CircTCF4 Suppresses Proliferation and Differentiation of Goat Skeletal Muscle Satellite Cells Independent from AGO2 Binding
Abstract
1. Introduction
2. Results
2.1. Characterization of Goat CircTCF4 Sequence
2.2. CircTCF4 Is Enriched in Developmental Skeletal Muscles and MuSCs
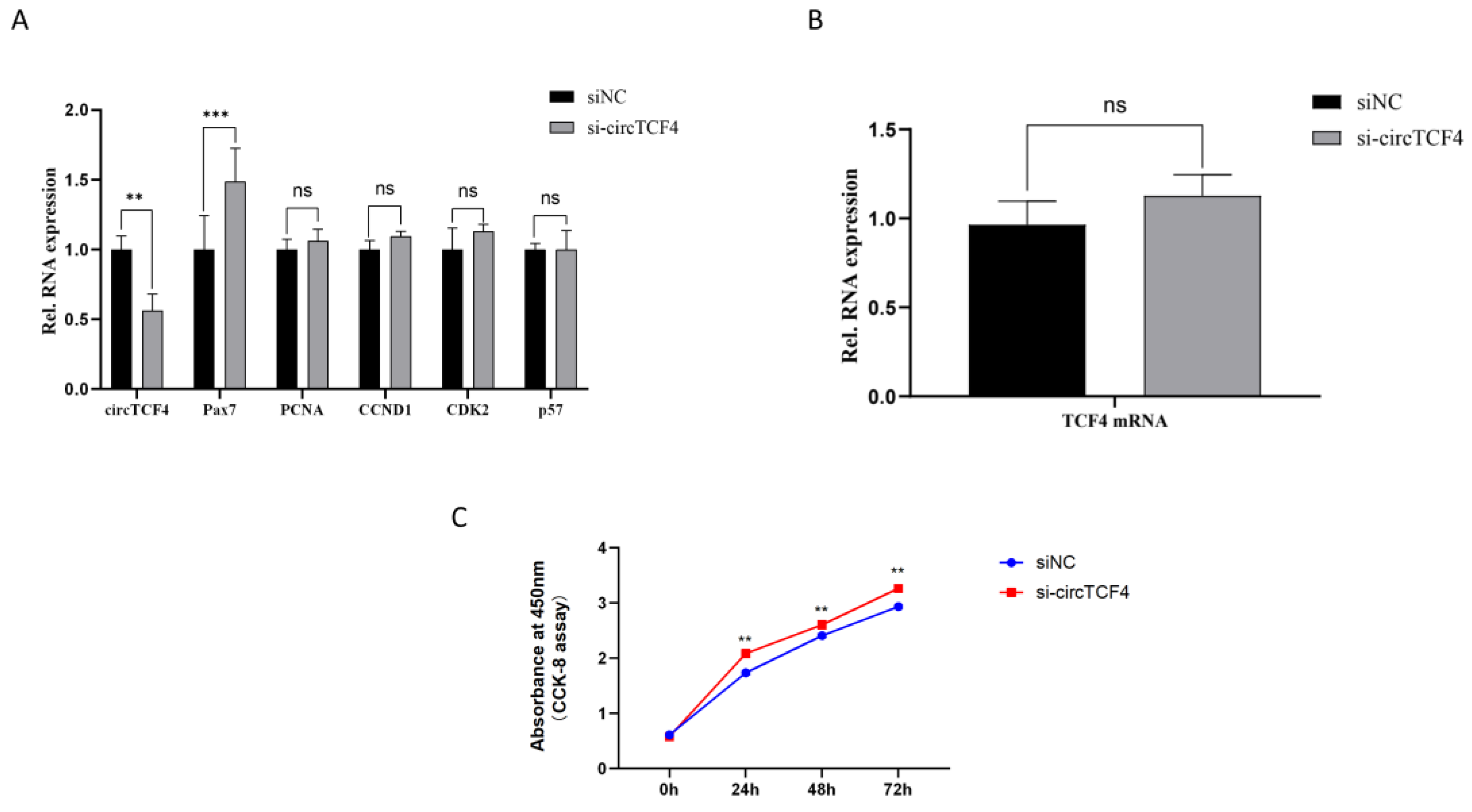
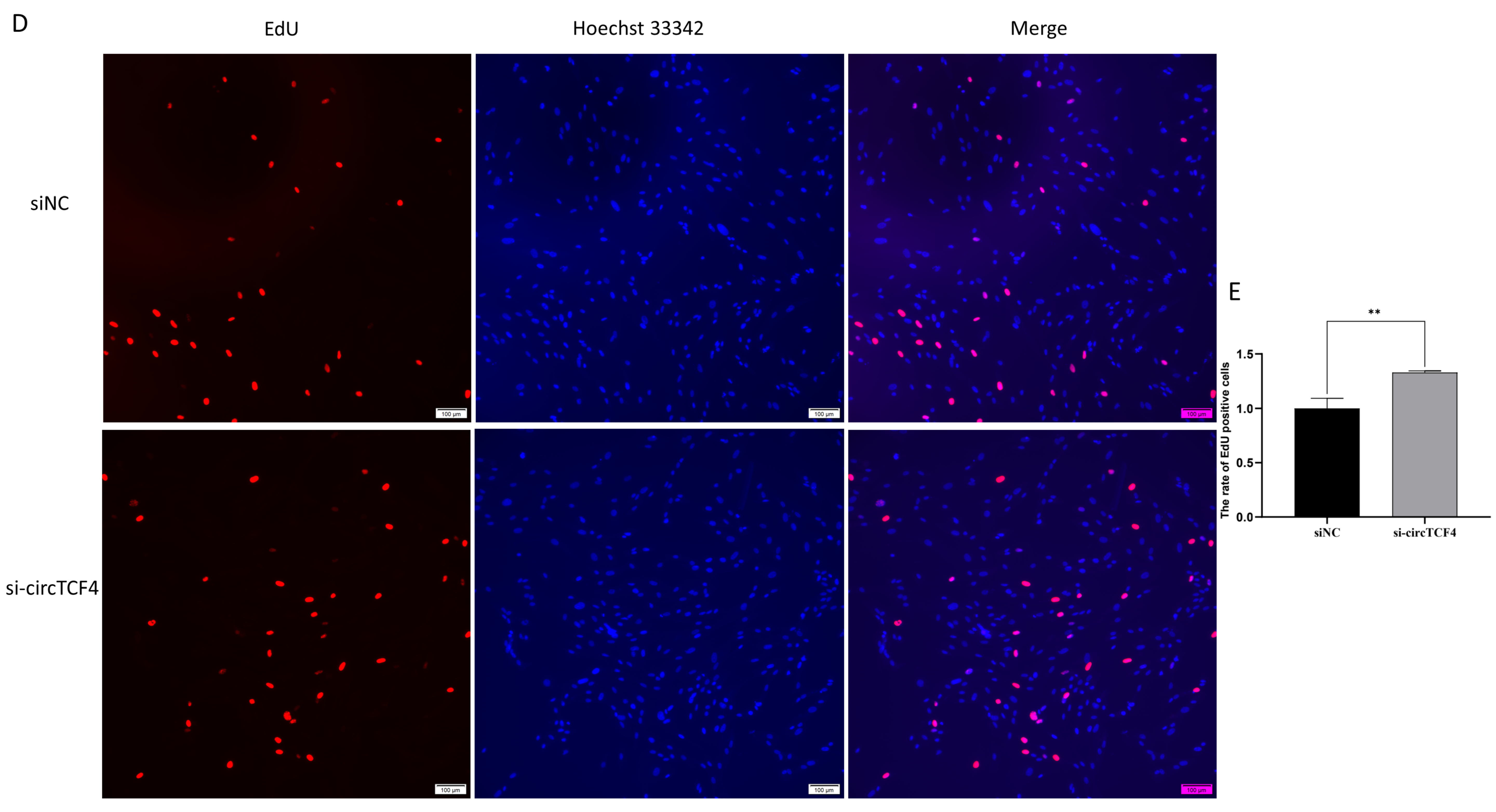
2.3. CircTCF4 Retained Goat MuSCs Proliferation
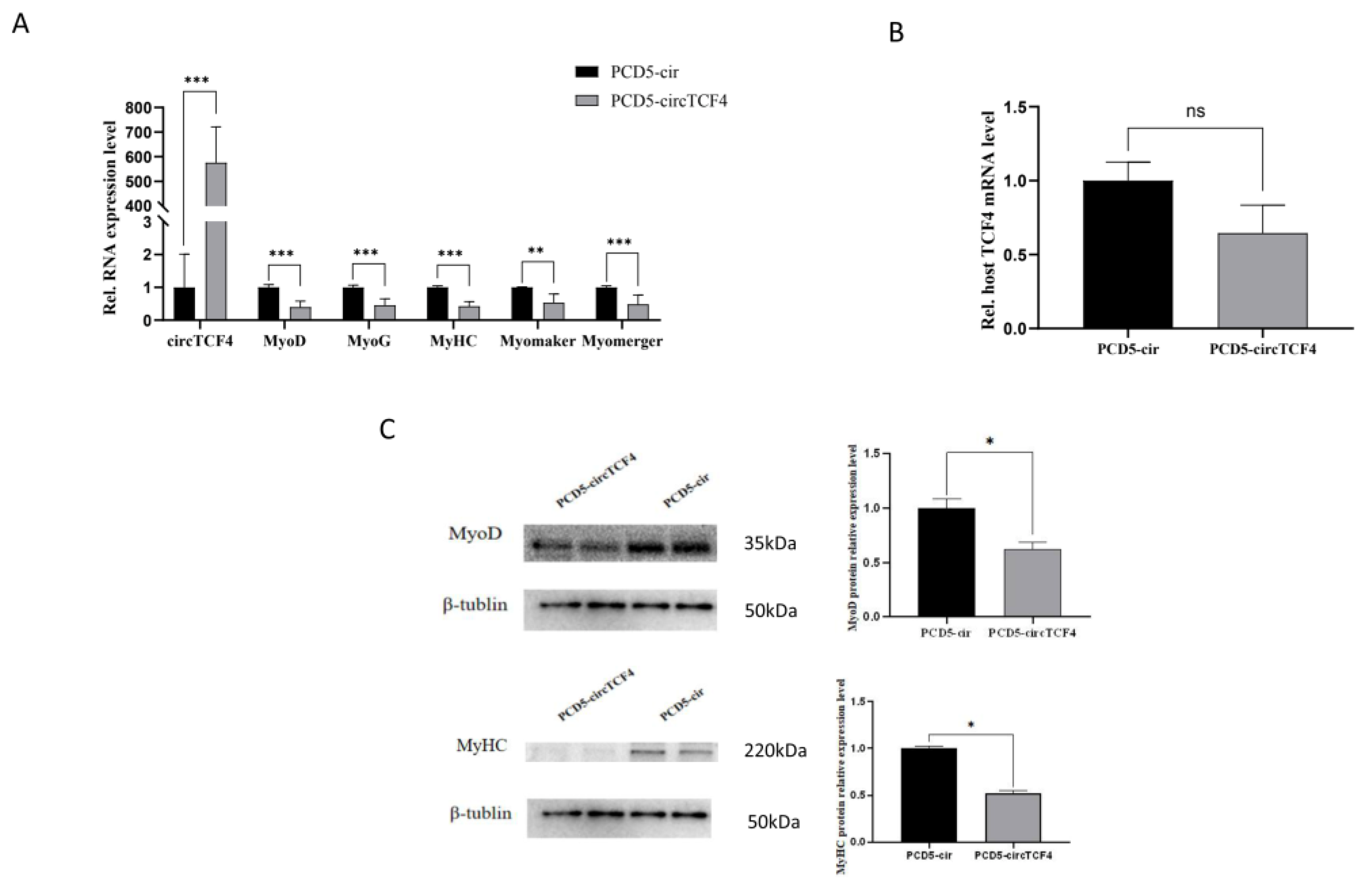
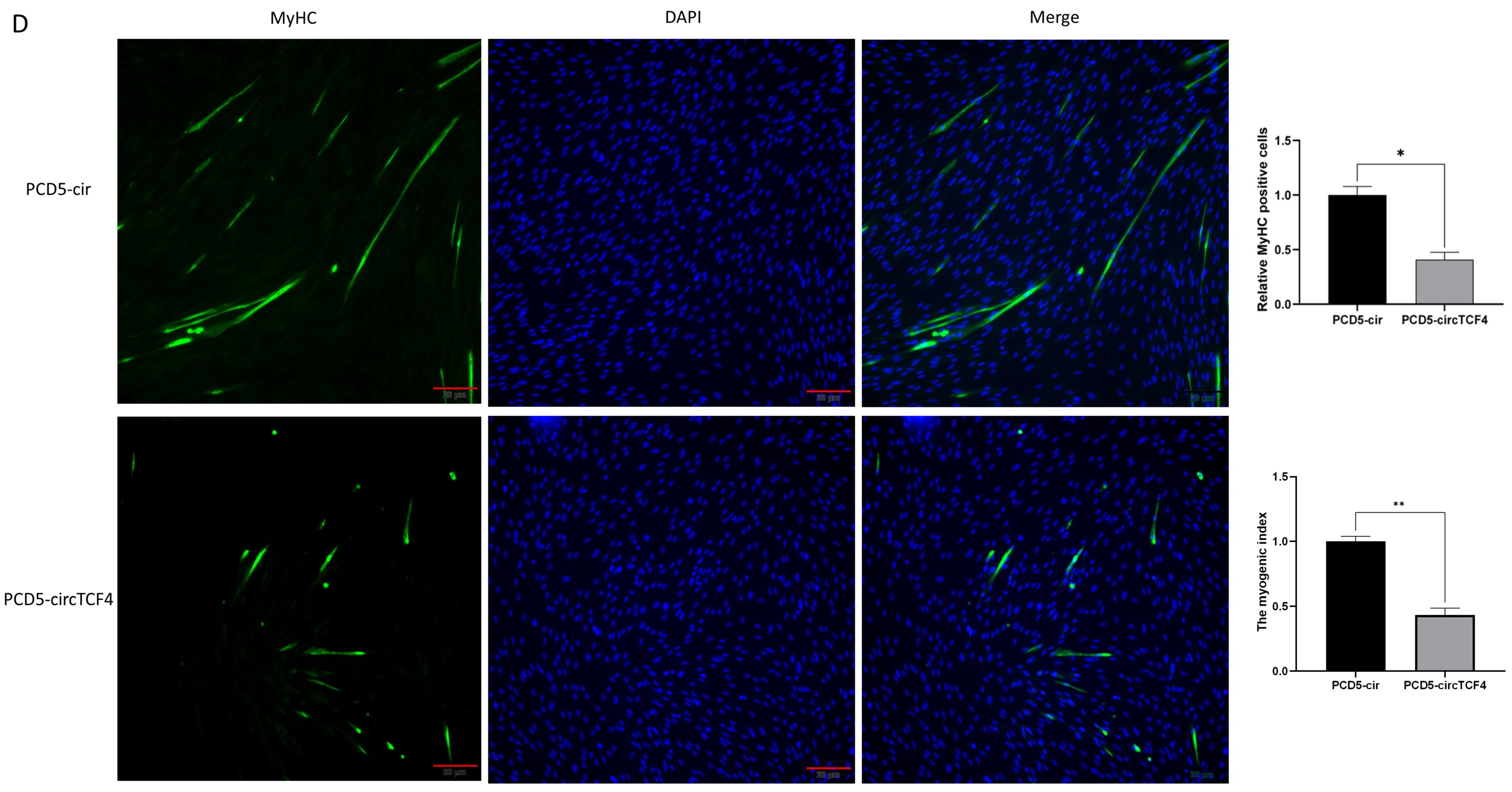
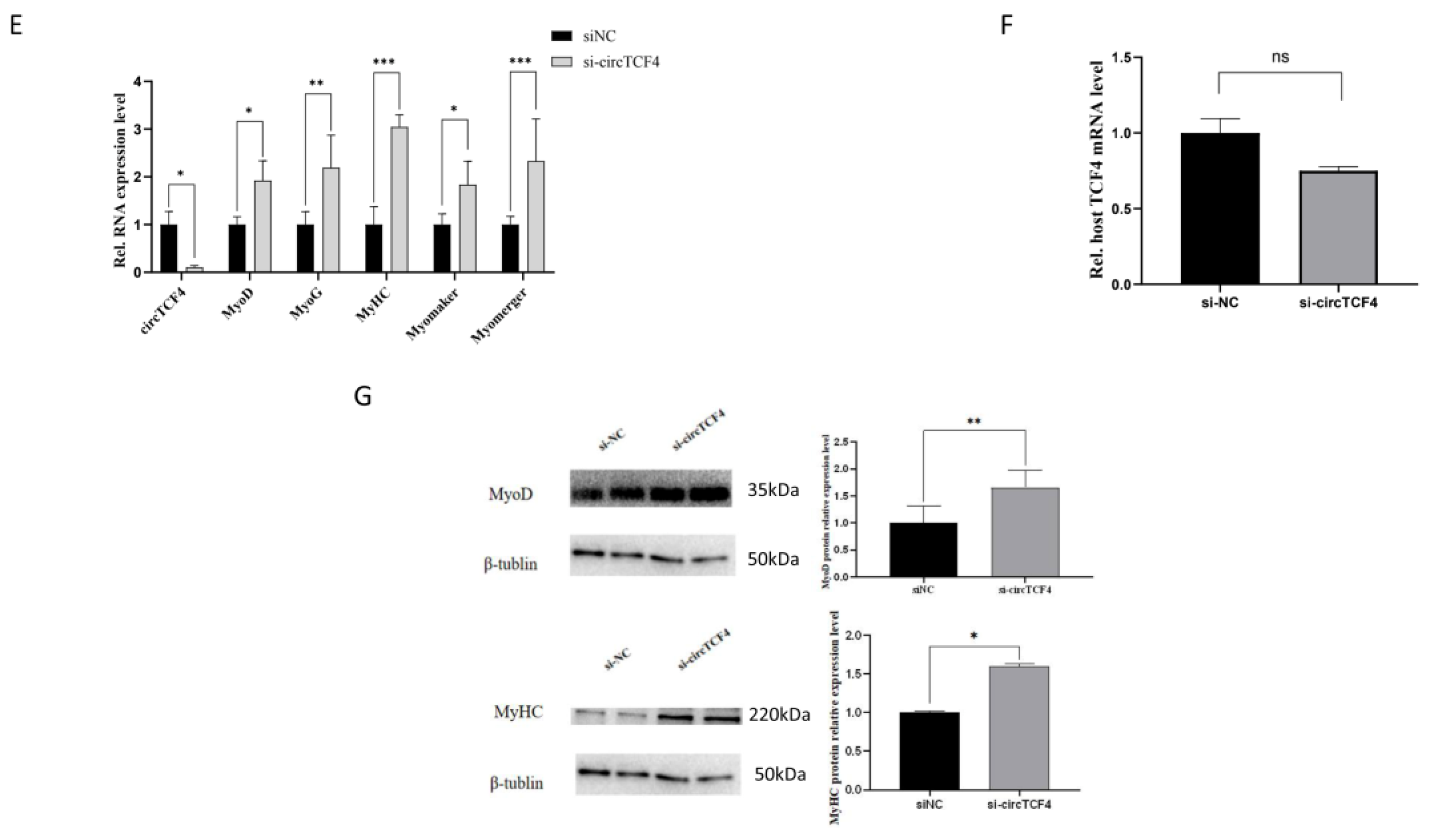
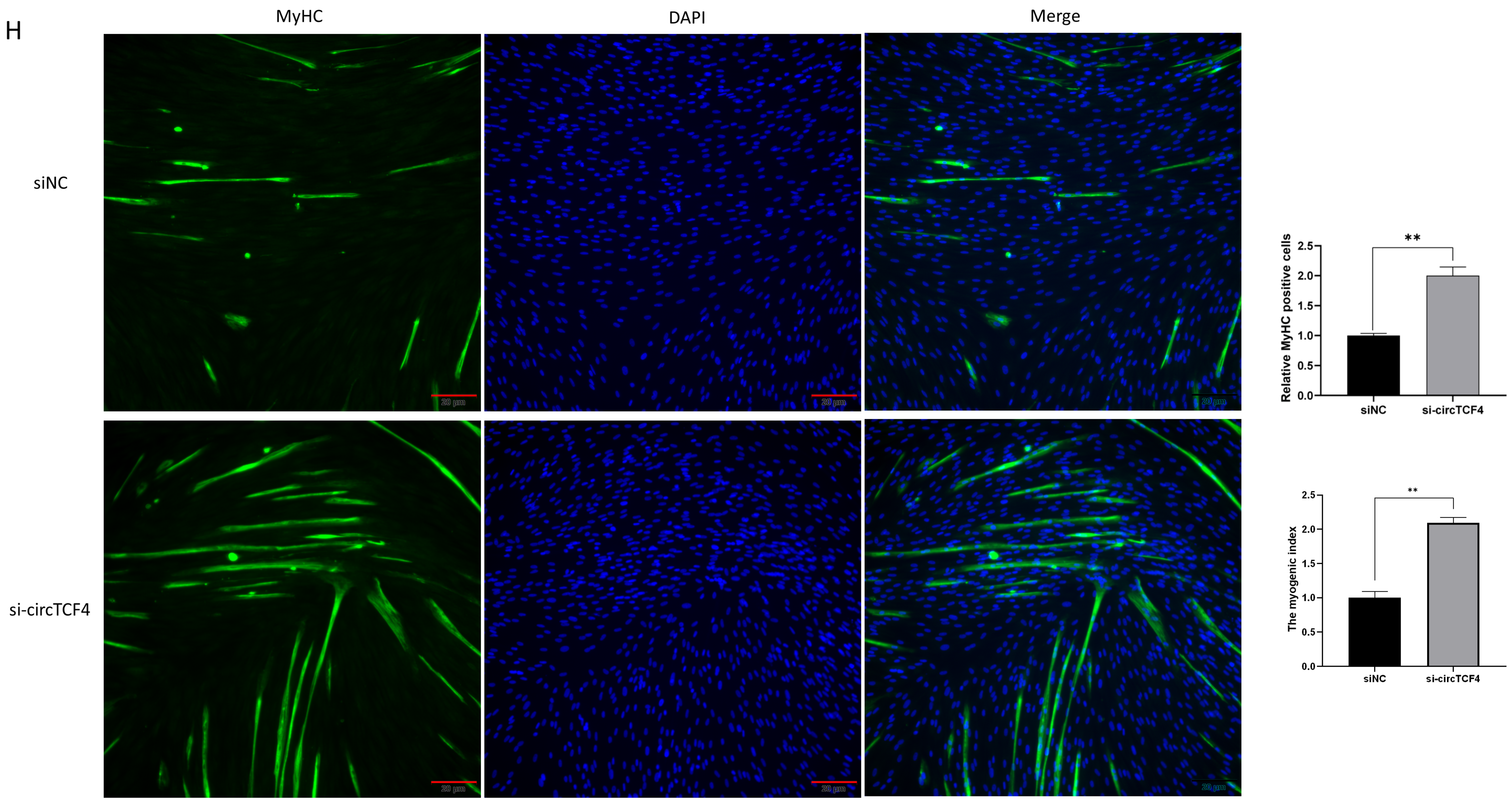
2.4. CircTCF4 Suppressed Goat MuSCs Differentiation
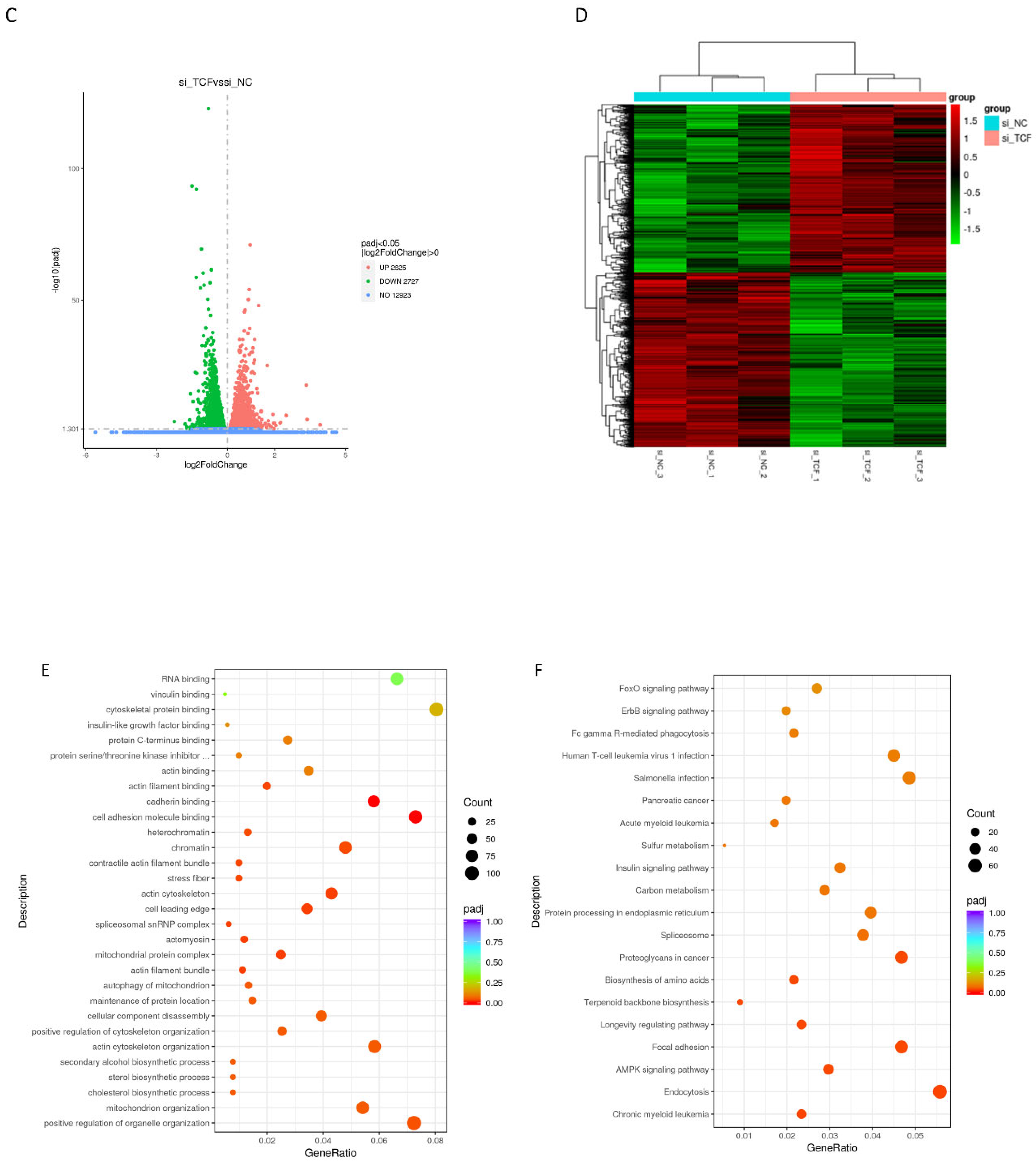
2.5. Signaling Pathways Involved in CircTCF4-Related MuSCs Differentiation
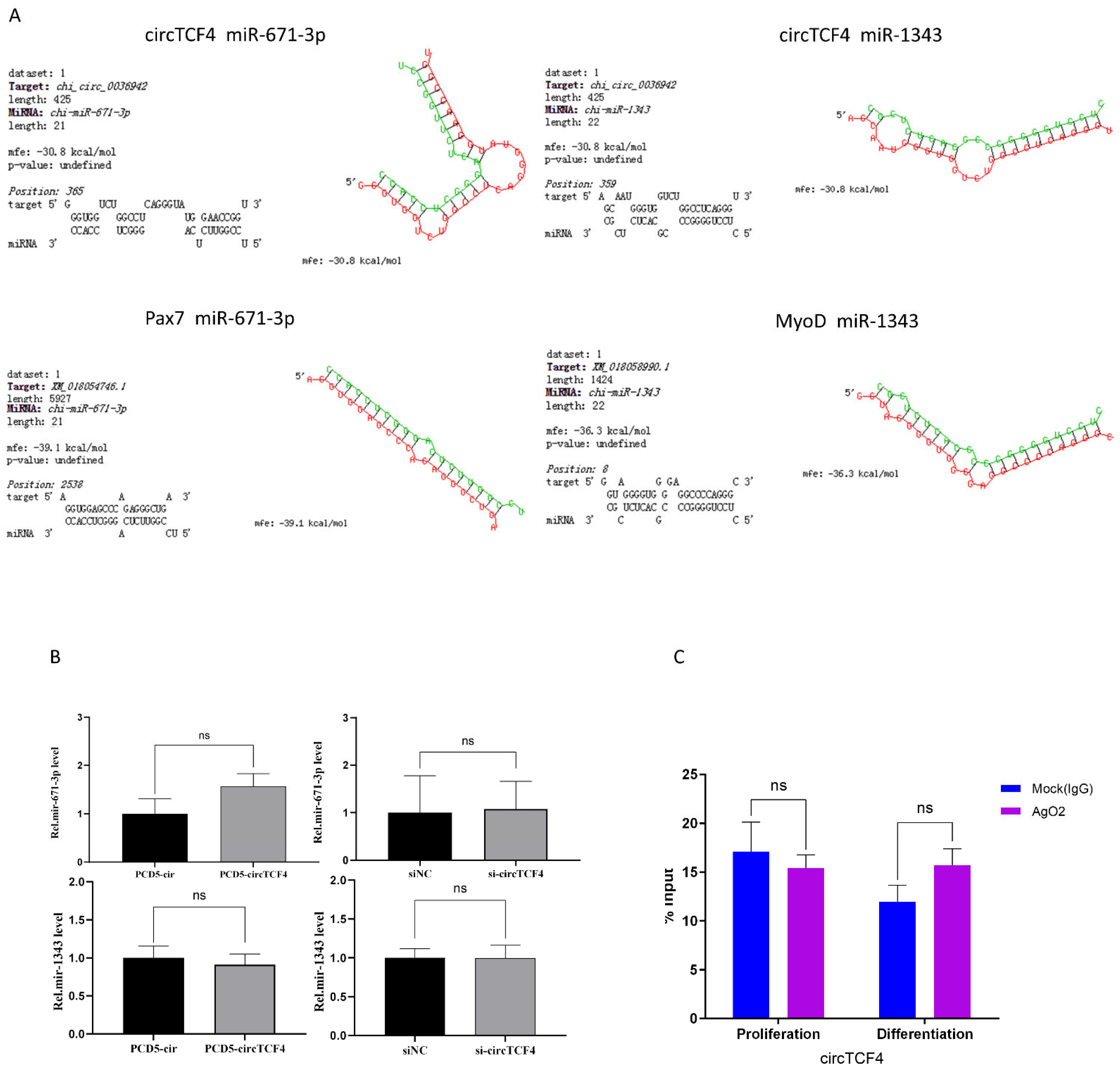
2.6. CircTCF4 Regulated the Proliferation and Differentiation of MuSCs Independent from AGO2 Binding
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Animals and Samples Collection
4.3. Skeletal Muscle Satellite Cells (MuSCs) Isolation
4.4. MuSCs Culture and Transfection
4.5. RNA Extraction and qPCR
4.6. RNase R Treatment
4.7. Plasmids Construct and Interfere RNA Design
4.8. Cell Counting Kit-8 (CCK-8)
4.9. EdU Assay
4.10. Western Blotting (WB) Assay
4.11. MyHC Immunofluorescence Assay
4.12. RNA-Binding Protein Immunoprecipitation (RIP) Analysis
4.13. mRNA-Seq and Bioinformatic Analyses
4.14. Bioinformatic Analysis
4.15. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BMP4 | bone morphogenetic protein 4 |
| CCK-8 | cell-counting kit-8 |
| ceRNA | competing endogenous RNAs |
| circRNA | circular RNA |
| DM | differentiation medium |
| EdU | 5-ethynyl-2′-deoxyuridine |
| GM | growth medium |
| IGF1R | insulin-like growth factor receptor 1 |
| MAPK | mitogen-activated protein kinase |
| miRNA | microRNAs |
| MuSCs | skeletal muscle satellite cells |
