Introgression of a Complex Genomic Structural Variation Causes Hybrid Male Sterility in GJ Rice (Oryza sativa L.) Subspecies
Abstract
1. Introduction
2. Results
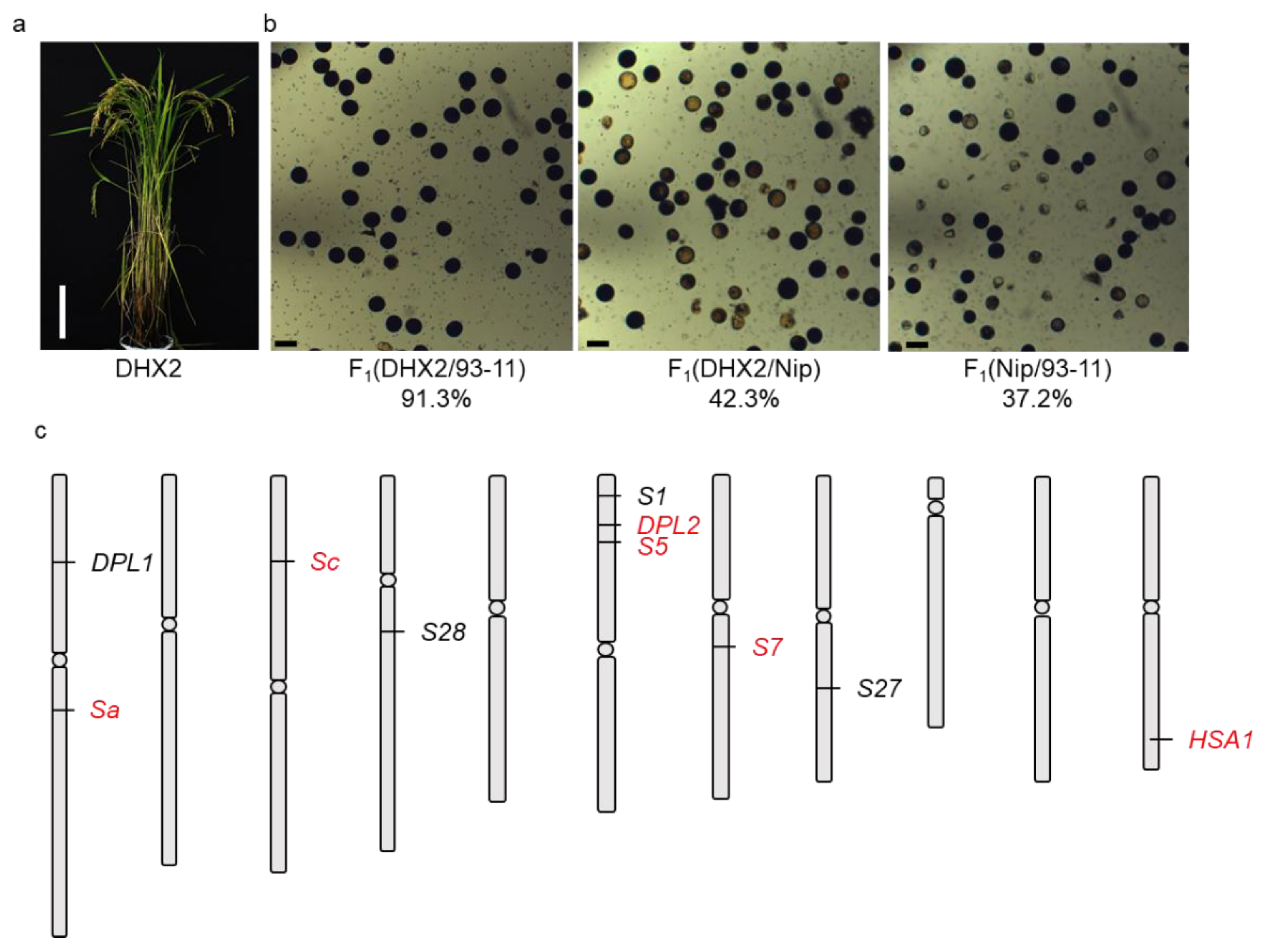
2.1. F1 Hybrid Sterility between Geng Varieties
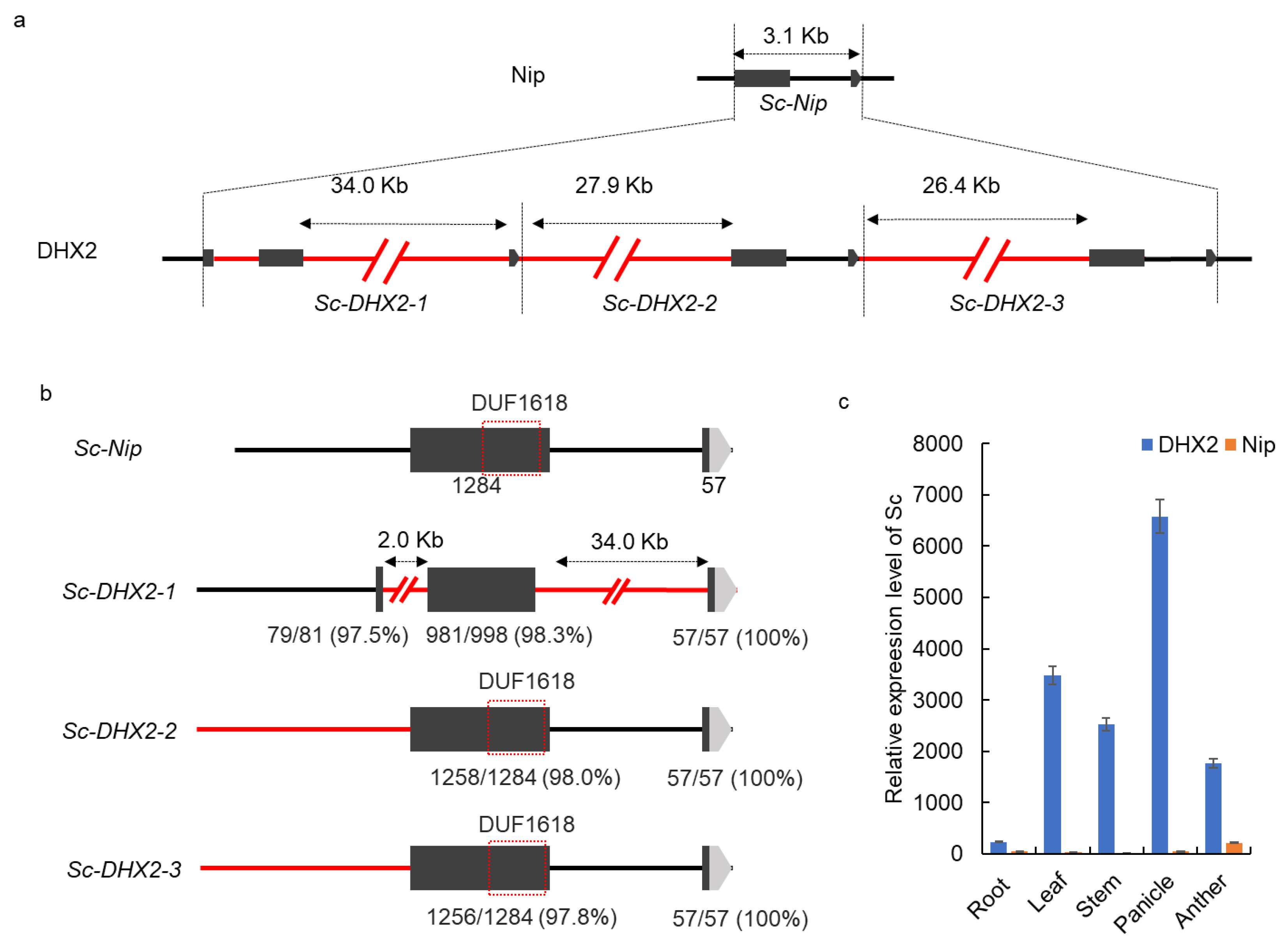
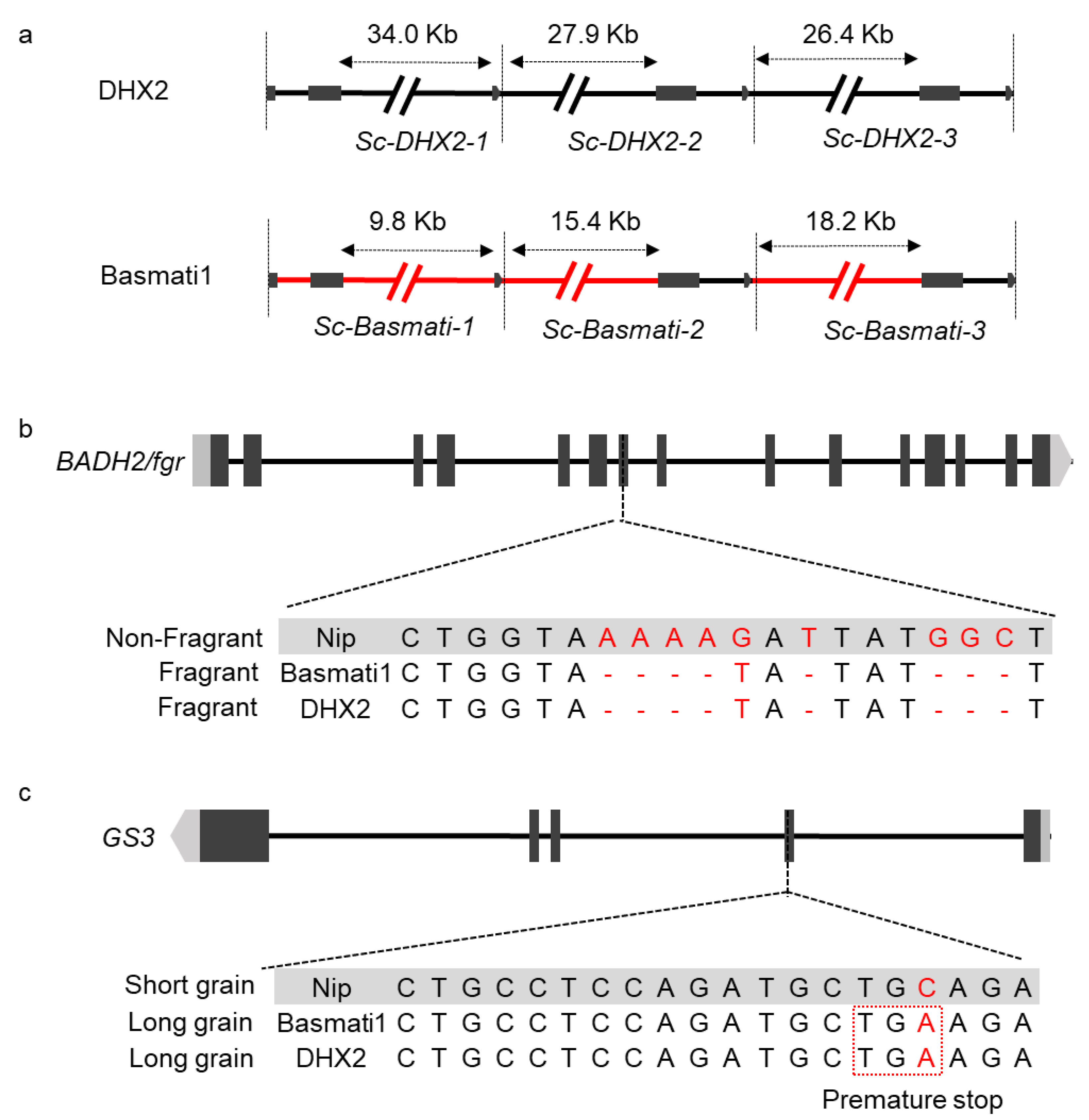
2.2. Complex Genomic Structural Variation in the Sc-DHX2 Alleles
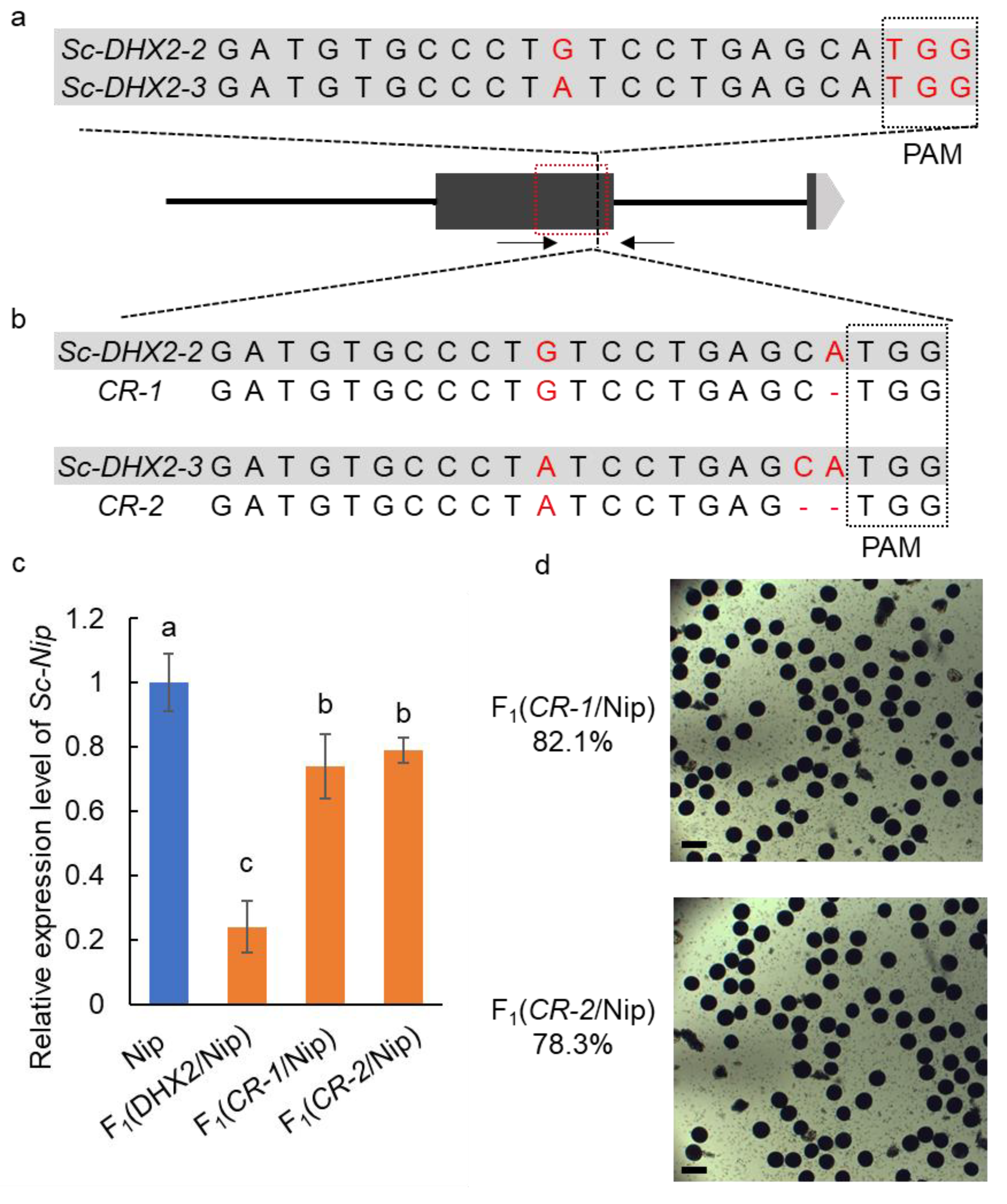
2.3. Knockout of Sc-DHX2-2 or Sc-DHX2-3 Rescues the F1 Hybrid Sterility
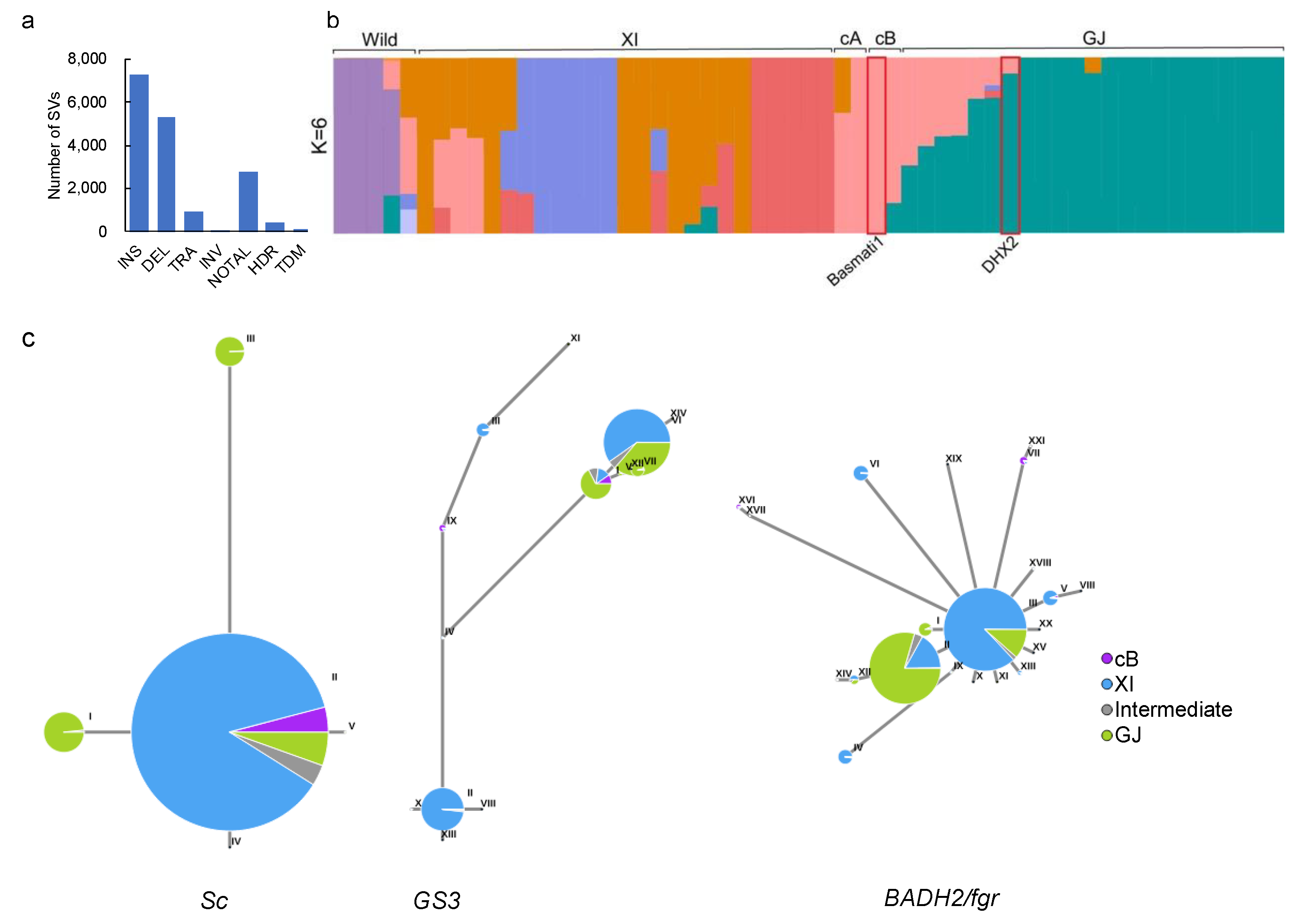
2.4. Introgression from Basmati Variety
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Pollen Fertility Test
4.3. Expression Analysis
4.4. Vector Construction and Plant Transformation
4.5. Population Structure Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, W.; Mauleon, R.; Hu, Z.; Chebotarov, D.; Tai, S.; Wu, Z.; Li, M.; Zheng, T.; Fuentes, R.R.; Zhang, F.; et al. Genomic variation in 3010 diverse accessions of Asian cultivated rice. Nature 2018, 557, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Yang, S.; Gong, J.; Zhao, Q.; Feng, Q.; Zhan, Q.; Zhao, Y.; Li, W.; Cheng, B.; Xia, J. Genomic architecture of heterosis for yield traits in rice. Nature 2016, 537, 629–633. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, Y.; Liu, Y.G.; Zhang, Q. Hybrid sterility in plant: Stories from rice. Curr. Opin. Plant Biol. 2010, 13, 186–192. [Google Scholar] [CrossRef] [PubMed]
