Classification of Subgroups with Immune Characteristics Based on DNA Methylation in Luminal Breast Cancer
Abstract
1. Introduction
2. Results
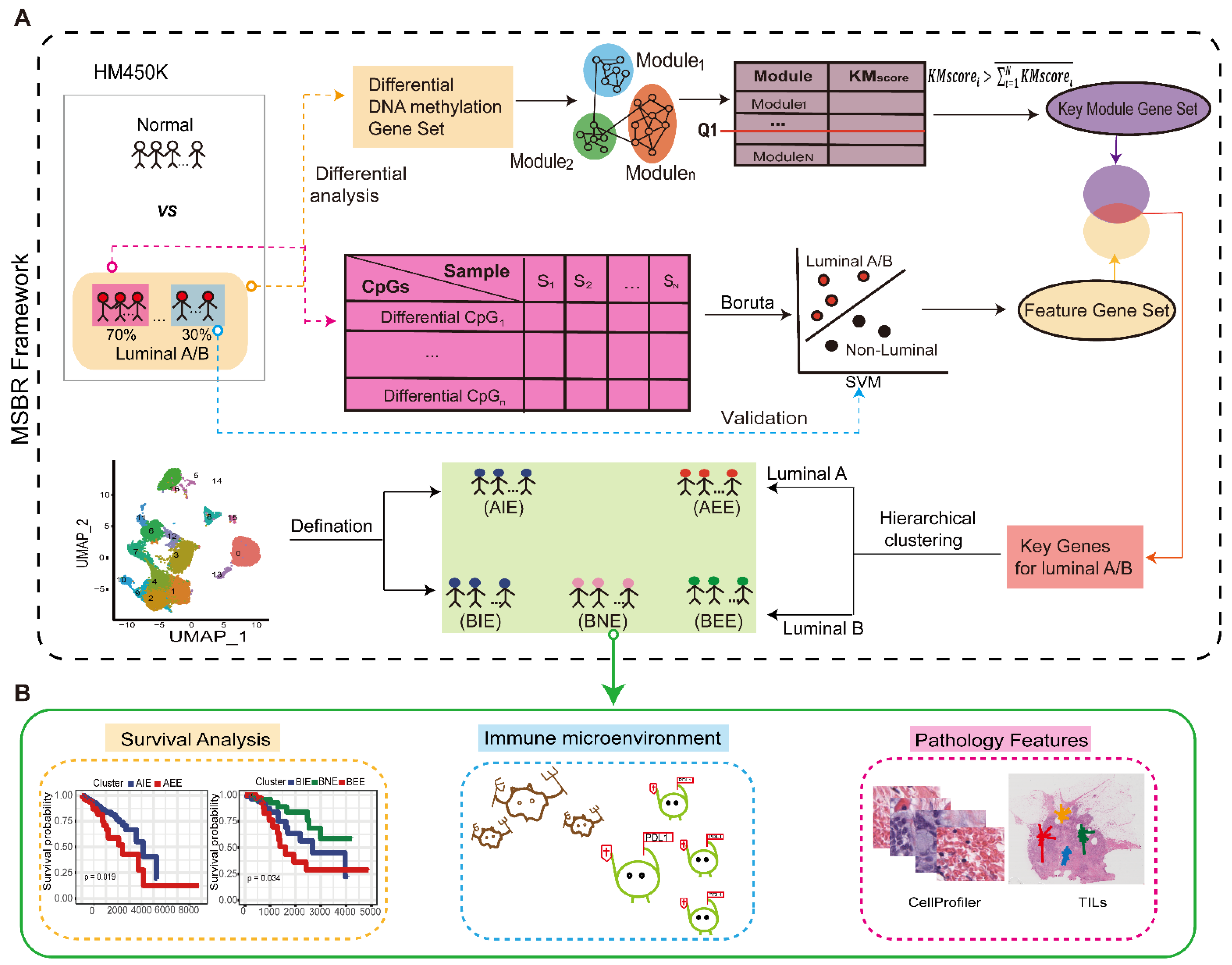
2.1. Overview of the Study
2.2. Acquisition of “Key Genes” from MSBR Framework
| Algorithm 1: Selection of key genes for subgrouping. | ||
| Input: KEGG pathways results (kegg pathway , p-value , genes sets , module number N, gene sets in module , feature DNA methylation genes . | ||
| Output: key genes | ||
| 1 | Take all the kegg pathways with p < 0.05 as set | |
| 2 | Obtain all genes in set as | |
| 3 | for to do | |
| 4 | Count the intersection of and , then divided by the number of elements of set as . | |
| 5 | end | |
| 6 | Define an empty set | |
| 7 | for to do | |
| 8 | if > the mean of {, , …, } and ≥ upper quartile of set {, , …, } then | |
| 9 | Add set into set | |
| 10 | end | |
| 11 | end | |
| 12 | Acquire set as the intersection of and | |
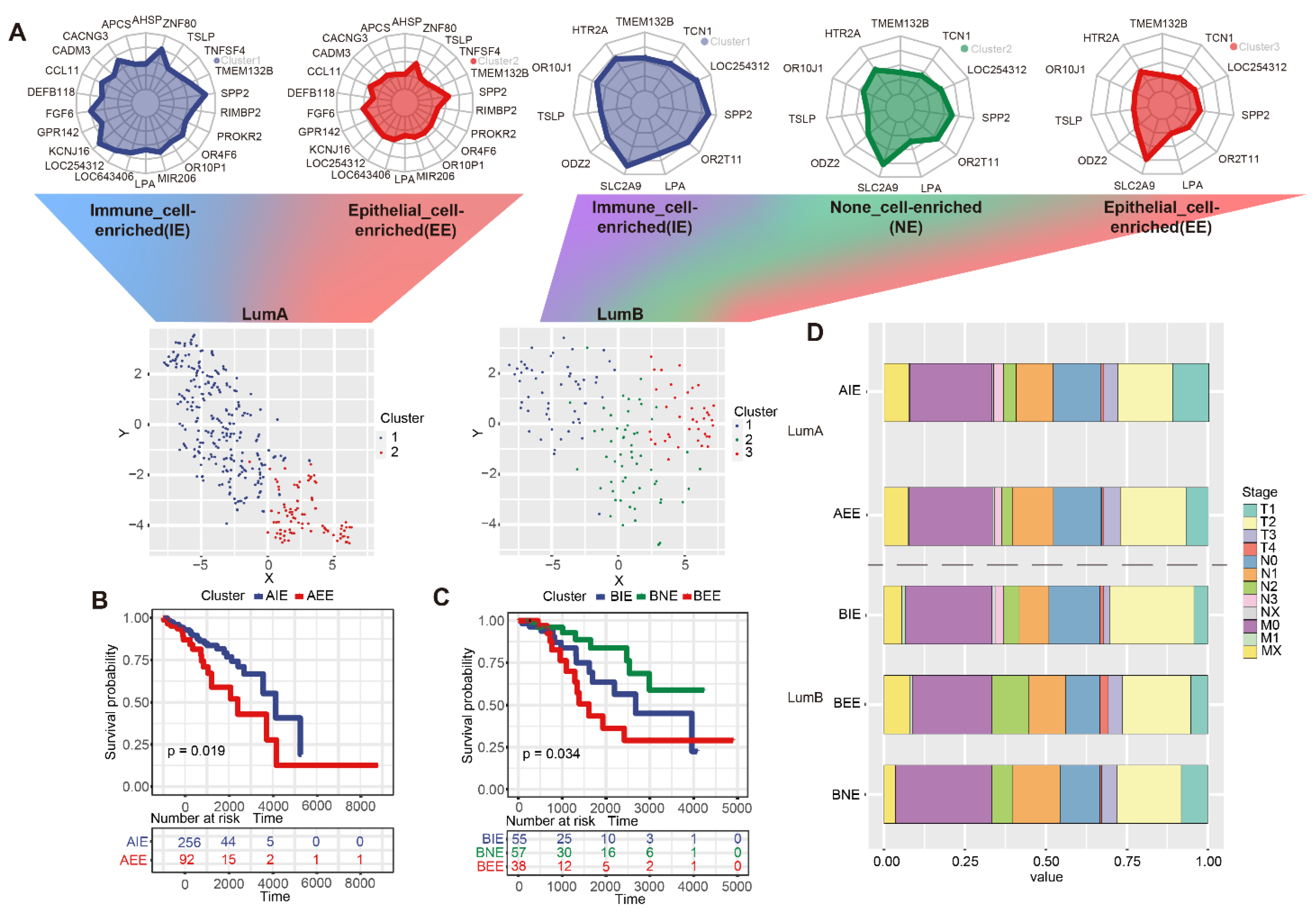
2.3. The Definition of Subgroups in Luminal A/B
2.4. The Immune Microenvironment in Subgroups
2.5. Differences in Pathological Characteristics of Subgroups
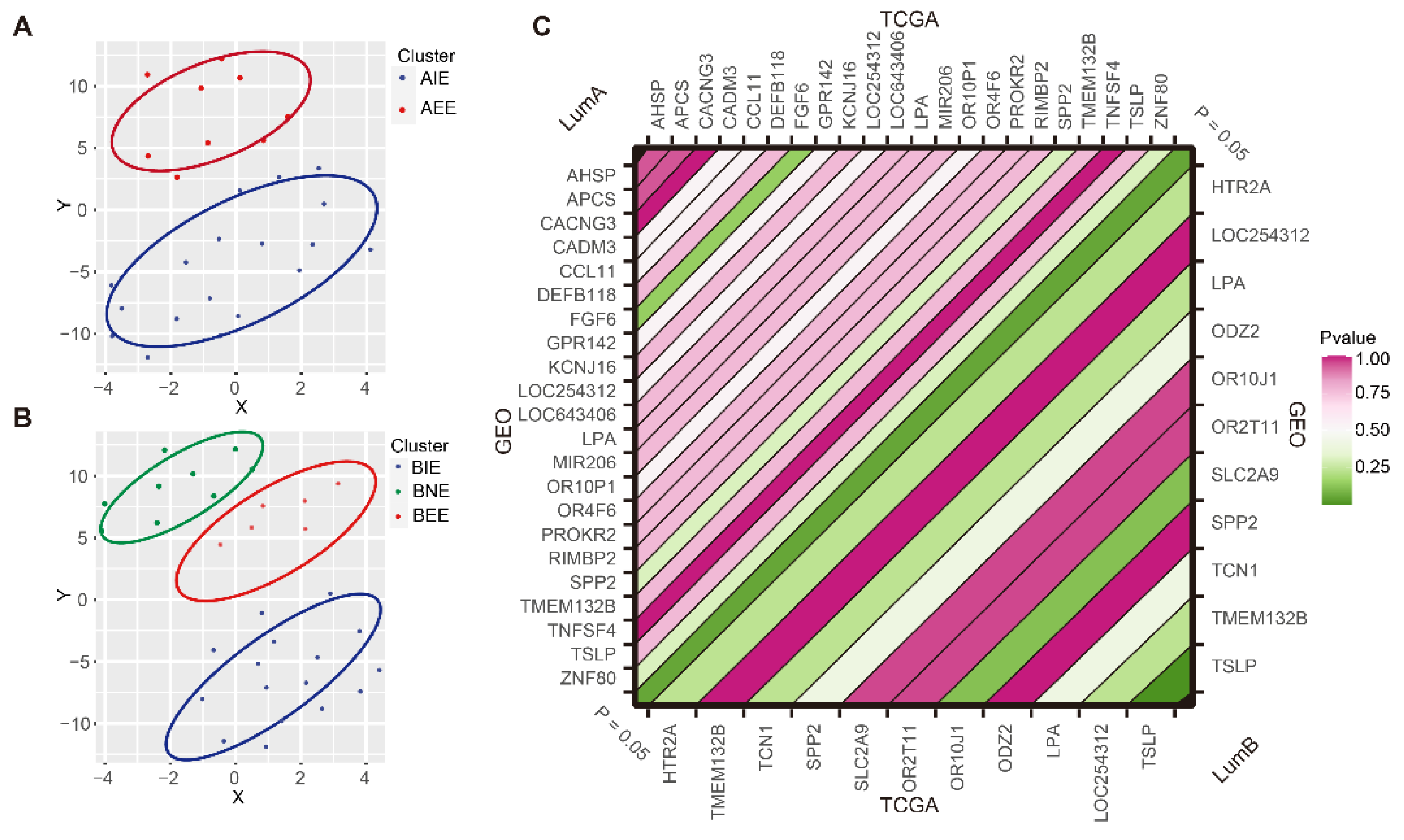
2.6. Subgroup Verification of GEO Data
3. Discussion
4. Materials and Methods
4.1. Data Source
4.2. Identify Differential DNA Methylation Sites and Genes
4.3. Functional Annotation of Differential DNA Methylation Genes
4.4. The Construction of MSBR Framework for Subgrouping Luminal BC
4.5. Single-Cell Signature Definition of Subgroups
4.6. Compare the Clinical Stage Differences between the Subgroups
4.7. Survival Analysis between Subgroups
4.8. Analysis of Immune Gene Methylation Levels and Immune Cell Infiltration between Subgroups
4.9. Calculation of Immune Cell Lysis Activity (CYT)
