Abstract
Nonalcoholic fatty liver disease (NAFLD) is the most prevalent liver pathology worldwide. Meanwhile, liver cancer represents the sixth most common malignancy, with hepatocellular carcinoma (HCC) as the primary, most prevalent subtype. Due to the rising incidence of metabolic disorders, NAFLD has become one of the main contributing factors to HCC development. However, although NAFLD might account for about a fourth of HCC cases, there is currently a significant gap in HCC surveillance protocols regarding noncirrhotic NAFLD patients, so the majority of NAFLD-related HCC cases were diagnosed in late stages when survival chances are minimal. However, in the past decade, the focus in cancer genomics has shifted towards the noncoding part of the genome, especially on the microRNAs (miRNAs) and long noncoding RNAs (lncRNAs), which have proved to be involved in the regulation of several malignant processes. This review aims to summarize the current knowledge regarding some of the main dysregulated, noncoding RNAs (ncRNAs) and their implications for NAFLD and HCC development. A central focus of the review is on miRNA and lncRNAs that can influence the progression of NAFLD towards HCC and how they can be used as potential screening tools and future therapeutic targets.
1. Introduction
Liver cancer is a frequent and aggressive cancer type, with 905,677 newly diagnosed cases and over 830,180 deaths worldwide in 2020 alone. The annual incidence of hepatic cancers is expected to exceed one million cases by 2025 [1,2]. Hepatocellular carcinoma (HCC) accounts for roughly 90% of all primary hepatic tumors, which makes it the sixth most common type of cancer worldwide and the third leading cause of cancer-associated mortality. Such a discrepancy between incidence and mortality emphasizes the aggressiveness and poor prognosis associated with this malignancy [3,4,5,6,7,8]. HCC prognosis is strongly linked with tumor stage, the best survival rates being reported in early diagnosed patients, whereas there is currently no curative option for advanced HCC stages [9,10].
NAFLD includes a heterogeneous variety of progressive conditions that range from steatosis (intrahepatic fat accumulation exceeding 5–10% by weight) to nonalcoholic steatohepatitis (NASH), which may progress for some patients to liver cirrhosis, HCC, and, ultimately, liver failure [11,12]. NAFLD remains the most common liver disease, with a worldwide prevalence of 25% [13,14]. This complex metabolic disorder has recently emerged as one of the main contributing factors to HCC development. The link between human cancer and viral infections has already been well documented [15,16]. However, although cirrhosis and chronic hepatitis B infection remain the main risk factors linked with HCC (in 50% of all cases), a growing amount of clinical evidence suggests that approximately 24% of the HCC cases are a consequence of neglected NAFLD [17,18,19,20,21,22]. This constant rise in NAFLD-related HCC relies on two distinct epidemiologic events. First, the growing incidence of other metabolic-syndrome-associated conditions, such as hyperlipidemia and excessive body weight, obesity, type 2 diabetes mellitus (T2DM), and insulin resistance [23,24,25], are jointly contributing towards hepatocarcinogenesis; thus, NAFLD is expected to become the leading cause of HCC [26,27,28,29,30,31,32,33,34] and the ultimate indication for HCC-related liver transplant candidates as well [35,36,37,38]. Second, with the successful implementation of anti-hepatitis B/C virus campaigns, the risk of viral-driven HCC has significantly decreased, NAFLD emerging as a primary risk factor for HCC [39]. Altogether, these two epidemiological events are promoting NAFLD as one of the primary causes of HCC.
Hepatocarcinogenesis can occur in both cirrhotic and noncirrhotic livers. However, since the presence of a pre-existing cirrhotic state has been reported in up to 80–90% of HCCs, screening is currently recommended for cirrhotic patients only, which leaves uncovered a significant part of early-stage HCC cases, especially since noncirrhotic patients are at elevated risk compared to the general population being frequently clinically silent. Therefore, noncirrhotic, nonviral, NAFLD-related HCC cases are usually diagnosed late, with limited therapeutic options. This emphasizes the need for a clearer understanding of the mechanisms underlying the steatosis–hepatocarcinogenesis transition, which could aid in discovering novel genetic biomarkers and in developing better screening methods [40,41,42,43].
Previous studies on liver tumorigenesis focused on exploring the genome’s protein-coding regions due to their central role in protein synthesis. However, less than 2% of the human genome encodes proteins, whereas, in the remaining 98% of the DNA, noncoding sequences and their RNA transcripts, generally known as noncoding RNAs (ncRNAs) can be found. These transcripts are valuable regulators of gene expression and are involved in modulating different biological processes [44,45]. Thus, increasing evidence indicates that multiple evolutionarily conserved ncRNAs, such as microRNA (miRNA) and long noncoding RNA (lncRNA), are highly involved in different molecular processes, including those associated with the pathogenic transition of steatosis to hepatocarcinoma [46,47].
In recent decades, ncRNAs have become one of the central focuses in cancer research. MiRNAs are endogenous, 19–24 nucleotides long, single-stranded RNA molecules that can modulate, at the post-transcriptional level, different complementary target messenger RNAs involved in many pathophysiological processes, including those associated with steatosis and hepatocarcinogenesis [48,49,50]. From the same group of transcripts but distinct through their structure and functions, lncRNAs are a highly conserved subgroup of ncRNAs, exceeding 200 nucleotides in length, with limited known protein-coding potential, that have recently emerged as significant contributors in the pathophysiology of various human conditions, entangling chronic liver diseases, such as NAFLD and HCC [51,52].
As these ncRNAs are differentially expressed in dependence on the hepatic state of the organism, exploring the miRNAs and lncRNAs’ involvement in the NAFLD-related HCC development is currently of primary interest, yet not a fully understood topic, so extensive research will provide a better view on the molecular mechanism behind the steatosis–hepatocarcinogenesis transition [53,54]. In this regard, many researchers are now conducting intensive studies designed to identify novel and reliable biomarkers that could be further validated and translated into screening panels [55,56]. This review aims to provide an updated view on the critical roles of miRNAs and lncRNAs in the pathologic transition of steatosis to hepatocarcinogenesis, highlighting the potential use of such ncRNAs as diagnostic biomarkers and possible future therapeutic targets for NAFLD-related HCC.
2. Current Epidemiologic Status of NAFLD-Related HCC
Within the NAFLD spectrum, NASH is the main risk factor for HCC development, as it can lead to advanced fibrosis and cirrhosis [38,57]. The cumulative annual incidence for developing NASH-related HCC is about 2.4–12.8% [58]. However, in the absence of NASH or cirrhosis, NAFLD remains the main underlying condition for hepatocarcinoma, which is usually less likely to be diagnosed by surveillance compared to HCC that develops in the setting of NASH, cirrhosis, or viral hepatitis [59,60]. Furthermore, implementing anti-hepatitis B/C virus campaigns decreased the risk of virally driven HCC. Along with the global rise in obesity and T2DM, a constant increase in NAFLD-related HCC incidence has been noted. Thus, NAFLD has rapidly emerged as a primary risk factor for HCC [39].
One quarter of the global population currently suffers from NAFLD [61,62]. This silent liver condition is the fastest growing cause of HCC in several regions worldwide, including the USA, China, Germany, France, and the UK [63], while being closely and bidirectionally connected with different metabolic-syndrome-associated conditions [64,65]. NAFLD is an underlying disorder in up to 68% of T2DM patients and between 60 and 95% of obese individuals [66,67].