- Kato, A. On the affinity of rice varieties as shown by the fertility of rice plants. Centr. Agric. Inst. Kyushu Imp. Univ. 1928, 2, 241–279. [Google Scholar]
- Liu, K.D.; Zhou, Z.Q.; Xu, C.G.; Zhang, Q.; Maroof, M.A.S. An analysis of hybrid sterility in rice using a diallel cross of 21 parents involving indica, japonica and wide compatibility varieties. Euphytica 1996, 90, 275–280. [Google Scholar] [CrossRef]
- Yang, S.; Shen, X.; Gu, W.; Cao, D. The breeding research of cross breeding between indica and japonica. Acta Agron. Sin. 1962, 1, 97–102. [Google Scholar]
- Yuan, L. The strategy of hybrid rice breeding. Hybrid Rice 1987, 1, 1–3. [Google Scholar]
- Zhang, G.Q. Prospects of utilization of inter-subspecific heterosis between indica and japonica rice—ScienceDirect. J. Integr. Agric. 2020, 19, 1–10. [Google Scholar] [CrossRef]
- Guo, J.; Xu, X.; Li, W.; Zhu, W.; Zhu, H.; Liu, Z.; Luan, X.; Dai, Z.; Liu, G.; Zhang, Z. Overcoming inter-subspecific hybrid sterility in rice by developing indica-compatible japonica lines. Sci. Rep. 2016, 6, 26878. [Google Scholar] [CrossRef]