| MyoD | myogenic differentiation protein 1 |
| MyoG | myogenin |
| MyHC | myosin heavy chain |
| Pax7 | paired box 7 |
| PCNA | proliferating cell nuclear antigen |
| PCR | polymerase chain reaction |
References
- Jin, W.; Peng, J.; Jiang, S. The epigenetic regulation of embryonic myogenesis and adult muscle regeneration by histone methylation modification. Biochem. Biophys. Rep. 2016, 6, 209–219. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, S.; Asahara, H. The myogenic transcriptional network. Cell Mol. Life Sci. 2011, 68, 1843–1849. [Google Scholar] [CrossRef] [PubMed]
- Braun, T.; Gautel, M. Transcriptional mechanisms regulating skeletal muscle differentiation, growth and homeostasis. Nat. Rev. Mol. Cell Biol. 2011, 12, 349–361. [Google Scholar] [CrossRef] [PubMed]
- Megeney, L.A.; Rudnicki, M.A. Determination versus differentiation and the MyoD family of transcription factors. Biochem. Cell Biol. 1995, 73, 723–732. [Google Scholar] [CrossRef]
- Lang, D.; Powell, S.K.; Plummer, R.S.; Young, K.P.; Ruggeri, B.A. PAX genes: Roles in development, pathophysiology, and cancer. Biochem. Pharmacol. 2007, 73, 24. [Google Scholar] [CrossRef] [PubMed]
- Potthoff, M.J.; Olson, E.N. MEF2: A central regulator of diverse developmental programs. Development 2007, 134, 4131–4140. [Google Scholar] [CrossRef] [PubMed]
- Geetha-Loganathan, P.; Nimmagadda, S.; Scaal, M.; Huang, R.; Christ, B. Wnt signaling in somite development. Ann. Anat. 2008, 190, 208–222. [Google Scholar] [CrossRef]
- Pourquie, O.; Fan, C.M.; Coltey, M.; Hirsinger, E.; Watanabe, Y.; Breant, C.; Francis-West, P.; Brickell, P.; Tessier-Lavigne, M.; Le Douarin, N.M. Lateral and axial signals involved in avian somite patterning: A role for BMP4. Cell 1996, 84, 461–471. [Google Scholar] [CrossRef]