In the past seven decades, a comprehensive, systematic review study by Orci et al. focused on assessing the incidence of HCC in patients with NAFLD. The results indicated, with a 95% confidence interval, that the noncirrhotic NAFLD-related HCC rate was 0.03 per 100 person-years, while the rate of cirrhotic NAFLD-related HCC was higher, reaching 3.78 per 100 person-years. Moreover, in cirrhotic patients undergoing regular HCC screening, the incidence rate was higher, at 4.62 per 100 person-years [68]. In another literature study, Pinyopornpanish et al. found that, out of all the 1110 NAFLD-related HCCs included in their research, 15% occurred in noncirrhotic patients [40], highlighting the importance of HCC screening in all NAFLD individuals, regardless of the cirrhotic status. Overall, the incidence of HCC has increased throughout the past several decades. Although viral HBV/HCV infections and cirrhosis appear to remain the most significant risk factors, the connection with metabolic-syndrome-related disorders, the clinically silent symptoms, and the rapidly growing incidence of NAFLD make this liver etiology one of the important causes of HCC development [5,69,70,71].
However, the current clinical guidelines do not include abdominal ultrasound screening for noncirrhotic patients. However, about half of the NAFLD-related HCC cases occur without an established cirrhosis [41,60,72,73]. This unmet need in the screening and surveillance of NAFLD-related HCC patients is a clinical management disruption that could be prevented. Nonetheless, despite all the advances in surgical procedures and chemotherapeutic formulations, NAFLD-related HCCs have a lower receipt of curative therapy with poorer survival rates. Therefore, although several targeted therapies have been previously approved [74] or investigated in different human cancers [75], due to its high prevalence, suboptimal screening and surveillance clinical guidelines, poor prognosis, and limited treatment response, NAFLD-related HCC remains a major global health problem [76,77].
3. Understanding the Pathogenesis of NAFLD-Related HCC
Histologically, NAFLD encompasses a broad spectrum of progressive clinicopathological states that include steatosis, a benign form of nonalcoholic fatty liver (NAFL), followed by a necroinflammation subtype called nonalcoholic steatohepatitis (NASH), which is characterized by hepatic inflammation, the presence of hepatocellular injury (hepatocyte ballooning), and fibrosis, which could further progress to liver cirrhosis, HCC, and eventually liver failure [39,56,61,78]. Hepatocarcinogenesis triggered on a lipotoxic, chronically inflamed liver has an elevated incidence in patients without cirrhosis, as up to 50% of NAFLD-related HCCs occur in noncirrhotic conditions [38,79]. Several research articles reported that patients with NAFLD-related HCC usually have a worse prognosis when compared with HCC developed based on other liver etiologies. This is due to corroboration of different factors, including patients usually being diagnosed at old age when they are already experiencing various other comorbidities and the lack of screening surveillance in noncirrhotic NAFLD, which leads to late-stage diagnosis when therapeutic options are limited [80,81,82,83,84,85].
The development of NAFLD-related HCC is a complex, multistep, multifactorial, and not yet fully understood process. NAFLD is described as a hepatic metabolic disease triggered by excessive fat accumulation in the liver due to a fat-rich diet, disrupted metabolic pathways, and genetic and epigenetic factors (ncRNAs and DNA methylation) [86,87]. Various pathophysiological alterations, such as insulin resistance, precise cytokine release, oxidative stress, and mitochondrial damage, are involved in the transition of NAFLD to HCC. In the background of a pre-existent NAFLD, obesity and lipotoxicity-mediated insulin resistance are significant contributors to HCC pathogenesis through systemic inflammation and the promotion of oncogenic pathways [88]. Moreover, the increased hepatic lipid storage, especially of free fatty acids (FFA) and diglycerol, can facilitate endoplasmic reticulum (ER) stress and reactive oxygen-species-mediated DNA damage, which leads to a chronically inflamed hepatic environment, resulting in NASH and liver fibrosis, which will further drive the oncogenesis of NAFLD-related HCC [86,89].
It is well established that IL-6, JAK, and STAT signaling pathways are essential drivers in the development and progression of the HCC [90,91,92]. In a recent study by Grohmann et al., it was shown that, in a hepatic oxidative environment in the context of obesity, STAT-1 and STAT-3 signaling are enhanced via the inactivation of STAT-1 and STAT-3 phosphatase T cell protein tyrosine phosphate. They also showed that STAT-1 signaling leads to T cell recruitment and the development of both NASH and fibrosis but not HCC. In contrast, STAT-3 signaling was responsible for NAFLD-related HCC development independent of NASH and fibrosis [93]. Finally, genetic factors are also involved in the pathogenesis of NAFLD-related HCC. Primarily, the genetic variants identified so far, PNPLA3, TM6SF2, and MBOAT7, increase the severity of NAFLD via elevating hepatic lipid storage levels by hydrolyzing triglycerides and retinyl esters, hampering lipid mobilization, and decreasing lipoprotein export, thus supporting the progression and development of HCC [94,95,96,97,98].
4. The Implications of ncRNAs in the Pathogenesis of NAFLD-Related HCC
The advances made in genome sequencing achieved over the past decades revealed that potentially between 97 and 98% of the human genome is transcribed into ncRNAs, which do not code for proteins but are somewhat involved in DNA replication, RNA splicing, translation, and epigenetic regulation of various biological processes, including hepatocarcinogenesis [99]. Based on their sequence length, these noncoding transcripts are divided into two groups: short ncRNAs containing less than ~200 nucleotides and long noncoding RNAs exceeding this threshold. In recent years, the expression of ncRNAs, especially miRNAs and lncRNAs, has been extensively studied and linked with several pathological changes, including the transition of steatosis to hepatocarcinoma [100,101,102].
Since their discovery, miRNAs have augmented the research on NAFLD and HCC pathogenesis, especially as they proved to be involved in the molecular events underlying the transition between the two. In recent years, alteration of specific hepatic miRNAs has been linked with several key hallmarks of both NAFLD and HCC. In contrast, few studies have investigated their implication in NAFLD-related HCC progression. Hence, specific hepatic miRNAs have been shown to regulate lipid metabolism, glucose homeostasis, cell proliferation, apoptosis, migration, and differentiation in NAFLD, HCC, or NAFLD-related HCC, thus being considered novel potential molecular biomarkers associated with these liver diseases [103,104,105,106,107].
4.1. miRNA Biogenesis
The biogenesis of these miRNAs is a multistep process that starts in the nuclei of hepatic cells [108,109]. Firstly, RNA polymerase II transcribes primary miRNAs (pri-miRNAs) transcripts within the canonical/mirtron pathway. Thus, the 5′ ends of pri-miRNAs are capped, while the 3′ ends are polyadenylated [110]. Then, a nuclear Rnase III enzyme named Drosha cleaves the pri-miRNAs into 70-100-nucleotide hairpin-structured precursors called pre-miRNAs [111]. Next, pre-miRNAs are exported into the cytoplasm, where they bind with exportin-5 and Ran-GTP to be further cleaved by an endoribonuclease, known as Dicer, into double-stranded pre-miRNAs [112,113]. Subsequently, these newly formed double-stranded pre-miRNAs undergo a fast unwinding by loading onto the AGO protein. Only one strand serves as a guide to target mRNAs, remaining bounded. Finally, the RNA-induced silencing complexes (RISC) and the AGO proteins form the mature miRNAs [50,114]. These mature noncoding transcripts, of which expression is often dysregulated, are ready to modulate the mRNA degradation and translational inhibition, regulating several targets involved in the pathogenesis of NAFLD-related HCC [115].