- Long, Y.; Zhao, L.; Niu, B.; Su, J.; Wu, H.; Chen, Y.; Zhang, Q.; Guo, J.; Zhuang, C.; Mei, M.; et al. Hybrid male sterility in rice controlled by interaction between divergent alleles of two adjacent genes. Proc. Natl. Acad. Sci. USA 2008, 105, 18871–18876. [Google Scholar] [CrossRef]
- Mizuta, Y.; Harushima, Y.; Kurata, N. Rice pollen hybrid incompatibility caused by reciprocal gene loss of duplicated genes. Proc. Natl. Acad. Sci. USA 2010, 107, 20417–20422. [Google Scholar] [CrossRef] [PubMed]
- Yamagata, Y.; Yamamoto, E.; Aya, K.; Win, K.T.; Doi, K.; Sobrizal; Ito, T.; Kanamori, H.; Wu, J.; Matsumoto, T.; et al. Mitochondrial gene in the nuclear genome induces reproductive barrier in rice. Proc. Natl. Acad. Sci. USA 2010, 107, 1494–1499. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhao, X.; Cheng, K.; Du, H.; Ouyang, Y.; Chen, J.; Qiu, S.; Huang, J.; Jiang, Y.; Jiang, L.; et al. A killer-protector system regulates both hybrid sterility and segregation distortion in rice. Science 2012, 337, 1336–1340. [Google Scholar] [CrossRef] [PubMed]
- Kubo, T.; Takashi, T.; Ashikari, M.; Yoshimura, A.; Kurata, N. Two Tightly Linked Genes at the hsa1 Locus Cause Both F1 and F2 Hybrid Sterility in Rice. Mol. Plant 2016, 9, 221–232. [Google Scholar] [CrossRef]
