Metabolomics Profiling of Nephrotic Syndrome towards Biomarker Discovery
Abstract
1. Introduction
2. Results
2.1. Demographics, Clinical and Molecular Features in NS Patients
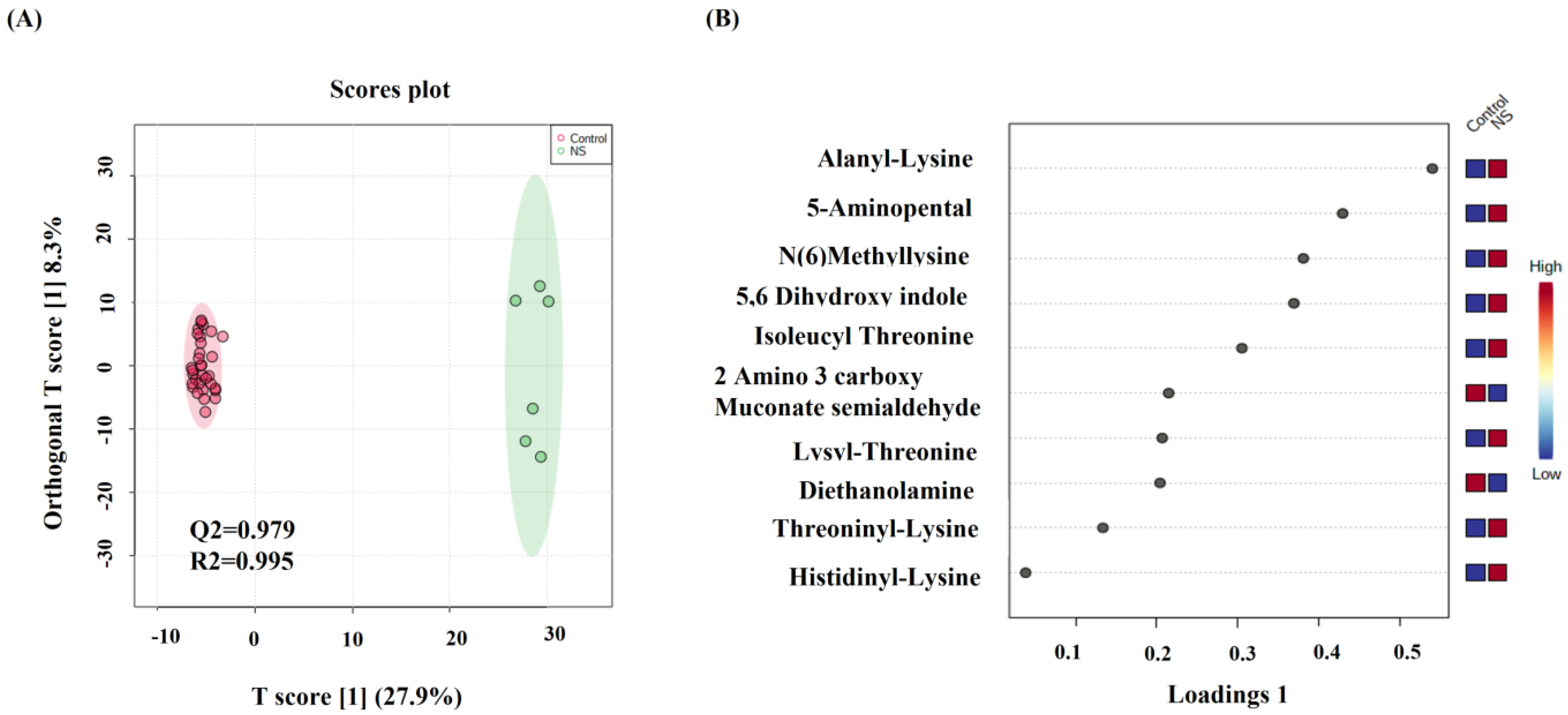
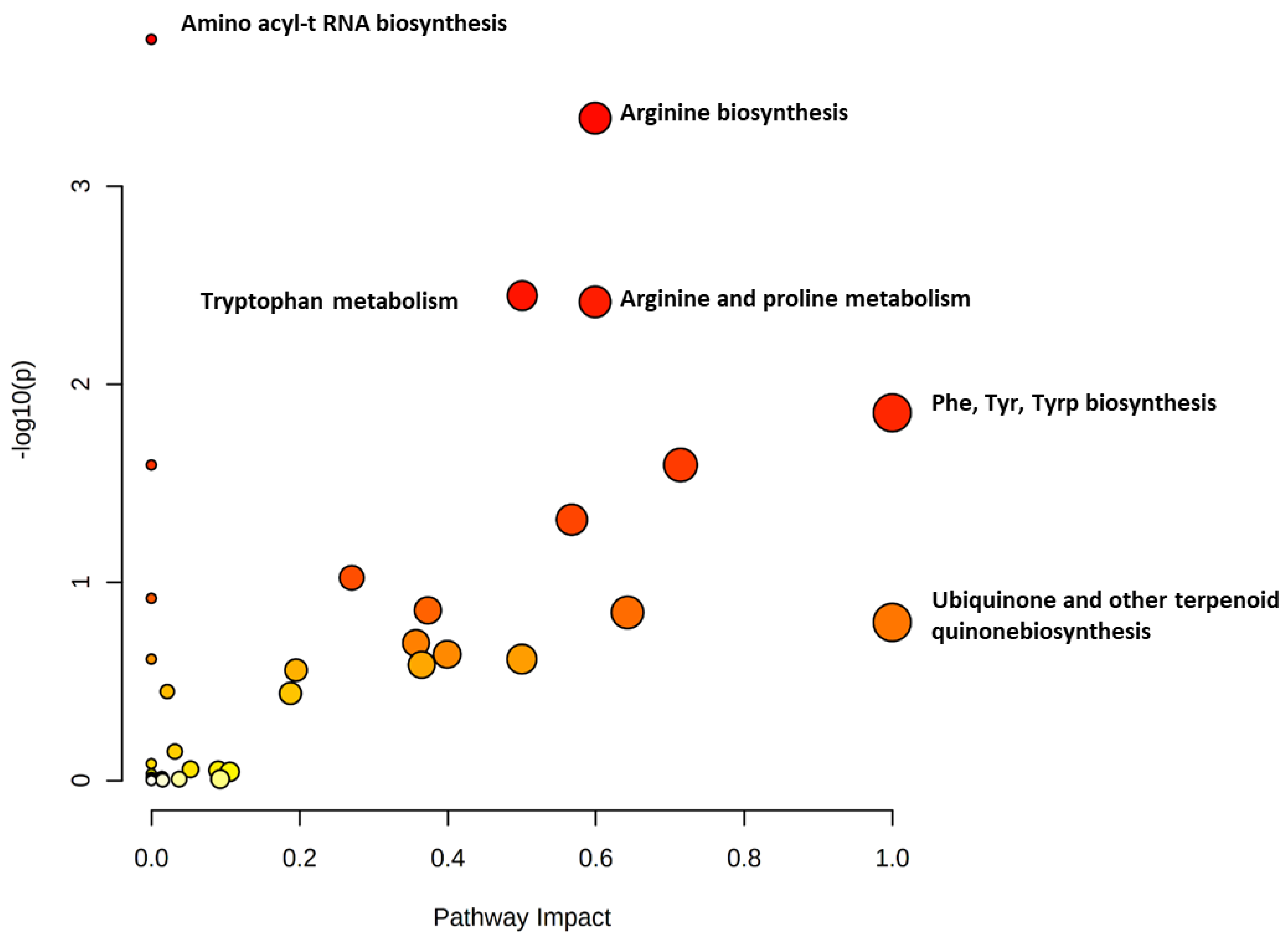
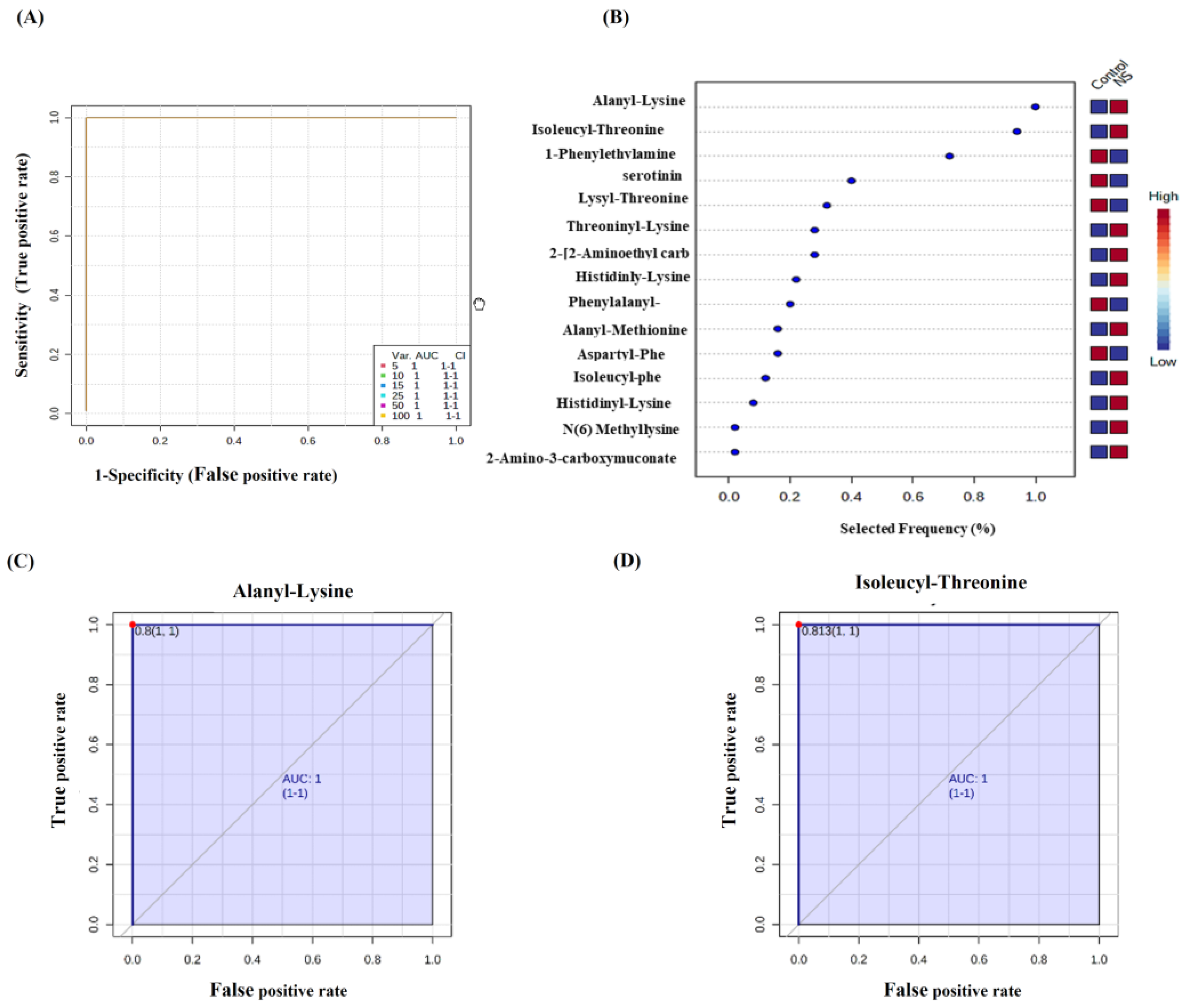
2.2. Metabolomics Profiling for NS Compared to Control
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Characteristics of the Study Population
4.3. CIL LC–MS Metabolomics Profiling
4.4. Data Processing and Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gooding, J.R.; Agrawal, S.; McRitchie, S.; Acuff, Z.; Merchant, M.L.; Klein, J.B.; Smoyer, W.E.; Sumner, S.J.; Mahan, J.; Patel, H. Predicting and defining steroid resistance in pediatric nephrotic syndrome using plasma metabolomics. Kidney Int. Rep. 2020, 5, 81–93. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.S.; Greenbaum, L.A. Nephrotic Syndrome. Pediatr. Clin. N. Am. 2019, 66, 73–85. [Google Scholar] [CrossRef] [PubMed]
- Larkins, N.G.; Liu, I.D.; Willis, N.S.; Craig, J.C.; Hodson, E.M. Non-corticosteroid immunosuppressive medications for steroid-sensitive nephrotic syndrome in children. Cochrane Database Syst. Rev. 2020, 4, CD002290. [Google Scholar] [CrossRef] [PubMed]
- Thalgahagoda, S.; Webb, N.J.; Roberts, D.; Birch, A.; Milford, D.V.; Tavakoli, A.; Shenoy, M. Successful ABO incompatible renal transplantation following rituximab and DFPP after failed immunoadsorption. Pediatr. Transplant. 2014, 18, E74–E76. [Google Scholar] [CrossRef] [PubMed]
- Feltran, L.S.; Watanabe, A.; Guaragna, M.S.; Machado, I.C.; Casimiro, F.M.; Neves, P.D.; Palma, L.M.; Varela, P.; Vaisbich, M.H.; Marie, S.K. Brazilian network of pediatric nephrotic syndrome (REBRASNI). Kidney Int. Rep. 2020, 5, 358–362. [Google Scholar] [CrossRef] [PubMed]