- Keren, A.; Tamir, Y.; Bengal, E. The p38 MAPK signaling pathway: A major regulator of skeletal muscle development. Mol. Cell. Endocrinol. 2006, 252, 224–230. [Google Scholar] [CrossRef]
- Hawke, T.J.; Garry, D.J. Myogenic satellite cells: Physiology to molecular biology. J. Appl. Physiol. 1985 2001, 91, 534–551. [Google Scholar] [CrossRef]
- Zammit, P.S.; Golding, J.P.; Nagata, Y.; Hudon, V.; Partridge, T.A.; Beauchamp, J.R. Muscle satellite cells adopt divergent fates: A mechanism for self-renewal? J. Cell Biol. 2004, 166, 347–357. [Google Scholar] [CrossRef] [PubMed]
- Manzano, R.; Toivonen, J.M.; Calvo, A.C.; Miana-Mena, F.J.; Zaragoza, P.; Munoz, M.J.; Montarras, D.; Osta, R. Sex, fiber-type, and age dependent in vitro proliferation of mouse muscle satellite cells. J. Cell Biochem. 2011, 112, 2825–2836. [Google Scholar] [CrossRef] [PubMed]
- Gnocchi, V.F.; White, R.B.; Ono, Y.; Ellis, J.A.; Zammit, P.S. Further characterisation of the molecular signature of quiescent and activated mouse muscle satellite cells. PLoS ONE 2009, 4, e5205. [Google Scholar] [CrossRef] [PubMed]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157. [Google Scholar] [CrossRef]
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.W.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019, 20, 675–691. [Google Scholar] [CrossRef]
- Salzman, J.; Gawad, C.; Wang, P.L.; Lacayo, N.; Brown, P.O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE 2012, 7, e30733. [Google Scholar] [CrossRef]
- Li, Z.; Huang, C.; Bao, C.; Chen, L.; Lin, M.; Wang, X.; Zhong, G.; Yu, B.; Hu, W.; Dai, L.; et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015, 22, 256–264. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, X.O.; Chen, T.; Xiang, J.F.; Yin, Q.F.; Xing, Y.H.; Zhu, S.; Yang, L.; Chen, L.L. Circular intronic long noncoding RNAs. Mol. Cell 2013, 51, 792–806. [Google Scholar] [CrossRef]