4.2. LncRNAs
LncRNAs represent a large and heterogeneous group of >200 nucleotide-length transcripts that lack an open reading frame, are frequently poorly expressed in comparison with the levels of protein-coding genes, and are less conserved than mRNAs having a high cell/tissue-specific expression [116,117,118]. Currently, the involvement of lncRNAs in the pathogenesis of NAFLD-related HCC is an evolving subject with promising results [51,119]. In terms of functionality, there are currently four molecular patterns describing the mechanism behind the regulatory activity of lncRNAs in NAFLD-related HCC. Hence, lncRNAs can act as: (a) a molecular guide that binds to the target directing their localization; (b) a molecular scaffold by mediating the protein–RNA interactions; (c) a molecular decoy that directly binds to the targeted proteins/miRNAs suppressing their functions; (d) a miRNA sponge; (e) a miRNA precursor; and as (f) a molecular signal regulating gene expression by interacting with transcription factors or chromatin-modifying enzymes [120,121,122,123]. Nonetheless, and often in direct relation to different miRNAs, lncRNAs are currently investigated for playing a regulatory role in the multistep process of steatosis–hepatocarcinoma progression [46,52]. A visual representation of the lncRNAs mechanisms of action is illustrated below in Figure 1.
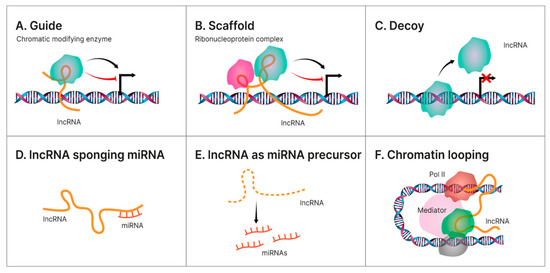
Figure 1.
Different mechanisms of action for lncRNAs. Illustration of the main regulatory mechanism endorsed by hepatic lncRNAs in the pathology transition of NAFLD towards HCC. (A). Guide—some lncRNAs aid specific proteins to reach their target, thus accomplishing their biological functions. Oftentimes, some of these proteins are transcription factors located on specific DNA regions and this guidance becomes an indirect way of regulating gene transcription; (B). Scaffold—LncRNA can facilitate the interaction of numerous molecules and proteins, enabling the assembly of different macromolecular complexes, thus promoting the conjunction and integration of molecular information within different signaling pathways; (C). Decoy—some lncRNAs bind directly to some protein molecules impairing their functions; (D). lncRNA sponging miRNA—some lncRNAs can act as ceRNAs, or sponges, reducing their effect upon the target mRNA, thus regulating gene expression; (E). lncRNA as miRNA precursor—some lncRNAs can act as precursors of miRNAs to directly modulate their regulatory activity; (F). Chromatin looping—as lncRNAs are long and mobile, they could serve as bridges to drive the inter- or intra-chromosomal interactions.
Recently, many findings emerged supporting the implication of miRNAs and lncRNAs in the pathogenesis of NAFLD-related HCC [124,125]. Dysregulated expression profiles of such ncRNAs could improve our understanding of this complex disease and hopefully provide the means for the development of noninvasive and cost-effective methods of diagnosis. In addition, most miRNAs and lncRNAs have either tumor-promoting or suppressive roles that can be exploited as therapeutic targets.
5. Dysregulated miRNAs and Their Role in NAFLD-Related HCC
NAFLD is associated with a dysregulation of hepatic metabolism, and, in the past decade, ncRNAs have been positioned among the primary regulators of these metabolic alterations [126]. The epigenetic mechanisms underlying several deregulations in the expression and function of specific miRNAs were involved in NAFLD development and its progression towards HCC [127]. Thus, several miRNAs were reported as critical regulators of the insulin signaling pathway, carbohydrate metabolism and hepatic glucose output, cholesterol and fatty acid metabolism (pathological accumulation of cholesterol and fatty acids), ER stress and autophagy, and the proinflammatory responses [50,128]. The cellular and tissue changes that occur in the hepatic architecture throughout the pathologic transition of healthy liver to NAFLD and further towards HCC [46,129], together with the main dysregulated miRNAs that modulate these disease-promoting processes, are illustrated in Figure 2.
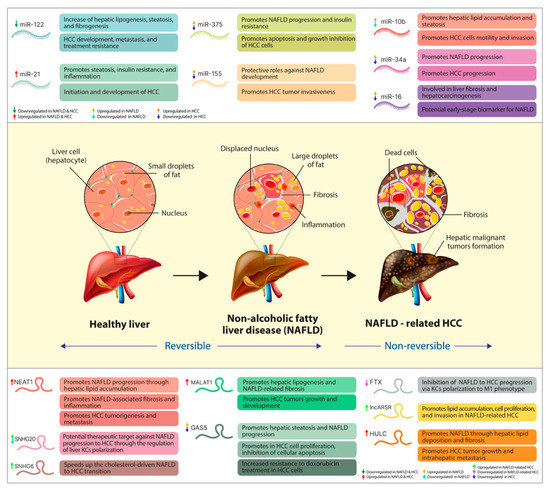
Figure 2.
The pathologic transition of healthy liver towards NAFLD-related HCC, together with the main dysregulated ncRNAs and the disease-promoting processes they regulate. In the upper part of the figure, we illustrated the primary miRNAs and their expression pattern throughout the transition of NAFLD towards HCC, together with the pathological processes of which regulation they play a part in. Centrally, we summarize the main changes in the cellular and tissue architecture of the liver in both NAFLD (reversible) and NAFLD-related HCC (nonreversible). At the bottom, we presented the main lncRNAs involved in NAFLD transition towards HCC, their dysregulated expression level, and the main disease-promoting process in which they play a regulatory role.
Chronic liver inflammation underlying different hepatic etiologies, including NAFLD, is associated with reduced miR-122 expression in hepatocytes [130,131]. The relatively low levels of miR-122 expression remained consistent in NAFLD-HCC compared to healthy tissues [132,133]. Several studies link this miR-122 downregulation with an aggressive HCC phenotype. For example, in vitro analysis of Coulouarn et al. on HCC-derived cells (PLC/PRF/5, Huh-1, and Hep40 exhibited very high levels of miR-122, whereas Hep3B and HepG2 expressed little miR-122, and SNU387 and SNU398 did not express it at all) and on primary liver tumors and 28 matched nontumor surrounding liver tissues concluded that loss of miR-122 expression coincided with the acquisition of an invasive phenotype and suppression of hepatic functions, while being associated with a poor prognosis [134]. Independently, Tsai et al. found that liver-specific miR-122 is significantly downregulated in metastatic Mahlavu and SK-HEP-1 HCC cells and negatively regulates tumorigenesis. It appears that ADAM17, a direct target of miR-122, plays a role in HCC metastatic progression. Silencing of ADAM17 resulted in an apparent reduction in in vitro migration, invasion, tumorigenesis, and angiogenesis in the hepatic models of nude mice, similar to the restoration of miR-122 expression [135].