- Adams, D.; Tollinche, L.; Yeoh, C.; Artman, J.; Mehta, M.; Phillips, D.; Fischer, G.; Quinlan, J.; Sakai, T. Short-term safety and effectiveness of sugammadex for surgical patients with end-stage renal disease: A two-centre retrospective study. Anaesthesia 2020, 75, 348–352. [Google Scholar] [CrossRef]
- Kodner, C. Diagnosis and management of nephrotic syndrome in adults. Am. Fam. Phys. 2016, 93, 479–485. [Google Scholar]
- Stone, H.; Magella, B.; Bennett, M.R. The search for biomarkers to aid in diagnosis, differentiation, and prognosis of childhood idiopathic nephrotic syndrome. Front. Pediatr. 2019, 7, 404. [Google Scholar] [CrossRef]
- Guo, J.; Zhao, J.; Liu, R.; Yu, J.; Zhang, M.; Wang, H.; Liu, L. Metabolomics analysis of serum in pediatric nephrotic syndrome based on targeted and non-targeted platforms. Metabolomics 2021, 17, 38. [Google Scholar] [CrossRef]
- Gagnebin, Y.; Julien, B.; Belén, P.; Serge, R. Metabolomics in chronic kidney disease: Strategies for extended metabolome coverage. J. Pharm. Biomed. Anal. 2018, 161, 313–325. [Google Scholar] [CrossRef]
- Kalim, S.; Rhee, E.P. An overview of renal metabolomics. Kidney Int. 2017, 91, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Huang, X.; Ärnlöv, J.; Cederholm, T.; Stenvinkel, P.; Lindholm, B.; Risérus, U.; Carrero, J.J. Clinical correlates of insulin sensitivity and its association with mortality among men with CKD stages 3 and 4. Clin. J. Am. Soc. Nephrol. 2014, 9, 690–697. [Google Scholar] [CrossRef] [PubMed]
- Rhee, E.P. A Systems-Level View of Renal Metabolomics. Semin. Nephrol. 2018, 38, 142–150. [Google Scholar] [CrossRef] [PubMed]