- Suzuki, H.; Tsukahara, T. A view of pre-mRNA splicing from RNase R resistant RNAs. Int. J. Mol. Sci. 2014, 15, 9331–9342. [Google Scholar] [CrossRef]
- Zheng, Q.; Bao, C.; Guo, W.; Li, S.; Chen, J.; Chen, B.; Luo, Y.; Lyu, D.; Li, Y.; Shi, G.; et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 2016, 7, 11215. [Google Scholar] [CrossRef]
- Veno, M.T.; Hansen, T.B.; Veno, S.T.; Clausen, B.H.; Grebing, M.; Finsen, B.; Holm, I.E.; Kjems, J. Spatio-temporal regulation of circular RNA expression during porcine embryonic brain development. Genom. Biol. 2015, 16, 245. [Google Scholar] [CrossRef] [PubMed]
- Rybak-Wolf, A.; Stottmeister, C.; Glazar, P.; Jens, M.; Pino, N.; Giusti, S.; Hanan, M.; Behm, M.; Bartok, O.; Ashwal-Fluss, R.; et al. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol. Cell 2015, 58, 870–885. [Google Scholar] [CrossRef] [PubMed]
- Holdt, L.M.; Kohlmaier, A.; Teupser, D. Molecular roles and function of circular RNAs in eukaryotic cells. Cell Mol. Life Sci. 2018, 75, 1071–1098. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Zhang, X.; Odame, E.; Xu, X.; Chen, Y.; Ye, J.; Zhou, H.; Dai, D.; Kyei, B.; Zhan, S.; et al. CircRNA-Protein Interactions in Muscle Development and Diseases. Int. J. Mol. Sci. 2021, 22, 3262. [Google Scholar] [CrossRef]
- Li, L.; Chen, Y.; Nie, L.; Ding, X.; Zhang, X.; Zhao, W.; Xu, X.; Kyei, B.; Dai, D.; Zhan, S.; et al. MyoD-induced circular RNA CDR1as promotes myogenic differentiation of skeletal muscle satellite cells. Biochim. Biophys. Acta Gene Regul. Mech. 2019, 1862, 807–821. [Google Scholar] [CrossRef]
- Shi, Y.; Jia, X.; Xu, J. The new function of circRNA: Translation. Clin. Transl. Oncol. 2020, 22, 2162–2169. [Google Scholar] [CrossRef]
- Mesman, S.; Wever, I.; Smidt, M.P. Tcf4 Is Involved in Subset Specification of Mesodiencephalic Dopaminergic Neurons. Biomedicines 2021, 9, 317. [Google Scholar] [CrossRef]
