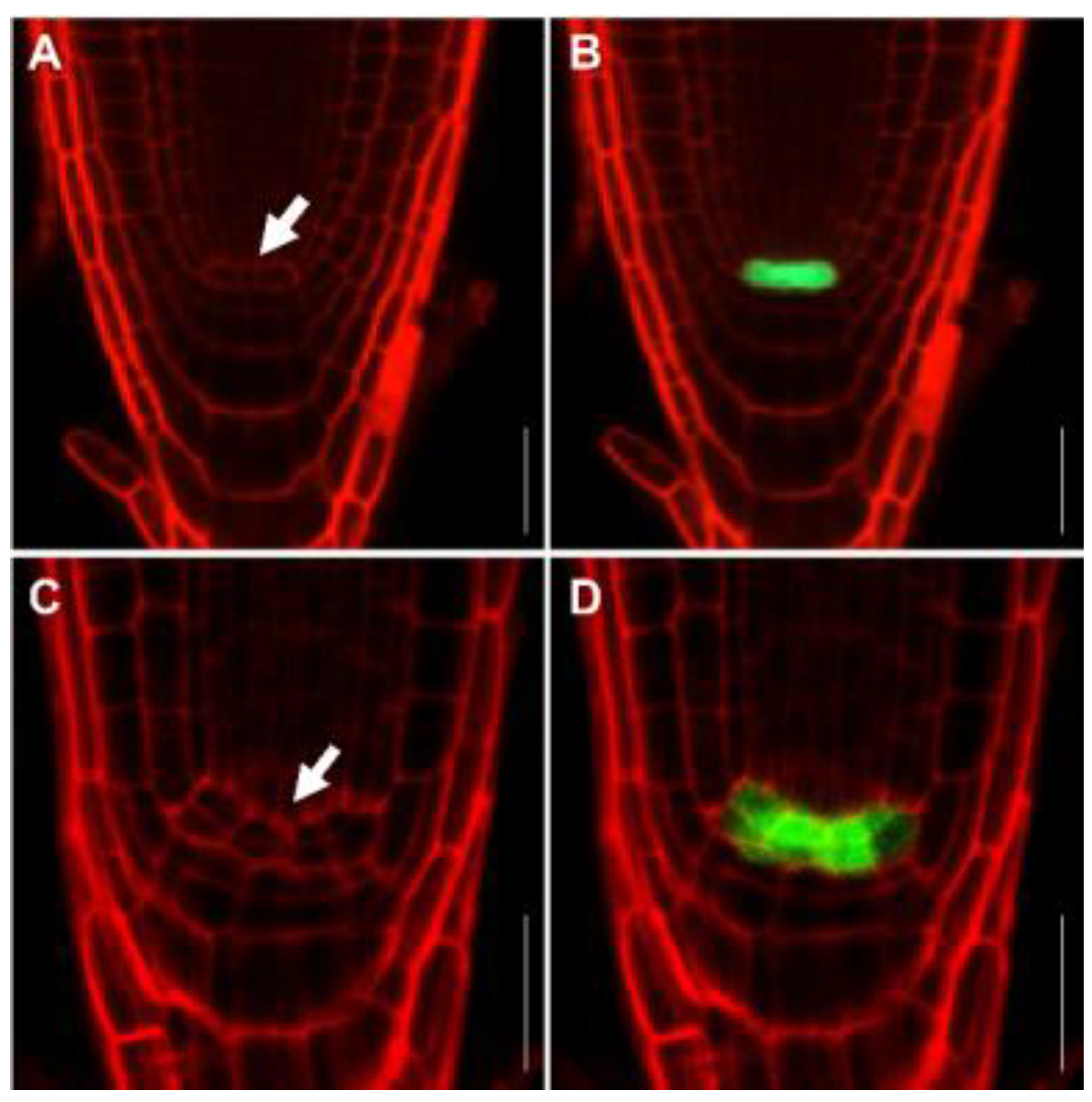
2.4. FB1 Induced Cell Death in the Root Meristem
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Chemical Treatments and Root Growth Analysis
4.3. Confocal Microscopy
4.4. Reverse Transcription Quantitative PCR Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Aichinger, E.; Kornet, N.; Friedrich, T.; Laux, T. Plant stem cell niches. Annu. Rev. Plant Biol. 2012, 63, 615–636. [Google Scholar] [CrossRef] [PubMed]
- Bennett, T.; van den Toorn, A.; Willemsen, V.; Scheres, B. Precise control of plant stem cell activity through parallel regulatory inputs. Development 2014, 141, 4055–4064. [Google Scholar] [CrossRef] [PubMed]
- Sozzani, R.; Iyer-Pascuzzi, A. Postembryonic control of root meristem growth and development. Curr. Opin. Plant Biol. 2014, 17, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Aida, M.; Beis, D.; Heidstra, R.; Willemsen, V.; Blilou, I.; Galinha, C.; Nussaume, L.; Noh, Y.S.; Amasino, R.; Scheres, B. The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 2004, 119, 109–120. [Google Scholar] [CrossRef]
- Dharmasiri, N.; Dharmasiri, S.; Weijers, D.; Lechner, E.; Yamada, M.; Hobbie, L.; Ehrismann, J.S.; Jurgens, G.; Estelle, M. Plant development is regulated by a family of auxin receptor F box proteins. Dev. Cell. 2005, 9, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Vanneste, S.; Friml, J. Auxin: A trigger for change in plant development. Cell 2009, 136, 1005–1016. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.; Friml, J. Auxin regulates distal stem cell differentiation in Arabidopsis roots. Proc. Natl. Acad. Sci. USA 2010, 107, 12046–12051. [Google Scholar] [CrossRef]
- Blilou, I.; Xu, J.; Wildwater, M.; Willemsen, V.; Paponov, I.; Friml, J.; Heidstra, R.; Aida, M.; Palme, K.; Scheres, B. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 2005, 433, 39–44. [Google Scholar] [CrossRef]
- Fulcher, N.; Sablowski, R. Hypersensitivity to DNA damage in plant stem cell niches. Proc. Natl. Acad. Sci. USA 2009, 106, 20984–20988. [Google Scholar] [CrossRef]
- Ubogoeva, E.V.; Zemlyanskaya, E.V.; Xu, J.; Mironova, V. Mechanisms of stress response in the root stem cell niche. J. Exp. Bot. 2021, 72, 6746–6754. [Google Scholar] [CrossRef]
- Pedroza-Garcia, J.A.; Xiang, Y.; De Veylder, L. Cell cycle checkpoint control in response to DNA damage by environmental stresses. Plant J. 2022, 109, 490–507. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, N.; Ogita, N.; Takahashi, T.; Taniguchi, S.; Tanaka, M.; Seki, M.; Umeda, M. A regulatory module controlling stress-induced cell cycle arrest in Arabidopsis. Elife 2019, 8, e43944. [Google Scholar] [CrossRef] [PubMed]