4.10. Features Extraction of Pathology Images
4.11. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Perou, C.M.; Sorlie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; Rees, C.A.; Pollack, J.R.; Ross, D.T.; Johnsen, H.; Akslen, L.A.; et al. Molecular portraits of human breast tumours. Nature 2000, 406, 747–752. [Google Scholar] [CrossRef] [PubMed]
- Howlader, N.; Altekruse, S.F.; Li, C.I.; Chen, V.W.; Clarke, C.A.; Ries, L.A.; Cronin, K.A. US incidence of breast cancer subtypes defined by joint hormone receptor and HER2 status. J. Natl. Cancer Inst. 2014, 106, dju055. [Google Scholar] [CrossRef]
- Ignatiadis, M.; Sotiriou, C. Luminal breast cancer: From biology to treatment. Nat. Rev. Clin. Oncol. 2013, 10, 494–506. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Tse, L.A.; Wang, D.; Koka, H.; Zhang, T.; Abubakar, M.; Lee, P.; Wang, F.; Wu, C.; Tsang, K.H.; et al. Immune gene expression profiling reveals heterogeneity in luminal breast tumors. Breast Cancer Res. 2019, 21, 147. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Wang, Y.; Gu, Y.; Zhu, J.; Ci, C.; Guo, Z.; Chen, C.; Wei, Y.; Lv, W.; Liu, H.; et al. Specific breast cancer prognosis-subtype distinctions based on DNA methylation patterns. Mol. Oncol. 2018, 12, 1047–1060. [Google Scholar] [CrossRef]
- Lehmann, B.D.; Pietenpol, J.A.; Tan, A.R. Triple-negative breast cancer: Molecular subtypes and new targets for therapy. Am. Soc. Clin. Oncol. Educ. Book 2015, 35, e31–e39. [Google Scholar] [CrossRef]
- Gao, J.J.; Swain, S.M. Luminal A Breast Cancer and Molecular Assays: A Review. Oncologist 2018, 23, 556–565. [Google Scholar] [CrossRef]