- Lennertz, L.; Quednow, B.B.; Benninghoff, J.; Wagner, M.; Maier, W.; Mossner, R. Impact of TCF4 on the genetics of schizophrenia. Eur. Arch. Psychiatr. Clin. Neurosci. 2011, 261 (Suppl. 2), S161–S165. [Google Scholar] [CrossRef]
- Mesman, S.; Bakker, R.; Smidt, M.P. Tcf4 is required for correct brain development during embryogenesis. Mol. Cell. Neurosci. 2020, 106, 103502. [Google Scholar] [CrossRef]
- Salzman, J.; Chen, R.E.; Olsen, M.N.; Wang, P.L.; Brown, P.O. Cell-type specific features of circular RNA expression. PLoS Genet. 2013, 9, e1003777. [Google Scholar] [CrossRef]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef] [PubMed]
- You, X.; Vlatkovic, I.; Babic, A.; Will, T.; Epstein, I.; Tushev, G.; Akbalik, G.; Wang, M.; Glock, C.; Quedenau, C.; et al. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat. Neurosci. 2015, 18, 603–610. [Google Scholar] [CrossRef] [PubMed]
- Zhan, S.; Dong, Y.; Zhao, W.; Guo, J.; Zhong, T.; Wang, L.; Li, L.; Zhang, H. Genome-wide identification and characterization of long non-coding RNAs in developmental skeletal muscle of fetal goat. BMC Genom. 2016, 17, 666. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Zhu, M.J.; Dodson, M.V.; Du, M. Developmental programming of fetal skeletal muscle and adipose tissue development. J. Genom. 2013, 1, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Sanger, H.L.; Klotz, G.; Riesner, D.; Gross, H.J.; Kleinschmidt, A.K. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. USA 1976, 73, 3852–3856. [Google Scholar] [CrossRef]
- Obi, P.; Chen, Y.G. The design and synthesis of circular RNAs. Methods 2021, 196, 85–103. [Google Scholar] [CrossRef]