Kojima et al. studied the correlation between miR-122 expression level and α-fetoprotein (AFP), a widely used biomarker in HCC surveillance. They demonstrated that the miR-122/CUX1/miR-214/ZBTB20 pathway regulates AFP expression, and the miR-122/CUX1/RhoA pathway regulates the aggressive characteristics of such hepatic malignancy [136]. Wu et al. reported that miR-122-knockout mice display increased lipogenesis, changes in lipid secretion, IL-6 and TNF-α production, and upregulation of chemokine ligand 2, thus rapidly developing NASH. They also noted that miR-122 expression enhances fibrogenesis by inducing HIF1α and MAPK1, facilitating HCC development [137]. An extensive and comprehensive study by Akuta et al. investigated the impact of serum miR-122 on the histological features of HCC in a 305 NAFLD patient cohort. The research group saw a clear bidirectional relationship between serum levels of miR-122 and the severity of steatosis, ballooning, lobular inflammation, and disease stage of NAFLD-related HCC. In fact, during the longitudinal evaluation, the scientists observed a decreased tendency in the serum levels of miR-122, even before the fibrotic stage [138]. Tsai et al. found that miR-122-deficient mice exhibit inflammation and extensive lipid accumulation (steatosis), highlighting the anti-inflammatory role of this abundant hepatic miRNA [139]. The aberrant expression of many other miRNAs, including the hepatic abundant miR-29, miR-155, and miR-21 have been found associated with HCC-related chronic inflammation [140]. Overall, hepatic miR-122 deregulation appears to be involved in NAFLD-HCC progression, with its circulating levels correlated with disease stages. However, although several attempts were already made to integrate this miRNA into various HCC diagnostic panels, further validation of these miRNAs is still required as a valid biomarker [141].
The clinical utility of miR-21 in human cancer has been well studied [142]. Hepatic miR-21 expression is elevated in patients and animal models with confirmed NAFLD [130,143,144] and livers of HCC individuals [145,146]. Still, its circulating levels are inconsistent in serum or plasma samples [147,148]. MiR-21 was found upregulated in many hepatic-associated processes of NAFLD patients, including physiological ones, such as inflammation, fibrosis, and carcinogenesis, being a potent promoter of HCC [149]. In NAFLD, miR-21 modulates glucose and lipid metabolism in hepatocytes through a complex transcription network. In a study conducted in 2016, Calo et al. reported that miR-21, an oncomiRNA overexpressed in HCC and other liver etiologies characterized by the presence of steatosis, promotes hepatic insulin resistance and lipogenesis in obesogenic-diet-fed mice through a fine regulation of Foxo1, Insig2, STAT3, and HNF4-α. The research group concluded that hepatic miR-21/miR-21* deficiency is a viable approach for preventing glucose intolerance and steatosis in mice, thus revealing the potential of miR-21/miR-21* as a therapeutic target for NAFLD [150]. The same year, Wu et al. confirmed the presence of elevated miR-21 levels in high-fat diet-fed mice, where it promoted hepatic lipid accumulation and cancer progression by interacting with the Hbp1-p53-Srebp1c pathway, thus endorsing miR-21 as a link between NAFLD and HCC but also as a potential therapeutic target for both disorders [151]. Su et al. provided experimental data supporting the involvement of miR-21 in regulating triglyceride and cholesterol metabolism in an in vitro model of NAFLD HepG2 cells via the inhibition of HMGCR expression [147]. Loyer et al. revealed that, through the inhibition of the PPARα signaling pathway, hepatic miR-21 contributes to hepatocyte injury, inflammation, and fibrosis [143]. These results indicate that miR-21 could be a viable biomarker for diagnosing and treating NAFLD.
Several studies confirmed that miR-21 induces hepatic fibrosis by simultaneously activating hepatic stellate cells (HSCs) via PTEN/Akt signaling [152], hepatocyte EMT by targeting SPRY2 or HNF4α [153], oxidation, and collagen synthesis through the enhanced activity of AngII upon the NLRP3 inflammasome via Spry1/ERK/NF-κB and Smad7/Smad2/3/NOX4 pathways [154,155]. As expected, several studies showed that loss of miR-21 expression results in decreased steatosis, inflammation, collagen storage, and impairment of fibrosis [156,157]. In patients with HCC, miR-21 can promote migration, invasion, and progression through the miR-21-PDCD4-AP-1 feedback loop [158] and the activation of PTEN, which further triggers AKT by interacting with phosphatidylinositol 3-kinase signaling pathway [145]. In addition, Wang et al. reported that overexpressed hepatic miR-21 is responsible for the inhibition of KLF5 w, which leads to HCC migration and invasion [158]. At the same time, Zhou et al. showed that exosomal miR-21 directly targeted PTEN, activating PDK1/AKT signaling in HSCs and promoting HCC progression by secreting a complex repertoire of angiogenic cytokines, including VEGF, MMP2, MMP9, bFGF, and TGF-β [159]. Finally, Wagenaar et al. demonstrated that treatment with specific single-stranded oligonucleotide inhibitors of miR-21 led to suppression of HCC growth in two separate xenograft models, highlighting the role of miR-21 in the maintenance of a tumorigenic phenotype in HCC [146]. Furthermore, multiple clinical data confirm the overexpression of miR-21 in tissue and serum samples, noticing a significant correlation between this oncomiRNA and tumor progression, thus suggesting its potential role as a biomarker for HCC prognosis [160,161,162,163]. Taken together, overexpression of miR-21 was confirmed in both NAFLD and HCC patients, with an apparent involvement in their pathogenesis, thus considered a potential link between the two diseases. [151,164].
Another highly conserved transcript, miR-375, was overexpressed in serum samples of NAFLD-confirmed patients compared with healthy individuals and was significantly associated with disease severity, thus being considered a potential biomarker for NAFLD progression [165]. In 2018, Lei et al. confirmed the genuine overexpression of miR-375 in serum samples of HFD-fed mice and correlated it with the decreased expression of AdipoR2, a direct target of miR-375. In this regard, the researchers noted that inhibition of miR-375 in human HCC cells HepG2 caused an apparent upregulation in adiponectin expression, while downregulating leptin, TNF-α, and IL-6 levels. Hence, miR-375 is involved in NAFLD progression and was proposed as a novel therapeutic target [166].