- Bundock, P.; Hooykaas, P. Severe Developmental Defects, Hypersensitivity to DNA-Damaging Agents, and Lengthened Telomeres in Arabidopsis MRE11 Mutants. Plant Cell 2002, 14, 2451–2462. [Google Scholar] [CrossRef]
- Yoshiyama, K.; Conklin, P.A.; Huefner, N.D.; Britt, A.B. Suppressor of gamma response 1 (SOG1) encodes a putative transcription factor governing multiple responses to DNA damage. Proc. Natl. Acad. Sci. USA 2009, 106, 12843–12848. [Google Scholar] [CrossRef]
- Hashimura, Y.; Ueguchi, C. The Arabidopsis MERISTEM DISORGANIZATION 1 gene is required for the maintenance of stem cells through the reduction of DNA damage. Plant J. 2011, 68, 657–669. [Google Scholar] [CrossRef] [PubMed]
- Gelderblom, W.C.A.; Jaskiewicz, K.; Marasas, W.F.O.; Thiel, P.G.; Kriek, N.P.J. Fumonisins--novel mycotoxins with cancer-promoting activity produced by Fusarium moniliforme. Appl. Environ. Microbiol. 1988, 54, 1806–1811. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wu, Q.; Wan, D.; Liu, Q.; Chen, D.; Liu, Z.; Martínez-Larrañaga, M.R.; Martínez, M.A.; Anadón, A.; Yuan, Z. Fumonisins: Oxidative stress-mediated toxicity and metabolism in vivo and in vitro. Arch Toxicol. 2015, 90, 81–101. [Google Scholar] [CrossRef] [PubMed]
- Voss, K.A.; Norred, W.P.; Meredith, F.I.; Riley, R.T.; Stephen Saunders, D. Fumonisin concentration and ceramide synthase inhibitory activity of corn, masa, and tortilla chips. J. Toxicol. Environ. Health A 2006, 69, 1387–1397. [Google Scholar] [CrossRef]
- Cingolani, F.; Futerman, A.H.; Casas, J. Ceramide synthases in biomedical research. Chem. Phys. Lipids 2016, 197, 25–32. [Google Scholar] [CrossRef]
- Markham, J.E.; Molino, D.; Gissot, L.; Bellec, Y.; Hematy, K.; Marion, J.; Belcram, K.; Palauqui, J.C.; Satiat-JeuneMaitre, B.; Faure, J.D. Sphingolipids Containing Very-Long-Chain Fatty Acids Define a Secretory Pathway for Specific Polar Plasma Membrane Protein Targeting in Arabidopsis. Plant Cell 2011, 23, 2362–2378. [Google Scholar] [CrossRef]
- Luttgeharm, K.D.; Kimberlin, A.N.; Cahoon, E.B. Plant Sphingolipid Metabolism and Function. Sub-Cell. Biochem. 2016, 86, 249–286. [Google Scholar]
- Riley, R.T.; Merrill, A.H. Ceramide synthase inhibition by fumonisins: A perfect storm of perturbed sphingolipid metabolism, signaling and disease. J. Lipid Res. 2019, 60, 1183–1189. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.-Y.; Li, C.-Y.; Yao, N. Fumonisin B1: A Tool for Exploring the Multiple Functions of Sphingolipids in Plants. Front. Plant Sci. 2020, 11, 600458. [Google Scholar] [CrossRef] [PubMed]