- Garikipati, V.N.S.; Verma, S.K.; Cheng, Z.; Liang, D.; Truongcao, M.M.; Cimini, M.; Yue, Y.; Huang, G.; Wang, C.; Benedict, C.; et al. Circular RNA CircFndc3b modulates cardiac repair after myocardial infarction via FUS/VEGF-A axis. Nat. Commun. 2019, 10, 4317. [Google Scholar] [CrossRef]
- Zhang, L.; Zhou, C.; Jiang, X.; Huang, S.; Li, Y.; Su, T.; Wang, G.; Zhou, Y.; Liu, M.; Xu, D. Circ0001470 Acts as a miR-140-3p Sponge to Facilitate the Progression of Embryonic Development through Regulating PTGFR Expression. Cells 2022, 11, 1746. [Google Scholar] [CrossRef]
- Girardi, F.; Le Grand, F. Wnt Signaling in Skeletal Muscle Development and Regeneration. Prog. Mol Biol. Transl. Sci. 2018, 153, 157–179. [Google Scholar] [CrossRef]
- Jing, Y.; Ren, Y.; Witzel, H.R.; Dobreva, G. A BMP4-p38 MAPK signaling axis controls ISL1 protein stability and activity during cardiogenesis. Stem Cell Rep. 2021, 16, 1894–1905. [Google Scholar] [CrossRef]
- Bengal, E.; Aviram, S.; Hayek, T. p38 MAPK in Glucose Metabolism of Skeletal Muscle: Beneficial or Harmful? Int. J. Mol. Sci. 2020, 21, 6480. [Google Scholar] [CrossRef] [PubMed]
- Thomson, D.M. The Role of AMPK in the Regulation of Skeletal Muscle Size, Hypertrophy, and Regeneration. Int. J. Mol. Sci. 2018, 19, 3125. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Cao, X.; Dong, D.; Shen, X.; Cheng, J.; Jiang, R.; Yang, Z.; Peng, S.; Huang, Y.; Lan, X.; et al. Circular RNA TTN Acts As a miR-432 Sponge to Facilitate Proliferation and Differentiation of Myoblasts via the IGF2/PI3K/AKT Signaling Pathway. Mol. Ther. Nucl. Acids 2019, 18, 966–980. [Google Scholar] [CrossRef] [PubMed]
- Legnini, I.; Di Timoteo, G.; Rossi, F.; Morlando, M.; Briganti, F.; Sthandier, O.; Fatica, A.; Santini, T.; Andronache, A.; Wade, M.; et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol. Cell 2017, 66, 22–37.e9. [Google Scholar] [CrossRef]