- Abbiss, H.; Maker, G.L.; Trengove, R.D. Metabolomics approaches for the diagnosis and understanding of kidney diseases. Metabolites 2019, 9, 34. [Google Scholar] [CrossRef]
- Hao, X.; Liu, X.; Wang, W.; Ren, H.; Xie, J.; Shen, P.; Lin, D.; Chen, N. Distinct metabolic profile of primary focal segmental glomerulosclerosis revealed by NMR-based metabolomics. PLoS ONE 2013, 8, e78531. [Google Scholar] [CrossRef]
- Gao, X.; Chen, W.; Li, R.; Wang, M.; Chen, C.; Zeng, R.; Deng, Y. Systematic variations associated with renal disease uncovered by parallel metabolomics of urine and serum. BMC Syst. Biol. 2012, 6 (Suppl. 1), S14. [Google Scholar] [CrossRef]
- Jia, L.; Wang, C.; Zhao, S.; Lu, X.; Xu, G. Metabolomic identification of potential phospholipid biomarkers for chronic glomerulonephritis by using high performance liquid chromatography–mass spectrometry. J. Chromatogr. B 2007, 860, 134–140. [Google Scholar] [CrossRef]
- Lee, J.E.; Lee, Y.H.; Kim, S.Y.; Kim, Y.G.; Moon, J.Y.; Jeong, K.H.; Lee, T.W.; Ihm, C.G.; Kim, S.; Kim, K.H.; et al. Systematic biomarker discovery and coordinative validation for different primary nephrotic syndromes using gas chromatography-mass spectrometry. J. Chromatogr. A 2016, 1453, 105–115. [Google Scholar] [CrossRef]
- Guo, K.; Li, L. Differential 12C-/13C-isotope dansylation labeling and fast liquid chromatography/mass spectrometry for absolute and relative quantification of the metabolome. Anal. Chem. 2009, 81, 3919–3932. [Google Scholar] [CrossRef]
- Han, W.; Li, L. Chemical Isotope Labeling LC-MS for Human Blood Metabolome Analysis. Methods Mol. Biol. 2018, 1730, 213–225. [Google Scholar] [CrossRef]