Regarding the expression and roles in HCC pathogenesis, Li et al. showed that miR-375 is significantly downregulated in human HCC tissues and cell lines (Huh-7, SK-HEP-1, MHCC97-H, MHCC97-L, and Hep3B2.1–7). At the same time, its induction resulted in the in vitro promotion of apoptosis and the inhibition of HCC proliferation. Furthermore, the scientists identified that ErbB2, a member of the epidermal growth factor receptors, is a direct target of miR-375 and is believed to play an essential role in the development and progression of HCC. Induction of miR-375 decreased the expression of ErbB2, while downregulation of ErbB2 was found to inhibit the growth of the HCC cell [167]. In a similar study, He et al. confirmed the significant downregulation of miR-375 in HCC tissues (60 pairs of HCC and matched adjacent nontumor tissues) and cell lines (SK-HEP-1, HepG2, Hep3B, Huh-7, MHCC97-H, and MHCC97-L compared with normal hepatocytes). By Liu et al., the induced overexpression of miR-375 was associated with an in vitro decrease in hepatocyte proliferation, clonogenicity, and invasion, and also with induced G1 arrest and apoptosis. AEG-1, which is a downstream regulator of oncogenic Ha-Ras and c-Myc, was found to be negatively regulated by miR-375. Thus, downregulation of miR-375 leads to AEG-1 overexpression, which activates the PI3K/Akt, NF-κB, and Wnt/β-catenin signaling pathways, reversing the antitumor effects of miR-375 deprivation in HCC [168,169]. Another direct and negatively regulated target of miR-375 is YAP, an oncogene involved in the HCC development [169]. Therefore, miR-375 may represent an important molecular link between NAFLD-associated insulin resistance and hepatocarcinogenesis, playing a part in the pathogenesis of both conditions.
Zheng et al. investigated a different mechanism for the pathogenesis of NAFLD and HCC through miR-10b, another ncRNA involved in the hepatic lipid metabolism and an active contributor to liver steatosis by modulating PPAR-α expression [170]. Celikbilek et al. reported the overexpression of miR-10b in steatosis hepatocytes while promoting intracellular lipids and triglyceride accumulation. However, serum levels of miR-10b were found to decrease in NAFLD patients and were inversely associated with the degree of liver inflammation [171]. In 2014, Liao et al. found the expression levels of miR-10b to be increased in HCC tissues and cell lines (MHCC-97L), which promoted HCC cell motility and invasion through the regulation of HOXD10/RhoC/uPAR/MMPs axis [172]. Jiang et al. noticed that serum levels of miR-10b progressively increased throughout the transition from the healthy liver to chronic hepatic disease and towards HCC. This progressive increase in miR-10b expression across the NAFLD-HCC change could imply its use in the surveillance of NAFLD patients in the HCC [173].
Another miRNA thought to play a part in the NAFLD-HCC progression is miR-34a, which was investigated by Castro et al. and found to be upregulated in NAFLD patients where it reflected the severity of the associated steatohepatitis. In addition, the researchers noted an inverse correlation between SIRT1 and disease severity, and a positive one between p53 acetylation and NAFLD progression, representing direct targets of miR-34a [174]. The expression level of miR-34a was also upregulated in serum samples of NAFLD-confirmed patients [175]. However, regarding HCC, miR-34a was downregulated in liver tissues of HCC patients and associated with tumor invasiveness and metastasis [176]. In a recent study by Sun et al., the lower expression level of miR-34a in comparison with nontumoral tissues was confirmed. Furthermore, miR-34a expression was negatively associated with the regulation of HDAC1, one of its direct targets, which is generally found to be overexpressed in HCC tissues and correlated with cancer-specific mortality [177,178,179]. Hence, miR-34a was involved in the pathogenesis of both NAFLD and HCC; additional investigation is required to understand the mechanisms that make miR-34a a link between these two hepatic diseases.
Research on murine models conducted by Miller et al. showed that miR-155 expression is elevated and exhibits protective effects upon the development of NAFLD. In part, these findings were correlated with the interaction between hepatic miR-155 and its target LXRα, which prevented the excessive lipid accumulation in the liver of murine NAFLD mice [180]. Lin et al. further confirmed the protective roles of miR-155 in a study conducted on liver samples from transgenic mice where the hepatic overexpression of miR-155 was found to alleviate NASH development through one of its direct targets Ces3/TGH [181]. In the context of steatosis to hepatocarcinogenesis progression, miR-155 was significantly upregulated in the early stages of hepatocarcinogenesis, alongside the reduced expression of its target C/EBPβ, a tumor suppressor frequently inhibited in HCC. In addition, the research group reported an association between the ectopic expression of miR-155 and the growth of HCC cells, while its downregulation inhibited their growth [182]. The oncogenic behavior of hepatic miR-155 was also confirmed by Yan et al. They correlated its elevation with the downregulation of SOCS1, which activated STAT3, previously confirmed as an important prognostic biomarker in other cancers, such as glioblastoma [183], and further downregulated MMP9 expression, consequently promoting HCC tumor invasiveness [184]. Research on animal models shows that hepatic miR-155 overexpression has a protective role in NAFLD and, paradoxically, a tumor-promoting one in HCC [185]. Therefore, although extensive data are required to understand its implication in NAFLD-HCC progression, miR-155 was found to be preferentially accumulated in the liver after the administration of exosomes containing synthetic miR-155 mimics into miR-155-knockout mice. This preferential distribution of miR-155 in the liver shows its potential as a disease modulator in both conditions [186].
Multiple studies reported increased circulating levels of miR-16, a transcript known for its involvement in liver fibrosis and hepatocarcinogenesis, in patients with histologically confirmed NAFLD [187,188,189]. However, it appears that plasma levels of miR-16 decrease with NAFLD progression, down to the point of being downregulated in HCC, mainly in patients with tumors over 5 cm in diameter [190]. Thus, lower tissue and serum levels of miR-16 were confirmed by several studies conducted on HCC patients and further proposed as reliable diagnostic biomarkers. The miR-16 level transition can be used as a monitoring biomarker to assess the NAFLD progression sequentially. A biomarker panel composed of miR-16 with AFP, AFP-L3%, and DCP was reported to offer more sensitivity and specificity for HCC patients with smaller tumors [191,192]. Taken together, alone, or in combination with other biomarkers, miR-16 could be a valuable biomarker. Still, extensive research is required to understand better its roles behind the transition from simple steatosis to hepatocarcinogenesis. A summary of other dysregulated miRNAs that could play a part in the NAFLD-related HCC pathogenesis is presented in the following table (Table 1).

Table 1.
Other dysregulated miRNAs involved in both NAFLD and HCC.
6. Dysregulated lncRNAs and Their Role in NAFLD-Related HCC
The other primary class of ncRNAs that has recently emerged as a significant contributor in the NAFLD-related HCC progression is the lncRNAs [51,119]. In recent years, several research studies explored the roles of lncRNAs in the pathogenesis of NAFLD and HCC and in some of the molecular events that occur during the transition between these two chronic hepatic conditions [54,214]. The cellular and tissue changes that occur in the hepatic architecture throughout the pathologic transition of healthy liver to NAFLD and further towards HCC [46,129], together with the main dysregulated lncRNAs that modulate these disease-promoting processes are illustrated in Figure 2.