- Liu, J.; Li, M.; Kong, L.; Cao, M.; Zhang, M.; Wang, Y.; Song, C.; Fang, X.; Chen, H.; Zhang, C. CircARID1A regulates mouse skeletal muscle regeneration by functioning as a sponge of miR-6368. FASEB J. 2021, 35, e21324. [Google Scholar] [CrossRef]
- Zheng, Q.; Zhu, C.; Jing, J.; Ling, Y.; Qin, S.; Wang, J.; Zha, L.; Liu, Y.; Fang, F. Morphological changes and functional circRNAs screening of rabbit skeletal muscle development. BMC Genom. 2021, 22, 469. [Google Scholar] [CrossRef]
- Qi, A.; Ru, W.; Yang, H.; Yang, Y.; Tang, J.; Yang, S.; Lan, X.; Lei, C.; Sun, X.; Chen, H. Circular RNA ACTA1 Acts as a Sponge for miR-199a-5p and miR-433 to Regulate Bovine Myoblast Development through the MAP3K11/MAP2K7/JNK Pathway. J. Agric. Food Chem. 2022, 70, 3357–3373. [Google Scholar] [CrossRef]
- Shen, X.; Tang, J.; Jiang, R.; Wang, X.; Yang, Z.; Huang, Y.; Lan, X.; Lei, C.; Chen, H. CircRILPL1 promotes muscle proliferation and differentiation via binding miR-145 to activate IGF1R/PI3K/AKT pathway. Cell Death Dis. 2021, 12, 142. [Google Scholar] [CrossRef]
- Yang, Y.; Fan, X.; Mao, M.; Song, X.; Wu, P.; Zhang, Y.; Jin, Y.; Yang, Y.; Chen, L.L.; Wang, Y.; et al. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res. 2017, 27, 626–641. [Google Scholar] [CrossRef]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4, 5005. [Google Scholar] [CrossRef]
- Kruger, J.; Rehmsmeier, M. RNAhybrid: MicroRNA target prediction easy, fast and flexible. Nucl. Acids Res. 2006, 34, W451–W454. [Google Scholar] [CrossRef]
- Loher, P.; Rigoutsos, I. Interactive exploration of RNA22 microRNA target predictions. Bioinformatics 2012, 28, 3322–3323. [Google Scholar] [CrossRef] [PubMed]