- Pogue-Geile, K.L.; Kim, C.; Jeong, J.H.; Tanaka, N.; Bandos, H.; Gavin, P.G.; Fumagalli, D.; Goldstein, L.C.; Sneige, N.; Burandt, E.; et al. Predicting degree of benefit from adjuvant trastuzumab in NSABP trial B-31. J. Natl. Cancer Inst. 2013, 105, 1782–1788. [Google Scholar] [CrossRef]
- Curtis, C.; Shah, S.P.; Chin, S.F.; Turashvili, G.; Rueda, O.M.; Dunning, M.J.; Speed, D.; Lynch, A.G.; Samarajiwa, S.; Yuan, Y.; et al. The genomic and transcriptomic architecture of 2000 breast tumours reveals novel subgroups. Nature 2012, 486, 346–352. [Google Scholar] [CrossRef]
- Kroemer, G.; Senovilla, L.; Galluzzi, L.; Andre, F.; Zitvogel, L. Natural and therapy-induced immunosurveillance in breast cancer. Nat. Med. 2015, 21, 1128–1138. [Google Scholar] [CrossRef]
- Netanely, D.; Avraham, A.; Ben-Baruch, A.; Evron, E.; Shamir, R. Expression and methylation patterns partition luminal-A breast tumors into distinct prognostic subgroups. Breast Cancer Res. 2016, 18, 74. [Google Scholar] [CrossRef] [PubMed]
- Bediaga, N.G.; Acha-Sagredo, A.; Guerra, I.; Viguri, A.; Albaina, C.; Ruiz Diaz, I.; Rezola, R.; Alberdi, M.J.; Dopazo, J.; Montaner, D.; et al. DNA methylation epigenotypes in breast cancer molecular subtypes. Breast Cancer Res. 2010, 12, R77. [Google Scholar] [CrossRef] [PubMed]
- Stefansson, O.A.; Moran, S.; Gomez, A.; Sayols, S.; Arribas-Jorba, C.; Sandoval, J.; Hilmarsdottir, H.; Olafsdottir, E.; Tryggvadottir, L.; Jonasson, J.G.; et al. A DNA methylation-based definition of biologically distinct breast cancer subtypes. Mol. Oncol. 2015, 9, 555–568. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.A.; Takai, D. The role of DNA methylation in mammalian epigenetics. Science 2001, 293, 1068–1070. [Google Scholar] [CrossRef] [PubMed]