In 2017, Wang et al. used an in vivo HFD-induced NAFLD SD rat model and an in vitro FFA-exposed BRL3A rat cell line to study the involvement of NEAT1 in the development and progression of NAFLD. Their findings showed increased levels of NEAT1 and mTOR signaling-pathway-associated protein, consistent in both in vivo and in vitro models of NAFLD. To assess the effects of NEAT1, the researchers transfected the rat hepatic BRL3A cells with either pcDNA-NEAT1 lentivirus or si-NEAT1. Overexpression of NEAT1 was positively correlated with the upregulation of acetyl-CoA carboxylase (ACC) and fatty acid synthase (FAS), both of which take part in NAFLD pathogenesis. On the other hand, the FFA-exposed BRL3A cells transfected with si-NEAT1 presented a reversed phenotype. Similarly, overexpression of NEAT1 increased the levels of p-mTOR and p-p70S6K1, while inhibition of NEAT1 showed the contrary effect. Finally, after a 4-week treatment with si-NEAT1 lentivirus, the triglyceride and cholesterol levels were alleviated in the animal model. These findings support the involvement of lnc-NEAT1 in NAFLD through regulating the h mTOR/S6K1 signaling pathway in rats. At the same time, its downregulation is proposed as a potential treatment for NAFLD [215].
In 2019, the upregulation of NEAT1 in hepatic models of NAFLD was confirmed by two separate studies and implicated in the miRNA-based mechanisms involved in NAFLD development. Sun et al. found that upregulation of NEAT1 was positively associated with the expression levels of miR-140 in liver tissues of C57 NAFLD mice and FFA-treated HepG2 cells, both responsible for NAFLD progression by inactivating AMPK/SREBP-1 signaling pathway. Silencing of miR-140 inhibited NEAT1 expression, decreasing the lipid deposition in HepG2 cells [216]. The same year, Chen et al. found that upregulation of NEAT1 inactivates the AMPK/SREBP-1 signaling pathway by sponging miR-146a, thus liberating the expression of ROCK1, promoting NAFLD progression through hepatic lipid accumulation [217]. Nonetheless, NEAT1 was involved in regulating NAFLD-associated fibrosis and inflammation, besides regulating lipid metabolism. Jin et al. reported that knockdown of NEAT1 inhibited GLI3 and promoted miR-506 expression, while overexpression of miR-506 silenced both NEAT1 and GLI3 in an in vitro model (BRL3A rat cell line). Thus, the authors concluded that NEAT1 plays a critical role in hepatic fibrosis and inflammation by regulating the miR-506/GLI3 axis [218]. Regarding its involvement in HCC development and progression, lnc-NEAT1 has been extensively studied. In 2015, Guo et al. conducted one of the first such projects. They reported an elevated expression of NEAT1 in the liver tissue samples from 95 HCC patients, which was further associated with several hallmarks of HCC, including the number of tumor nodes, metastasis, clinical TNM stage, the status of portal vein tumor embolus, vascular invasion, and the infiltration of tumor cells. Thus, authors consider NEAT1 as a pivotal player in tumorigenesis and metastasis of HCC [219]. In 2017, Fang et al. reported that upregulated NEAT1 promotes HCC progression by regulating a miR-129-5p-VCP-IκB [220]. In 2020, Zhang et al. came to a similar conclusion, as NEAT1 expression was found to be significantly upregulated in Huh-7 and MHCC-97H HCC cell lines compared with the HSC LX-2 cells. MiR-230a, a direct target of NEAT1, was significantly downregulated in HCC cells, confirmed in an in vivo nude mouse model. Moreover, a dual-luciferase activity assay identified LAGE3, a prognostic biomarker associated with progression, as a direct target of miR-320a. The researchers concluded that NEAT1/miR-320a/LAGE3 axis participates in HCC development and that NEAT1 could be a potential therapeutic target [221].
In addition, the researchers observed that induced overexpression of NEAT1 abolished the inhibitory effects of miR-504 on HCC cell viability, migration, and invasion, thus promoting cellular apoptosis. The underlying mechanism proved to be the negative regulation of miR-504 on SMO expression levels. Hence, the NEAT1/miR-504/SMO axis deserves a closer look, especially since the downregulation of NEAT1 alleviated HCC hallmarks in vitro, which promotes NEAT1 as a valuable molecular target for HCC treatments [222]. Yeermaike et al. observed that m6A demethylase ALKBH5 is responsible for the upregulation of NEAT1 in human HCC cell lines SMMC-7721, Huh-7, and L02, which further regulates HCC cell proliferation and migration by sponging miR-214 [223]. Based on these studies, the relationship between NEAT1 and miRNAs appears to be involved at different levels in HCC pathogenesis. Therefore, it deserves a more in-depth analysis to better understand and benefit from these findings. Moreover, Sakaguchi et al. continued the in vitro studies on human HCC cell lines, Huh-7, HLF, and Huh-6, identifying a novel oncogenic role for NEAT1, that of inducing cellular autophagy through GABARAP expression, which promotes radioresistance. Hence, NEAT1 and GABARAP are attractive molecular targets that could improve radiation therapy [224].
In 2019, Shen et al. observed an increased hepatic expression of HULC lncRNA in rat models of NAFLD. Furthermore, the research group demonstrated that inhibition of HULC expression was associated with improved pathological state and liver-function-related indexes of hepatic lipid deposition, improved degree of hepatic fibrosis, reduced hepatocyte apoptosis but, also, the inhibition of MAPK signaling pathway, evidence that all supports a clear association between increased HULC expression and NAFLD progression [225]. A few years before, Li et al. reported an elevated expression of HULC in the liver tissue of 39 HCC patients compared with 21 healthy controls. This upregulation was positively correlated with their clinical stage, suggesting that HULC may also be involved in the development and progression of HCC. The increased expression of HULC was also confirmed in Huh-6, Huh-7, HepG2, BEL-7402, MHCC-97H, Sk-Hep1, and SMMC-7721 cell lines. At the same time, the knockdown of HULC resulted in an inhibition of cellular proliferation and in the induction of cellular apoptosis in vitro. Moreover, their research data indicate that HULC negatively regulates miR-200a-3p, upregulating ZEB1 expression in HCC cells, which enhances EMT. Finally, it was also observed that HULC is involved in tumor growth and intrahepatic metastasis, thus being considered a promoter of HCC pathogenesis through miR-200a-3p/ZEB1 EMT pathway [226]. Altogether, these findings highlight a promising research direction, but extensive studies are required for a complete understanding of HULC implication in NAFLD-related HCC.
Wang et al. aimed to investigate which part of SNHG20 lncRNA plays in the pathogenesis of NAFLD-related HCC and whether polarization of liver resident macrophages, Kupffer cells (KCs), is a contributive factor. Their findings showed that SNHG20 expression is decreased in human NAFLD but increased in human NAFLD-HCC livers, the results are also consistent in the mouse models. Both human and mouse NAFLD KCs displayed M1 polarization compared with NAFLD-HCC KCs, while silencing SNHG20 induced M1 polarization in RAW264.7 macrophages and delayed the progression of NAFLD to HCC in the animal model. However, SNHG20 overexpression induced M2 polarization by activating STAT6 in RAW264.7 macrophages. Based on these results, SNHG20 could be a viable therapeutic target against NAFLD progression to HCC, as it regulates liver KCs polarization [227].