- Stark, A.; Brennecke, J.; Russell, R.B.; Cohen, S.M. Identification of Drosophila MicroRNA targets. PLoS Biol. 2003, 1, E60. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucl. Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genom. Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Armaos, A.; Colantoni, A.; Proietti, G.; Rupert, J.; Tartaglia, G.G. catRAPID omics v2.0: Going deeper and wider in the prediction of protein-RNA interactions. Nucl. Acids Res. 2021, 49, W72–W79. [Google Scholar] [CrossRef]
- Wu, W.; Ji, P.; Zhao, F. CircAtlas: An integrated resource of one million highly accurate circular RNAs from 1070 vertebrate transcriptomes. Genom. Biol. 2020, 21, 101. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, S.; Li, L.; Zhou, H.; Zhang, X.; Xu, X.; Dai, D.; Zhan, S.; Cao, J.; Guo, J.; Zhong, T.; et al. CircTCF4 Suppresses Proliferation and Differentiation of Goat Skeletal Muscle Satellite Cells Independent from AGO2 Binding. Int. J. Mol. Sci. 2022, 23, 12868. https://doi.org/10.3390/ijms232112868
Zheng S, Li L, Zhou H, Zhang X, Xu X, Dai D, Zhan S, Cao J, Guo J, Zhong T, et al. CircTCF4 Suppresses Proliferation and Differentiation of Goat Skeletal Muscle Satellite Cells Independent from AGO2 Binding. International Journal of Molecular Sciences. 2022; 23(21):12868. https://doi.org/10.3390/ijms232112868
Chicago/Turabian StyleZheng, Shuailong, Li Li, Helin Zhou, Xujia Zhang, Xiaoli Xu, Dinghui Dai, Siyuan Zhan, Jiaxue Cao, Jiazhong Guo, Tao Zhong, and et al. 2022. "CircTCF4 Suppresses Proliferation and Differentiation of Goat Skeletal Muscle Satellite Cells Independent from AGO2 Binding" International Journal of Molecular Sciences 23, no. 21: 12868. https://doi.org/10.3390/ijms232112868
APA StyleZheng, S., Li, L., Zhou, H., Zhang, X., Xu, X., Dai, D., Zhan, S., Cao, J., Guo, J., Zhong, T., Wang, L., & Zhang, H. (2022). CircTCF4 Suppresses Proliferation and Differentiation of Goat Skeletal Muscle Satellite Cells Independent from AGO2 Binding. International Journal of Molecular Sciences, 23(21), 12868. https://doi.org/10.3390/ijms232112868