- Sriwi, D.; Alabdaljabar, M.S.; Jacob, M.; Mujamammi, A.H.; Gu, X.; Sabi, E.M.; Li, L.; Hussein, M.H.; Dasouki, M.; Abdel Rahman, A.M. Metabolomics Profiling of Cystic Renal Disease towards Biomarker Discovery. Biology 2021, 10, 770. [Google Scholar] [CrossRef] [PubMed]
- Polanco, N.; Gutiérrez, E.; Covarsí, A.; Ariza, F.; Carreño, A.; Vigil, A.; Baltar, J.; Fernández-Fresnedo, G.; Martín, C.; Pons, S.; et al. Spontaneous remission of nephrotic syndrome in idiopathic membranous nephropathy. J. Am. Soc. Nephrol. JASN 2010, 21, 697–704. [Google Scholar] [CrossRef] [PubMed]
- Hilmanto, D.; Mawardi, F.; Lestari, A.S.; Widiasta, A. Disease-Associated Systemic Complications in Childhood Nephrotic Syndrome: A Systematic Review. Int. J. Nephrol. Renov. Dis. 2022, 15, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Hollis, A.; Dart, A.; Morgan, C.; Mammen, C.; Zappitelli, M.; Chanchlani, R.; Samuel, S. Delays in diagnosis of nephrotic syndrome in children: A survey study. Paediatr. Child Health 2019, 24, 258–262. [Google Scholar] [CrossRef] [PubMed]
- Jacob, M.; Lopata, A.L.; Dasouki, M.; Abdel Rahman, A.M. Metabolomics toward personalized medicine. Mass Spectrom. Rev. 2019, 38, 221–238. [Google Scholar] [CrossRef]
- Frank, J.; Gupta, A.; Osadchiy, V.; Mayer, E.A. Brain-Gut-Microbiome Interactions and Intermittent Fasting in Obesity. Nutrients 2021, 13, 584. [Google Scholar] [CrossRef]
- Zhou, R.; Tseng, C.L.; Huan, T.; Li, L. IsoMS: Automated processing of LC-MS data generated by a chemical isotope labeling metabolomics platform. Anal. Chem. 2014, 86, 4675–4679. [Google Scholar] [CrossRef]
- Matyjek, A.; Literacki, S.; Niemczyk, S.; Rymarz, A. Protein energy-wasting associated with nephrotic syndrome—The comparison of metabolic pattern in severe nephrosis to different stages of chronic kidney disease. BMC Nephrol. 2020, 21, 346. [Google Scholar] [CrossRef]