- Desai, K.; Sullards, M.C.; Allegood, J.; Wang, E.; Merrill, A.H. Fumonisins and fumonisin analogs as inhibitors of ceramide synthase and inducers of apoptosis. Biochim. Biophys. Acta 2003, 1585, 188–192. [Google Scholar] [CrossRef]
- Kimberlin, A.N.; Majumder, S.; Han, G.; Chen, M.; Cahoon, R.E.; Stone, J.M.; Dunn, T.M.; Cahoon, E.B. Arabidopsis 56-amino acid serine palmitoyltransferase-interacting proteins stimulate sphingolipid synthesis, are essential, and affect mycotoxin sensitivity. Plant Cell 2013, 25, 4627–4639. [Google Scholar] [CrossRef]
- Gutierrez-Najera, N.A.; Saucedo-Garcia, M.; Noyola-Martinez, L.; Vazquez-Vazquez, C.; Palacios-Bahena, S.; Carmona-Salazar, L.; Plasencia, J.; El-Hafidi, M.; Gavilanes-Ruiz, M. Sphingolipid Effects on the Plasma Membrane Produced by Addition of Fumonisin B1 to Maize Embryos. Plants 2020, 9, 150. [Google Scholar] [CrossRef] [PubMed]
- Michaelson, L.V.; Napier, J.A.; Molino, D.; Faure, J.D. Plant sphingolipids: Their importance in cellular organization and adaption. Biochim. Biophys. Acta 2016, 1861, 1329–1335. [Google Scholar] [CrossRef]
- Sperling, P.; Franke, S.; Luthje, S.; Heinz, E. Are glucocerebrosides the predominant sphingolipids in plant plasma membranes? Plant Physiol. Biochem. 2005, 43, 1031–1038. [Google Scholar] [CrossRef] [PubMed]
- Markham, J.E.; Lynch, D.V.; Napier, J.A.; Dunn, T.M.; Cahoon, E.B. Plant sphingolipids: Function follows form. Curr. Opin. Plant Biol. 2013, 16, 350–357. [Google Scholar] [CrossRef]
- Aubert, A.; Marion, J.; Boulogne, C.; Bourge, M.; Abreu, S.; Bellec, Y.; Faure, J.-D.; Satiat-Jeunemaitre, B. Sphingolipids involvement in plant endomembrane differentiation: The BY2 case. Plant J. 2011, 65, 958–971. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, J.T.; Silverman, F.P.; Liang, H. Uncoupling Salicylic Acid-Dependent Cell Death and Defense-Related Responses From Disease Resistance in the Arabidopsis Mutant acd5. Genetics 2000, 156, 341–350. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.; Yao, N.; Song, J.T.; Luo, S.; Lu, H.; Greenberg, J.T. Ceramides modulate programmed cell death in plants. Gene Dev. 2003, 17, 2636–2641. [Google Scholar] [CrossRef] [PubMed]
- Asai, T.; Stone, J.M.; Heard, J.E.; Kovtun, Y.; Yorgey, P.; Ausubel, S.F.M. Fumonisin B1–Induced Cell Death in Arabidopsis Protoplasts Requires Jasmonate-, Ethylene-, and Salicylate-Dependent Signaling Pathways. Plant Cell 2000, 12, 1823–1835. [Google Scholar]
- Da Costa, M.; Bach, L.; Landrieu, I.; Bellec, Y.; Catrice, O.; Brown, S.; De Veylder, L.; Lippens, G.; Inzé, D.; Faure, J.-D. Arabidopsis PASTICCINO2 Is an Antiphosphatase Involved in Regulation of Cyclin-Dependent Kinase A. Plant Cell 2006, 18, 1426–1437. [Google Scholar] [CrossRef]