- Shen, R.; Wang, L.; Liu, X.; Wu, J.; Jin, W.; Zhao, X.; Xie, X.; Zhu, Q.; Tang, H.; Li, Q.; et al. Genomic structural variation-mediated allelic suppression causes hybrid male sterility in rice. Nat. Commun. 2017, 8, 1310. [Google Scholar] [CrossRef]
- Xie, Y.; Shen, R.; Chen, L.; Liu, Y.G. Molecular mechanisms of hybrid sterility in rice. Sci. China. Life Sci. 2019, 62, 737–743. [Google Scholar] [CrossRef]
- Qin, P.; Lu, H.; Du, H.; Wang, H.; Chen, W.; Chen, Z.; He, Q.; Ou, S.; Zhang, H.; Li, X.; et al. Pan-genome analysis of 33 genetically diverse rice accessions reveals hidden genomic variations. Cell 2021, 184, 3542–3558.e16. [Google Scholar] [CrossRef]
- Stein, J.C.; Yu, Y.; Copetti, D.; Zwickl, D.J.; Zhang, L.; Zhang, C.; Chougule, K.; Gao, D.; Iwata, A.; Goicoechea, J.L.; et al. Genomes of 13 domesticated and wild rice relatives highlight genetic conservation, turnover and innovation across the genus Oryza. Nat. Genet. 2018, 50, 285–296. [Google Scholar] [CrossRef]
- Zhang, F.; Xue, H.; Dong, X.; Li, M.; Zheng, X.; Li, Z.; Xu, J.; Wang, W.; Wei, C. Long-read sequencing of 111 rice genomes reveals significantly larger pan-genomes. Genome Res. 2022, 32, 853–863. [Google Scholar] [CrossRef]
- Singh, V.; Singh, A.K.; Mohapatra, T.; Krishnan, S.G.; Ellur, R.K. Pusa Basmati 1121—a rice variety with exceptional kernel elongation and volume expansion after cooking. Rice 2018, 11, 19. [Google Scholar] [CrossRef]