- Zanetti, M.; Barazzoni, R.; Garibotto, G.; Davanzo, G.; Gabelli, C.; Kiwanuka, E.; Piccoli, A.; Tosolini, M.; Tessari, P. Plasma protein synthesis in patients with low-grade nephrotic proteinuria. Am. J. Physiol. Endocrinol. Metab. 2001, 280, E591–E597. [Google Scholar] [CrossRef]
- Shouman, M.; Abdallah, N.; El Tablawy, N.; Rashed, L. Biochemical markers of endothelial dysfunction in pediatric nephrotic syndrome. Arch. Med. Sci. 2009, 5, 415–421. [Google Scholar]
- Hocher, B.; Adamski, J. Metabolomics for clinical use and research in chronic kidney disease. Nat. Rev. Nephrol. 2017, 13, 269–284. [Google Scholar] [CrossRef] [PubMed]
- Schefold, J.C.; Zeden, J.-P.; Fotopoulou, C.; von Haehling, S.; Pschowski, R.; Hasper, D.; Volk, H.-D.; Schuett, C.; Reinke, P. Increased indoleamine 2, 3-dioxygenase (IDO) activity and elevated serum levels of tryptophan catabolites in patients with chronic kidney disease: A possible link between chronic inflammation and uraemic symptoms. Nephrol. Dial. Transplant. 2009, 24, 1901–1908. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; LI, J.; Qiao, S.; Xia, Y.; Tang, F. Application of metabolomics in establishing primary nephrotic syndrome diagnosis model. Chin. J. Nephrol. 2016, 32, 334–338. [Google Scholar]
- Hu, X.; Ouyang, S.; Xie, Y.; Gong, Z.; Du, J. Characterizing the gut microbiota in patients with chronic kidney disease. Postgrad. Med. 2020, 132, 495–505. [Google Scholar] [CrossRef]
- National Center for Biotechnology Information. N(6)-Methyllysine. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/N_6_-Methyllysine (accessed on 12 August 2022.).