- Chen, M.; Markham, J.E.; Cahoon, E.B. Sphingolipid Delta8 unsaturation is important for glucosylceramide biosynthesis and low-temperature performance in Arabidopsis. Plant J. 2012, 69, 769–781. [Google Scholar] [CrossRef]
- Msanne, J.; Chen, M.; Luttgeharm, K.D.; Bradley, A.M.; Mays, E.S.; Paper, J.M.; Boyle, D.L.; Cahoon, R.E.; Schrick, K.; Cahoon, E.B. Glucosylceramide is Critical for Cell-Type Differentiation and Organogenesis, but not for Cell Viability in Arabidopsis. Plant J. 2015, 84, 188–201. [Google Scholar] [CrossRef]
- Luttgeharm, K.D.; Chen, M.; Mehra, A.; Cahoon, R.E.; Markham, J.E.; Cahoon, E.B. Overexpression of Arabidopsis Ceramide Synthases Differentially Affects Growth, Sphingolipid Metabolism, Programmed Cell Death, and Mycotoxin Resistance. Plant Physiol. 2015, 169, 1108–1117. [Google Scholar] [CrossRef]
- Sabatini, S.; Beis, D.; Wolkenfelt, H.; Murfett, J.; Guilfoyle, T.; Malamy, J.; Benfey, P.; Leyser, O.; Bechtold, N.; Weisbeek, P.; et al. An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 1999, 99, 463–472. [Google Scholar] [CrossRef]
- Friml, J.; Vieten, A.; Sauer, M.; Weijers, D.; Schwarz, H.; Hamann, T.; Offringa, R.; Jurgens, G. Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 2003, 426, 147–153. [Google Scholar] [CrossRef]
- Doerks, T.; Copley, R.R.; Schultz, J.; Ponting, C.P.; Bork, P. SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genome Res. 2002, 12, 47–56. [Google Scholar] [CrossRef]
- Helariutta, Y.; Fukaki, H.; Wysocka-Diller, J.; Nakajima, K.; Jung, J.; Sena, G.; Hauser, M.T.; Benfey, P.N. The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 2000, 101, 555–567. [Google Scholar] [CrossRef]
- Wildwater, M.; Campilho, A.; Perez-Perez, J.M.; Heidstra, R.; Blilou, I.; Korthout, H.; Chatterjee, J.; Mariconti, L.; Gruissem, W.; Scheres, B. The RETINOBLASTOMA-RELATED gene regulates stem cell maintenance in Arabidopsis roots. Cell 2005, 123, 1337–1349. [Google Scholar] [CrossRef]
- De Schutter, K.; Joubes, J.; Cools, T.; Verkest, A.; Corellou, F.; Babiychuk, E.; Van Der Schueren, E.; Beeckman, T.; Kushnir, S.; Inze, D.; et al. Arabidopsis WEE1 kinase controls cell cycle arrest in response to activation of the DNA integrity checkpoint. Plant Cell 2007, 19, 211–225. [Google Scholar] [CrossRef]
- Heyman, J.; Cools, T.; Vandenbussche, F.; Heyndrickx, K.S.; Van Leene, J.; Vercauteren, I.; Vanderauwera, S.; Vandepoele, K.; De Jaeger, G.; Van Der Straeten, D.; et al. ERF115 controls root quiescent center cell division and stem cell replenishment. Science 2013, 342, 860–863. [Google Scholar] [CrossRef]