- Keshet, I.; Schlesinger, Y.; Farkash, S.; Rand, E.; Hecht, M.; Segal, E.; Pikarski, E.; Young, R.A.; Niveleau, A.; Cedar, H.; et al. Evidence for an instructive mechanism of de novo methylation in cancer cells. Nat. Genet. 2006, 38, 149–153. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, M.M.; Bird, A. DNA methylation landscapes: Provocative insights from epigenomics. Nat. Rev. Genet. 2008, 9, 465–476. [Google Scholar] [CrossRef]
- Orlando, F.A.; Brown, K.D. Unraveling breast cancer heterogeneity through transcriptomic and epigenomic analysis. Ann. Surg. Oncol. 2009, 16, 2270–2279. [Google Scholar] [CrossRef][Green Version]
- Stirzaker, C.; Zotenko, E.; Song, J.Z.; Qu, W.; Nair, S.S.; Locke, W.J.; Stone, A.; Armstong, N.J.; Robinson, M.D.; Dobrovic, A.; et al. Methylome sequencing in triple-negative breast cancer reveals distinct methylation clusters with prognostic value. Nat. Commun. 2015, 6, 5899. [Google Scholar] [CrossRef]
- Xu, K.; Wang, R.T.; Xie, H.; Hu, L.F.; Wang, C.; Xu, J.L.; Zhu, C.J.; Liu, Y.Q.; Gao, F.Y.; Li, X.T.; et al. Single-cell RNA sequencing reveals cell heterogeneity and transcriptome profile of breast cancer lymph node metastasis. Oncogenesis 2021, 10, 66. [Google Scholar] [CrossRef]
- Takahashi, H.; Kawaguchi, T.; Yan, L.; Peng, X.; Qi, Q.; Morris, L.G.T.; Chan, T.A.; Tsung, A.; Otsuji, E.; Takabe, K. Immune Cytolytic Activity for Comprehensive Understanding of Immune Landscape in Hepatocellular Carcinoma. Cancers 2020, 12, 1221. [Google Scholar] [CrossRef]
- Chen, Z.; Wang, M.; De Wilde, R.L.; Feng, R.; Su, M.; Torres-de la Roche, L.A.; Shi, W. A Machine Learning Model to Predict the Triple Negative Breast Cancer Immune Subtype. Front. Immunol. 2021, 12, 749459. [Google Scholar] [CrossRef] [PubMed]