Previous studies have shown that hepatic cholesterol is one of the main lipotoxic molecules involved in the progression of NAFLD to HCC [228,229,230]. In a recently published article, Liu et al. orchestrated a comprehensive study design conducted on HCC patients, human liver cell lines HepG2, HEK293T, and Huh-7 cells, cholesterol-driven C57BL/6J NAFLD–HCC mice, liver orthotopic xenograft tumors, and on patient-derived xenograft (PDX) tumors with high expression of SNHG6. The researchers observed a dynamic interplay between cholesterol and the SNHG6, a putative cholesterol effector, and their engagement in a self-amplified cycle that accelerated the NAFLD-related HCC development. From a mechanistic point of view, it was shown that cholesterol binds to the ER-anchored FAF2 protein, which allows the formation of the SNHG6–FAF2–mTOR complex, through which the SNHG6 co-ordinates the mTORC1 kinase cascade activation and cellular cholesterol biosynthesis in a self-amplified cycle that speeds up the cholesterol-driven NAFLD to HCC transition. Moreover, the loss of SNHG6 expression was responsible for mTORC1 signaling inhibition and the growth impairment of PDX liver tumors, promoting SNHG6 as a promising biomarker for developing HCC targeted therapies [231].
In 2020, Wang et al. found that GAS5 lncRNA is downregulated in tumor tissue samples collected from HCC patients and in HCC HepG2 and HepB3 cell lines. This downregulation of GAS5 expression contributed to an elevated level of miR-21 and, later, to suppression of expression of PTEN, which finally resulted in HCC cell proliferation, inhibition of cellular apoptosis, and increased resistance to doxorubicin treatment. Hence, the researchers highlight the potential of restoring GAS5 expression as a new therapeutic approach for HCC [232]. One year later, Cui and his research group investigated the same lncRNA in a NAFLD C57BL/6 mice model and revealed that GAS5 and NOTCH2 expression is elevated, while miRNA-29a-3p expression is decreased. They concluded that GAS5 overexpression augmented NOTCH2 levels in liver cells and promoted NAFLD progression by sponging miR-29a-3p in vivo. To prove these findings, the researchers also demonstrated that GAS5 knockdown attenuated hepatic steatosis and lipid accumulation, thus reducing the NAFLD activity score in HFD mice. Moreover, GAS5 knockdown caused a reduction in serum triglyceride cholesterol levels while inhibiting alanine aminotransferase and aspartate aminotransferase activities in vivo. Their results suggest that GAS5 is a potent regulator of the miR-29a-3p/NOTCH2 axis while actively involved in NAFLD progression, which recommends it as a potential therapeutic target against NAFLD [125]. These findings reflect a possible role of GAS5 in the progression of NAFLD to HCC, but extensive research and validation are required.
Wu et al. performed a series of comprehensive investigations focused directly on a cohort of NAFLD-related HCC cases and in vivo and in vitro lab-grown study models. M2 to M1 phenotype is an essential transition in cancer development that can reverse the tumor-promoting effects of M2 macrophages [227,233,234]. The expression of lncRNA FTX and M1/M2 KCs ratio decreased during the NAFLD conversion to HCC. At the same time, upregulation of FTX inhibited this pathological transition via promoting KCs polarization to M1 phenotype [233]. Chi et al. investigated the role of lncARSR in NAFLD and its role in the progression towards HCC. They revealed an elevated expression status of lncARSR in HFD-fed mice and FFA-treated HepG2 cells. Moreover, they observed that lncARSR binds to YAP1, a crucial component of the Hippo pathway and a regulator of HCC progression, thus blocking its nuclear translocation while activating IRS2/AKT pathway further to increase lipid accumulation, proliferation, and invasion. LncARSR proves to be involved in NAFLD and HCC through the regulation of the YAP1/IRS2/AKT axis, while silencing its expression seems to reduce lipid accumulation in the NAFLD mice model [235].
MALAT1 lncRNA expression was elevated in liver tissue samples of NAFLD patients, serum samples of HFD-fed mice model of NAFLD, and inHepG2 cells treated with 1 mM of FFA (in vitro model of NAFLD). The researchers demonstrated that knockdown of MALAT1 upregulated the expression of PPARα and reduced CD36 levels, thus reversing fatty-acids-induced lipid accumulation in HepG2 cells. In addition, knockdown of MALAT1 was found to be responsible for the reduced levels of miR-206, while it also increased the expression of ARNT, which is known to be involved in NAFLD development. These findings pinpoint the PPARα/CD36-mediated hepatic lipogenesis roles of MALAT1 in NAFLD through regulating the miR-206/ARNT axis [236]. Wu et al. reported that MALAT1 is an active promoter of NAFLD-associated fibrosis as it was found to upregulate post-TGF-β1 exposure and was associated with a reduced expression of SIRT1 in CCL4-treated mice and in LX-2 HSCs [237]. At the same time, Malakar et al. described MALAT1 as a critical oncogene involved in the upregulation of SRSF1 and the activation of the Wnt pathway, thus promoting HCC growth and development in liver tumors of HCC mouse model [238]. Therefore, although the current lack of data limits understanding of the implications of MALAT1 in NAFLD-related HCC pathogenesis, these findings highlight a promising research avenue.
A summary of other dysregulated lncRNAs that could play a part in the NAFLD-related HCC pathogenesis and deserve further investigation is presented in the following table (Table 2).

Table 2.
Other dysregulated lncRNAs involved in both NAFLD and HCC.
7. Conclusions
The medical advances that lead to a decrease in hepatitis B and hepatitis C infection, combined with changes in the populational diet and disease prevalence with an increase in obesity and T2D, indicate that NAFLD will become the leading risk factor associated with the development of HCC.
In this regard, the need to reconsider HCC clinical guidelines to include clear screening and clinical surveillance recommendations for patients with noncirrhotic NAFLD. Progresses in genomic medicine led to the identification of multiple regulatory pathways involved in the development of NAFLD and the progression of NAFLD towards HCC. Among the newly discovered regulatory elements, a particular interest has been decoding the roles of ncRNAs in modulating these regulatory pathways. These ncRNA molecules, represented mainly by miRNAs and lncRNAs, play a central regulatory role.
A better understanding on the progression of NAFLD towards HCC and molecular alterations present in the tumor cells and surrounding tumor microenvironment can be achieved by the study of molecular markers, such as ncRNAs, that represent an important tool for decoding cellular dynamics during early-tumor progression. Functional ncRNA species are mainly represented by miRNA, lncRNAs, and, more recently, an important regulatory role seems to be associated with circularRNAs. These ncRNAs species were shown to be important regulators of the important processes involved in HCC development and progression, such as chronic inflammation, maintaining a hypoxic microenvironment, and angiogenesis.
Chronic inflammation is an important pathogenic factor in the development of NAFLD and further in the progression towards HCC. In addition to metabolic stressors that are known to be involved in the initiation and progression of inflammation, we showed that an important regulatory role at a molecular level is represented by miRNAs and lncRNAs that orchestrate a complex network. Evaluation of the dynamics of these molecules can identify at-risk patients that can be further screened or supposed to therapeutic interventions to limit further disease progression.
Recent evidence showed that ncRNAs are involved in the regulation of hypoxia in HCC via HIF-1α and are responsible for the development of an immunosuppressive tumor microenvironment, which facilitates immune evasion of tumor cells [249]. A hypoxic environment further promotes angiogenesis, which enhances tumor growth and dissemination. Therefore, decoding the regulatory ncRNA network involved in the promotion of angiogenesis can identify new approaches of limiting tumor growth and dissemination [250].