- Huen, M.S.; Sy, S.M.; Chen, J. BRCA1 and its toolbox for the maintenance of genome integrity. Nat. Rev. Mol. Cell Biol. 2010, 11, 138–148. [Google Scholar] [CrossRef]
- Song, J.; Keppler, B.D.; Wise, R.R.; Bent, A.F. PARP2 Is the Predominant Poly(ADP-Ribose) Polymerase in Arabidopsis DNA Damage and Immune Responses. PLoS Genet. 2015, 11, e1005200. [Google Scholar] [CrossRef]
- Inagaki, S. Arabidopsis TEBICHI, with Helicase and DNA Polymerase Domains, Is Required for Regulated Cell Division and Differentiation in Meristems. Plant Cell 2006, 18, 879–892. [Google Scholar] [CrossRef]
- Abbas, H.K.; Tanaka, T.; Duke, S.O.; Porter, J.K.; Wray, E.M.; Hodges, L.; Sessions, A.E.; Wang, E.; Merrill, A.H., Jr.; Riley, R.T. Fumonisin- and AAL-Toxin-Induced Disruption of Sphingolipid Metabolism with Accumulation of Free Sphingoid Bases. Plant Physiol. 1994, 106, 1085–1093. [Google Scholar] [CrossRef]
- Spassieva, S.D.; Markham, J.E.; Hille, J. The plant disease resistance gene Asc-1 prevents disruption of sphingolipid metabolism during AAL-toxin-induced programmed cell death. Plant J. 2002, 32, 561–572. [Google Scholar] [CrossRef]
- Sarkar, A.K.; Luijten, M.; Miyashima, S.; Lenhard, M.; Hashimoto, T.; Nakajima, K.; Scheres, B.; Heidstra, R.; Laux, T. Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 2007, 446, 811–814. [Google Scholar] [CrossRef]
- Manova, V.; Gruszka, D. DNA damage and repair in plants—From models to crops. Front Plant Sci. 2015, 6, 885. [Google Scholar] [CrossRef] [PubMed]
- Surova, O.; Zhivotovsky, B. Various modes of cell death induced by DNA damage. Oncogene 2013, 32, 3789–3797. [Google Scholar] [CrossRef]
- Yoshiyama, K.O.; Sakaguchi, K.; Kimura, S. DNA damage response in plants: Conserved and variable response compared to animals. Biology 2013, 2, 1338–1356. [Google Scholar] [CrossRef] [PubMed]
- Norbury, C.J.; Hickson, I.D. Cellular responses to DNA damage. Annu. Rev. Pharmacol. 2001, 41, 367–401. [Google Scholar] [CrossRef]
- Furukawa, T.; Curtis, M.J.; Tominey, C.M.; Duong, Y.H.; Wilcox, B.W.; Aggoune, D.; Hays, J.B.; Britt, A.B. A shared DNA-damage-response pathway for induction of stem-cell death by UVB and by gamma irradiation. DNA Repair 2010, 9, 940–948. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.H.; Savina, M.; Du, J.; Devendran, A.; Kannivadi Ramakanth, K.; Tian, X.; Sim, W.S.; Mironova, V.V.; Xu, J. A Sacrifice-for-Survival Mechanism Protects Root Stem Cell Niche from Chilling Stress. Cell 2017, 170, 102–113.e14. [Google Scholar] [CrossRef]
- Zhao, Y.; Wang, L.; Sun, X.; Bao, Y.; Liu, H.; Zhao, Y. Inhibition of ribosome biogenesis by actinomycin D affects Arabidopsis root development. Biochem. Biophys. Commun. 2022, 588, 61–67. [Google Scholar] [CrossRef]