- Di Blasi, R.; Blyuss, O.; Timms, J.F.; Conole, D.; Ceroni, F.; Whitwell, H.J. Non-histone protein methylation: Biological significance and bioengineering potential. ACS Chem. Biol. 2021, 16, 238–250. [Google Scholar] [CrossRef]
- Kaysen, G.; Carstensen, A.; Martin, V. Muscle protein synthesis is impaired in nephrotic rats. Miner. Electrolyte Metab. 1992, 18, 228–232. [Google Scholar]
- Trisrivirat, D.; Lawan, N.; Chenprakhon, P.; Matsui, D.; Asano, Y.; Chaiyen, P. Mechanistic insights into the dual activities of the single active site of l-lysine oxidase/monooxygenase from Pseudomonas sp. AIU 813. J. Biol. Chem. 2020, 295, 11246–11261. [Google Scholar] [CrossRef]
- Demir, S.; Kaplan, O.; Celebier, M.; Sag, E.; Bilginer, Y.; Lay, I.; Ozen, S. Predictive biomarkers of IgA vasculitis with nephritis by metabolomic analysis. Semin. Arthritis Rheum. 2020, 50, 1238–1244. [Google Scholar] [CrossRef]
- Lee, H.; Jang, H.B.; Yoo, M.G.; Park, S.I.; Lee, H.J. Amino Acid Metabolites Associated with Chronic Kidney Disease: An Eight-Year Follow-Up Korean Epidemiology Study. Biomedicines 2020, 8, 222. [Google Scholar] [CrossRef]
- Banday, A.A.; Lokhandwala, M.F. Renal dopamine oxidation and inflammation in high salt fed rats. J. Am. Heart Assoc. 2020, 9, e014977. [Google Scholar] [CrossRef]
- Stolzenberg-Solomon, R.; Derkach, A.; Moore, S.; Weinstein, S.J.; Albanes, D.; Sampson, J. Associations between metabolites and pancreatic cancer risk in a large prospective epidemiological study. Gut 2020, 69, 2008–2015. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Guo, A.; Oler, E.; Wang, F.; Anjum, A.; Peters, H.; Dizon, R.; Sayeeda, Z.; Tian, S.; Lee, B.L.; et al. HMDB 5.0: The Human Metabolome Database for 2022. Nucleic Acids Res. 2021, 50, D622–D631. [Google Scholar] [CrossRef] [PubMed]
- Fragiadaki, M.; Lannoy, M.; Themanns, M.; Maurer, B.; Leonhard, W.N.; Peters, D.J.; Moriggl, R.; Ong, A.C. STAT5 drives abnormal proliferation in autosomal dominant polycystic kidney disease. Kidney Int. 2017, 91, 575–586. [Google Scholar] [CrossRef] [PubMed]
- Greenbaum, L.A.; Benndorf, R.; Smoyer, W.E. Childhood nephrotic syndrome—Current and future therapies. Nat. Rev. Nephrol. 2012, 8, 445–458. [Google Scholar] [CrossRef]
- Lim, V.S.; Kopple, J.D. Protein metabolism in patients with chronic renal failure: Role of uremia and dialysis. Kidney Int. 2000, 58, 1–10. [Google Scholar] [CrossRef]
- Jacob, M.; Gu, X.; Luo, X.; Al-Mousa, H.; Arnaout, R.; Al-Saud, B.; Lopata, A.L.; Li, L.; Dasouki, M.; Rahman, A.M.A. Metabolomics distinguishes DOCK8 deficiency from atopic dermatitis: A Biomarker discovery. Front. Immunol. 2019, 9, 274. [Google Scholar] [CrossRef]
- Goedert, J.J.; Sampson, J.N.; Moore, S.C.; Xiao, Q.; Xiong, X.; Hayes, R.B.; Ahn, J.; Shi, J.; Sinha, R. Fecal metabolomics: Assay performance and association with colorectal cancer. Carcinogenesis 2014, 35, 2089–2096. [Google Scholar] [CrossRef]