- Yizhak, K.; Aguet, F.; Kim, J.; Hess, J.M.; Kubler, K.; Grimsby, J.; Frazer, R.; Zhang, H.; Haradhvala, N.J.; Rosebrock, D.; et al. RNA sequence analysis reveals macroscopic somatic clonal expansion across normal tissues. Science 2019, 364, eaaw0726. [Google Scholar] [CrossRef] [PubMed]
- Hinshelwood, R.A.; Clark, S.J. Breast cancer epigenetics: Normal human mammary epithelial cells as a model system. J. Mol. Med. 2008, 86, 1315–1328. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Liu, G.; Zhou, F.; Su, B.; Li, Y. DNA methylation profiles in cancer diagnosis and therapeutics. Clin. Exp. Med. 2018, 18, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Wu, L.; Shu, X.O.; Cai, Q.; Shu, X.; Li, B.; Guo, X.; Ye, F.; Michailidou, K.; Bolla, M.K.; et al. Genetically Predicted Levels of DNA Methylation Biomarkers and Breast Cancer Risk: Data from 228,951 Women of European Descent. J. Natl. Cancer Inst. 2020, 112, 295–304. [Google Scholar] [CrossRef] [PubMed]
- Doroshow, D.B.; Bhalla, S.; Beasley, M.B.; Sholl, L.M.; Kerr, K.M.; Gnjatic, S.; Wistuba, I.I.; Rimm, D.L.; Tsao, M.S.; Hirsch, F.R. PD-L1 as a biomarker of response to immune-checkpoint inhibitors. Nat. Rev. Clin. Oncol. 2021, 18, 345–362. [Google Scholar] [CrossRef] [PubMed]
- Roufas, C.; Georgakopoulos-Soares, I.; Zaravinos, A. Molecular correlates of immune cytolytic subgroups in colorectal cancer by integrated genomics analysis. NAR Cancer 2021, 3, zcab005. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Chen, L.; Zeng, H.; Liao, Q.; Ji, J.; Ma, X. Integrative Analysis of Histopathological Images and Genomic Data in Colon Adenocarcinoma. Front. Oncol. 2021, 11, 636451. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research, N.; Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.R.; Ozenberger, B.A.; Ellrott, K.; Shmulevich, I.; Sander, C.; Stuart, J.M. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 2013, 45, 1113–1120. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Lichtenberg, T.; Hoadley, K.A.; Poisson, L.M.; Lazar, A.J.; Cherniack, A.D.; Kovatich, A.J.; Benz, C.C.; Levine, D.A.; Lee, A.V.; et al. An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell 2018, 173, 400–416. [Google Scholar] [CrossRef] [PubMed]
- Jeschke, J.; Bizet, M.; Desmedt, C.; Calonne, E.; Dedeurwaerder, S.; Garaud, S.; Koch, A.; Larsimont, D.; Salgado, R.; Van den Eynden, G.; et al. DNA methylation-based immune response signature improves patient diagnosis in multiple cancers. J. Clin. Investig. 2017, 127, 3090–3102. [Google Scholar] [CrossRef] [PubMed]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Charrad, M.; Ghazzali, N.; Boiteau, V.; Niknafs, A. Nbclust: An R Package for Determining the Relevant Number of Clusters in a Data Set. J. Stat. Softw. 2014, 61, 1–36. [Google Scholar] [CrossRef]
- Aran, D.; Looney, A.P.; Liu, L.; Wu, E.; Fong, V.; Hsu, A.; Chak, S.; Naikawadi, R.P.; Wolters, P.J.; Abate, A.R.; et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 2019, 20, 163–172. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Hao, S.; Andersen-Nissen, E.; Mauck, W.M., 3rd; Zheng, S.; Butler, A.; Lee, M.J.; Wilk, A.J.; Darby, C.; Zager, M.; et al. Integrated analysis of multimodal single-cell data. Cell 2021, 184, 3573–3587. [Google Scholar] [CrossRef] [PubMed]
- Thorsson, V.; Gibbs, D.L.; Brown, S.D.; Wolf, D.; Bortone, D.S.; Ou Yang, T.H.; Porta-Pardo, E.; Gao, G.F.; Plaisier, C.L.; Eddy, J.A.; et al. The Immune Landscape of Cancer. Immunity 2018, 48, 812–830. [Google Scholar] [CrossRef] [PubMed]