- Xu, J.; Hofhuis, H.; Heidstra, R.; Sauer, M.; Friml, J.; Scheres, B. A molecular framework for plant regeneration. Science 2006, 311, 385–388. [Google Scholar] [CrossRef]
- Xu, J.; Scheres, B. Dissection of Arabidopsis ADP-RIBOSYLATION FACTOR 1 function in epidermal cell polarity. Plant Cell 2005, 17, 525–536. [Google Scholar] [CrossRef]
- Dello Ioio, R.; Nakamura, K.; Moubayidin, L.; Perilli, S.; Taniguchi, M.; Morita, M.T.; Aoyama, T.; Costantino, P.; Sabatini, S.A. genetic framework for the control of cell division and differentiation in the root meristem. Science 2008, 322, 1380–1384. [Google Scholar] [CrossRef]
- Heidstra, R.; Welch, D.; Scheres, B. Mosaic analyses using marked activation and deletion clones dissect Arabidopsis. SCARECROW action in asymmetric cell division. Gene Dev. 2004, 18, 1964–1969. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, K.; Sena, G.; Nawy, T.; Benfey, P.N. Intercellular movement of the putative transcription factor SHR in root. patterning. Nature 2001, 413, 307–311. [Google Scholar] [CrossRef] [PubMed]
- Kornet, N.; Scheres, B. Members of the GCN5 histone acetyltransferase complex regulate PLETHORA-mediated root stem cell niche maintenance and transit amplifying cell proliferation in Arabidopsis. Plant Cell 2009, 21, 1070–1079. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Guo, S.; Lu, M.; Zhang, Y.; Li, J.; Wang, W.; Wang, P.; Zhang, J.; Hu, Z.; Li, L.; et al. Biosynthesis of DHGA12 and its roles in Arabidopsis seedling establishment. Nat. Commun. 2019, 10, 1768. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Y.; Liu, Z.; Wang, L.; Liu, H. Fumonisin B1 as a Tool to Explore Sphingolipid Roles in Arabidopsis Primary Root Development. Int. J. Mol. Sci. 2022, 23, 12925. https://doi.org/10.3390/ijms232112925
Zhao Y, Liu Z, Wang L, Liu H. Fumonisin B1 as a Tool to Explore Sphingolipid Roles in Arabidopsis Primary Root Development. International Journal of Molecular Sciences. 2022; 23(21):12925. https://doi.org/10.3390/ijms232112925
Chicago/Turabian StyleZhao, Yanxue, Zhongjie Liu, Lei Wang, and Hao Liu. 2022. "Fumonisin B1 as a Tool to Explore Sphingolipid Roles in Arabidopsis Primary Root Development" International Journal of Molecular Sciences 23, no. 21: 12925. https://doi.org/10.3390/ijms232112925
APA StyleZhao, Y., Liu, Z., Wang, L., & Liu, H. (2022). Fumonisin B1 as a Tool to Explore Sphingolipid Roles in Arabidopsis Primary Root Development. International Journal of Molecular Sciences, 23(21), 12925. https://doi.org/10.3390/ijms232112925