- Mao, H.; Sun, S.; Yao, J.; Wang, C.; Yu, S.; Xu, C.; Li, X.; Zhang, Q. Linking differential domain functions of the GS3 protein to natural variation of grain size in rice. Proc. Natl. Acad. Sci. USA 2010, 107, 19579–19584. [Google Scholar] [CrossRef]
- Chen, S.; Yang, Y.; Shi, W.; Ji, Q.; He, F.; Zhang, Z.; Cheng, Z.; Liu, X.; Xu, M. Badh2, encoding betaine aldehyde dehydrogenase, inhibits the biosynthesis of 2-acetyl-1-pyrroline, a major component in rice fragrance. Plant Cell 2008, 20, 1850–1861. [Google Scholar] [CrossRef] [PubMed]
- Kovach, M.J.; Calingacion, M.N.; Fitzgerald, M.A.; McCouch, S.R. The origin and evolution of fragrance in rice (Oryza sativa L.). Proc. Natl. Acad. Sci. USA 2009, 106, 14444–14449. [Google Scholar] [CrossRef] [PubMed]
- Li, J.Y.; Wang, J.; Zeigler, R.S. The 3000 rice genomes project. GigaScience 2014, 3, 7. [Google Scholar] [CrossRef]
- Sun, J.; Liu, D.; Wang, J.; Ma, D.; Tang, L.; Gao, H.; Xu, Z.; Chen, W. The contribution of intersubspecific hybridization to the breeding of super-high-yielding japonica rice in northeast China. Theor. Appl. Genet. 2012, 125, 1149–1157. [Google Scholar] [CrossRef]
- Chen, Z.; Li, X.; Lu, H.; Gao, Q.; Du, H.; Peng, H.; Qin, P.; Liang, C. Genomic atlases of introgression and differentiation reveal breeding footprints in Chinese cultivated rice. J. Genet. Genom. = Yi Chuan Xue Bao 2020, 47, 637–649. [Google Scholar] [CrossRef]