- McQuin, C.; Goodman, A.; Chernyshev, V.; Kamentsky, L.; Cimini, B.A.; Karhohs, K.W.; Doan, M.; Ding, L.; Rafelski, S.M.; Thirstrup, D.; et al. CellProfiler 3.0: Next-generation image processing for biology. PLoS Biol. 2018, 16, e2005970. [Google Scholar] [CrossRef] [PubMed]
- Saltz, J.; Gupta, R.; Hou, L.; Kurc, T.; Singh, P.; Nguyen, V.; Samaras, D.; Shroyer, K.R.; Zhao, T.; Batiste, R.; et al. Spatial Organization and Molecular Correlation of Tumor-Infiltrating Lymphocytes Using Deep Learning on Pathology Images. Cell Rep. 2018, 23, 181–193. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, M.; Ma, T.; Wang, C.; Zhao, J.; Xing, J.; Liu, H.; Su, M.; Zhai, R.; Liu, T.; Sun, B.; et al. Classification of Subgroups with Immune Characteristics Based on DNA Methylation in Luminal Breast Cancer. Int. J. Mol. Sci. 2022, 23, 12747. https://doi.org/10.3390/ijms232112747
Zhang M, Ma T, Wang C, Zhao J, Xing J, Liu H, Su M, Zhai R, Liu T, Sun B, et al. Classification of Subgroups with Immune Characteristics Based on DNA Methylation in Luminal Breast Cancer. International Journal of Molecular Sciences. 2022; 23(21):12747. https://doi.org/10.3390/ijms232112747
Chicago/Turabian StyleZhang, Mengyan, Te Ma, Cong Wang, Jiyun Zhao, Jie Xing, Honghao Liu, Mu Su, Ruiyang Zhai, Ting Liu, Baoqing Sun, and et al. 2022. "Classification of Subgroups with Immune Characteristics Based on DNA Methylation in Luminal Breast Cancer" International Journal of Molecular Sciences 23, no. 21: 12747. https://doi.org/10.3390/ijms232112747
APA StyleZhang, M., Ma, T., Wang, C., Zhao, J., Xing, J., Liu, H., Su, M., Zhai, R., Liu, T., Sun, B., & Zhang, Y. (2022). Classification of Subgroups with Immune Characteristics Based on DNA Methylation in Luminal Breast Cancer. International Journal of Molecular Sciences, 23(21), 12747. https://doi.org/10.3390/ijms232112747