- Brown, D.G.; Rao, S.; Weir, T.L.; O’Malia, J.; Bazan, M.; Brown, R.J.; Ryan, E.P. Metabolomics and metabolic pathway networks from human colorectal cancers, adjacent mucosa, and stool. Cancer Metab. 2016, 4, 11. [Google Scholar] [CrossRef]
- Platten, M.; Nollen, E.A.A.; Röhrig, U.F.; Fallarino, F.; Opitz, C.A. Tryptophan metabolism as a common therapeutic target in cancer, neurodegeneration and beyond. Nat. Rev. Drug Discov. 2019, 18, 379–401. [Google Scholar] [CrossRef]
- Xia, J.; Psychogios, N.; Young, N.; Wishart, D.S. MetaboAnalyst: A web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 2009, 37, W652–W660. [Google Scholar] [CrossRef]

| Patient | NS Phenotype/ Renal Disease | Age (Yrs) | BMI | Body Weight (Kg) | SBP (mmHg) | DBP (mmHg) | eGFR (>60 mL/min/ 1.73 m2) | Serum Creatinine (64–115 μmol/L) | Urea (2.3–6.7 mmol/L) | Protein Dipstick | Protein Random Urine (g/L) | Pr/Cr Random Urine Ratio (mg/mmol) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NS-1 | Congenital | 15 | 14.6 | 29.5 | 138 | 87 | N/A | 412 | 12.8 | 3+ | 1.52 | 323.4 |
| NS-2 | MCD, Relapsing | 36 | 29.1 | 72 | 113 | 75 | >60 | 65 | 4.4 | trace | 0 | 5.65 |
| NS-3 | FSGS, Steroid-dependent | 19 | 17.6 | 45 | 131 | 91 | >60 | 57 | 4.6 | 2+ | 0.49 | 32.77 |
| NS-4 | Steroid-dependent, FSGS + MCD | 18 | 40.4 | 106 | 128 | 75 | >60 | 87 | 14.2 | 3+ | 4.26 | 132.3 |
| NS-5 | Steriod-dependent, MCD | 14 | 37 | 83.3 | 132 | 74 | N/A | 42 | 3.3 | Negative | 0.17 | 10.69 |
| NS-6 | Steriod-dependent, MCD | 14 | 34 | 72.5 | 108 | 62 | N/A | 98 | 5.2 | 2+ | 0.69 | 127.78 |
| Average ± SEM | 19.3 ± 3.44 | 28.8 ± 4.31 | 68.1 ± 11.14 | 125 ± 4.82 | 77.3 ± 4.23 | 126.8 ± 57.63 | 7.4 ± 1.95 | 1.2 ± 0.65 | 105.4 ± 49.31 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jacob, M.; Nimer, R.M.; Alabdaljabar, M.S.; Sabi, E.M.; Al-Ansari, M.M.; Housien, M.; Sumaily, K.M.; Dahabiyeh, L.A.; Abdel Rahman, A.M. Metabolomics Profiling of Nephrotic Syndrome towards Biomarker Discovery. Int. J. Mol. Sci. 2022, 23, 12614. https://doi.org/10.3390/ijms232012614
Jacob M, Nimer RM, Alabdaljabar MS, Sabi EM, Al-Ansari MM, Housien M, Sumaily KM, Dahabiyeh LA, Abdel Rahman AM. Metabolomics Profiling of Nephrotic Syndrome towards Biomarker Discovery. International Journal of Molecular Sciences. 2022; 23(20):12614. https://doi.org/10.3390/ijms232012614
Chicago/Turabian StyleJacob, Minnie, Refat M. Nimer, Mohamad S. Alabdaljabar, Essa M. Sabi, Mysoon M. Al-Ansari, Maged Housien, Khalid M. Sumaily, Lina A. Dahabiyeh, and Anas M. Abdel Rahman. 2022. "Metabolomics Profiling of Nephrotic Syndrome towards Biomarker Discovery" International Journal of Molecular Sciences 23, no. 20: 12614. https://doi.org/10.3390/ijms232012614
APA StyleJacob, M., Nimer, R. M., Alabdaljabar, M. S., Sabi, E. M., Al-Ansari, M. M., Housien, M., Sumaily, K. M., Dahabiyeh, L. A., & Abdel Rahman, A. M. (2022). Metabolomics Profiling of Nephrotic Syndrome towards Biomarker Discovery. International Journal of Molecular Sciences, 23(20), 12614. https://doi.org/10.3390/ijms232012614