- Cui, D.; Zhou, H.; Ma, X.; Lin, Z.; Sun, L.; Han, B.; Li, M.; Sun, J.; Liu, J.; Jin, G.; et al. Genomic insights on the contribution of introgressions from Xian/Indica to the genetic improvement of Geng/Japonica rice cultivars. Plant Commun. 2022, 3, 100325. [Google Scholar] [CrossRef]
- Lye, Z.N.; Purugganan, M.D. Copy Number Variation in Domestication. Trends Plant Sci. 2019, 24, 352–365. [Google Scholar] [CrossRef]
- Shang, L.; Li, X.; He, H.; Yuan, Q.; Song, Y.; Wei, Z.; Lin, H.; Hu, M.; Zhao, F.; Zhang, C.; et al. A super pan-genomic landscape of rice. Cell Res. 2022, 32, 878–889. [Google Scholar] [CrossRef]
- Wang, Y.; Li, F.; Zhang, F.; Wu, L.; Xu, N.; Sun, Q.; Chen, H.; Yu, Z.; Lu, J.; Jiang, K.; et al. Time-ordering japonica/geng genomes analysis indicates the importance of large structural variants in rice breeding. Plant Biotechnol. J. 2022. early view. [Google Scholar] [CrossRef]
- Wang, Y.; Xiong, G.; Hu, J.; Jiang, L.; Yu, H.; Xu, J.; Fang, Y.; Zeng, L.; Xu, E.; Xu, J. Copy number variation at the GL7 locus contributes to grain size diversity in rice. Nat. Genet. 2015, 47, 944–948. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Shen, R.; Chen, L.T.; Liu, Y.G. Characterization of a novel DUF1618 gene family in rice. J. Integr. Plant Biol. 2014, 56, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Chen, L.; Tian, S.; Lin, Y.; Tang, Q.; Zhou, X.; Li, D.; Yeung, C.K.L.; Che, T.; Jin, L.; et al. Comprehensive variation discovery and recovery of missing sequence in the pig genome using multiple de novo assemblies. Genome Res. 2017, 27, 865–874. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.D.; Scott, D.A.; Weinstein, J.A.; Ran, F.A.; Konermann, S.; Agarwala, V.; Li, Y.; Fine, E.J.; Wu, X.; Shalem, O. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013, 31, 827–832. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, A.; Aichi, I.; Matsuoka, M. A protocol for Agrobacterium-mediated transformation in rice. Nat. Protoc. 2006, 1, 2796. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, Q.; Zhu, Q.; Liu, W.; Chen, Y.; Qiu, R.; Wang, B.; Yang, Z.; Li, H.; Lin, Y. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 2015, 8, 1274–1284. [Google Scholar] [CrossRef]
- Marçais, G.; Delcher, A.L.; Phillippy, A.M.; Coston, R.; Salzberg, S.L.; Zimin, A. MUMmer4: A fast and versatile genome alignment system. PLoS Comput. Biol. 2018, 14, e1005944. [Google Scholar] [CrossRef]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef]
- Zhao, H.; Li, J.; Yang, L.; Qin, G.; Xia, C.; Xu, X.; Su, Y.; Liu, Y.; Ming, L.; Chen, L.L.; et al. An inferred functional impact map of genetic variants in rice. Mol. Plant 2021, 14, 1584–1599. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, N.; Xu, H.; Xu, Z.; Li, F.; Xu, Q. Introgression of a Complex Genomic Structural Variation Causes Hybrid Male Sterility in GJ Rice (Oryza sativa L.) Subspecies. Int. J. Mol. Sci. 2022, 23, 12804. https://doi.org/10.3390/ijms232112804
Xu N, Xu H, Xu Z, Li F, Xu Q. Introgression of a Complex Genomic Structural Variation Causes Hybrid Male Sterility in GJ Rice (Oryza sativa L.) Subspecies. International Journal of Molecular Sciences. 2022; 23(21):12804. https://doi.org/10.3390/ijms232112804
Chicago/Turabian StyleXu, Na, Hai Xu, Zhengjin Xu, Fengcheng Li, and Quan Xu. 2022. "Introgression of a Complex Genomic Structural Variation Causes Hybrid Male Sterility in GJ Rice (Oryza sativa L.) Subspecies" International Journal of Molecular Sciences 23, no. 21: 12804. https://doi.org/10.3390/ijms232112804
APA StyleXu, N., Xu, H., Xu, Z., Li, F., & Xu, Q. (2022). Introgression of a Complex Genomic Structural Variation Causes Hybrid Male Sterility in GJ Rice (Oryza sativa L.) Subspecies. International Journal of Molecular Sciences, 23(21), 12804. https://doi.org/10.3390/ijms232112804