Understanding their dynamics in NAFLD and HCC can identify new biomarkers and therapeutic targets that will allow a better future follow-up and management of these patients. Therefore, our review aims to provide an up-to-date analysis of recent advances in the field of ncRNA involvement in NAFLD development and progression, focusing on possible translational usage of these molecules.
Author Contributions
Conceptualization, I.R., I.B.-N. and N.A.H.; methodology, R.P. and P.C.; data curation, D.R.R.; writing—original draft preparation, R.P., P.C., V.R.P. and A.C.F.; writing—review and editing, I.R., I.B.-N. and N.A.H.; visualization, A.N.; supervision, I.B.-N. and N.A.H.; funding acquisition, I.B.-N. All authors have read and agreed to the published version of the manuscript.
Funding
This paper was supported by the following projects: PDI-PFECDI 2021, entitled Increasing the Performance of Scientific Research, Supporting Excellence in Medical Research and Innovation, PROGRES, no. 40PFE/30.12.2021.
Institutional Review Board Statement
Not applicable.
Acknowledgments
This paper was published under the frame of European Social Found, Human Capital Operational Programme 2014-2020, project no. POCU/380/6/13/125171.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Villanueva, A.; Longo, D.L. Hepatocellular Carcinoma. N. Engl. J. Med. 2019, 380, 1450–1462. [Google Scholar] [CrossRef] [PubMed]
- McGlynn, K.A.; Petrick, J.L.; El-Serag, H.B. Epidemiology of Hepatocellular Carcinoma. Hepatology 2021, 73 (Suppl. S1), 4–13. [Google Scholar] [CrossRef] [PubMed]
- Singal, A.G.; Lampertico, P.; Nahon, P. Epidemiology and surveillance for hepatocellular carcinoma: New trends. J. Hepatol. 2020, 72, 250–261. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.Q.; El-Serag, H.B.; Loomba, R. Global epidemiology of NAFLD-related HCC: Trends, predictions, risk factors and prevention. Nat. Rev. Gastroenterol. Hepatol. 2020, 18, 223–238. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.D.; Hainaut, P.; Gores, G.J.; Amadou, A.; Plymoth, A.; Roberts, L.R. A global view of hepatocellular carcinoma: Trends, risk, prevention and management. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 589–604. [Google Scholar] [CrossRef] [PubMed]
- Fitzmaurice, C.; Abate, D.; Abbasi, N.; Abbastabar, H.; Abd-Allah, F.; Abdel-Rahman, O.; Abdoli, A.; Abdollahpour, I.; Abdulle, A.S.M.; Abebe, N.D.; et al. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability-Adjusted Life-Years for 29 Cancer Groups, 1990 to 2017: A Systematic Analysis for the Global Burden of Disease Study. JAMA Oncol. 2019, 5, 1749–1768. [Google Scholar]
- Kudo, M.; Izumi, N.; Kubo, S.; Kokudo, N.; Sakamoto, M.; Shiina, S.; Tateishi, R.; Nakashima, O.; Murakami, T.; Matsuyama, Y.; et al. Report of the 20th Nationwide follow-up survey of primary liver cancer in Japan. Hepatol. Res. 2019, 50, 15–46. [Google Scholar] [CrossRef]
- Ha, J.; Yan, M.; Aguilar, M.; Bhuket, T.; Tana, M.M.; Liu, B.; Gish, R.G.; Wong, R.J. Race/ethnicity-specific disparities in cancer incidence, burden of disease, and overall survival among patients with hepatocellular carcinoma in the United States. Cancer 2016, 122, 2512–2523. [Google Scholar] [CrossRef]
- Fateen, W.; Ryder, S.D. Screening for hepatocellular carcinoma: Patient selection and perspectives. J. Hepatocell. Carcinoma 2017, 4, 71–79. [Google Scholar] [CrossRef]
- Akuta, N.; Kawamura, Y.; Arase, Y.; Saitoh, S.; Fujiyama, S.; Sezaki, H.; Hosaka, T.; Kobayashi, M.; Kobayashi, M.; Suzuki, Y.; et al. Hepatocellular carcinoma is the most common liver-related complication in patients with histopathologically-confirmed NAFLD in Japan 11 Medical and Health Sciences 1103 Clinical Sciences. BMC Gastroenterol. 2018, 18, 165. [Google Scholar]
- Duan, X.Y.; Zhang, L.; Fan, J.G.; Qiao, L. NAFLD leads to liver cancer: Do we have sufficient evidence? Cancer Lett. 2014, 345, 230–234. [Google Scholar] [CrossRef] [PubMed]
- Mitra, S.; De, A.; Chowdhury, A. Epidemiology of non-alcoholic and alcoholic fatty liver diseases. Transl. Gastroenterol. Hepatol. 2020, 5, 16. [Google Scholar] [CrossRef] [PubMed]
- Cotter, T.G.; Rinella, M. Nonalcoholic Fatty Liver Disease 2020: The State of the Disease. Gastroenterology 2020, 158, 1851–1864. [Google Scholar] [CrossRef] [PubMed]
- Budisan, L.; Zanoaga, O.; Braicu, C.; Pirlog, R.; Covaliu, B.; Esanu, V.; Korban, S.S.; Berindan-Neagoe, I. Links between Infections, Lung Cancer, and the Immune System. Int. J. Mol. Sci. 2021, 22, 9394. [Google Scholar] [CrossRef]
- Schiller, J.T.; Lowy, D.R. An Introduction to Virus Infections and Human Cancer. Viruses Hum. Cancer 2020, 217, 1. [Google Scholar]
- Yasui, K.; Hashimoto, E.; Komorizono, Y.; Koike, K.; Arii, S.; Imai, Y.; Shima, T.; Kanbara, Y.; Saibara, T.; Mori, T.; et al. Characteristics of Patients With Nonalcoholic Steatohepatitis Who Develop Hepatocellular Carcinoma. Clin. Gastroenterol. Hepatol. 2011, 9, 428–433. [Google Scholar] [CrossRef]
- Reeves, H.L.; Zaki, M.Y.W.; Day, C.P. Hepatocellular Carcinoma in Obesity, Type 2 Diabetes, and NAFLD. Am. J. Dig. Dis. 2016, 61, 1234–1245. [Google Scholar] [CrossRef]
- Liu, Y.; Li, H.; Ye, N.; Luo, C.J.; Hu, Y.Y.; Wu, H.; Gong, J.-P. Non-Cirrhotic Liver is Associated with Poor Prognosis of Hepatocellular Carcinoma: A Literature Review. Med. Sci. Monit. 2019, 25, 6615. [Google Scholar] [CrossRef]
- de Martel, C.; Maucort-Boulch, D.; Plummer, M.; Franceschi, S. World-wide relative contribution of hepatitis B and C viruses in hepatocellular carcinoma. Hepatology 2015, 62, 1190–1200. [Google Scholar] [CrossRef]
- Akinyemiju, T.; Abera, S.; Ahmed, M.; Alam, N.; Alemayohu, M.A.; Allen, C.; Al-Raddadi, R.; Alvis-Guzman, N.; Amoako, Y.; Artaman, A.; et al. The Burden of Primary Liver Cancer and Underlying Etiologies from 1990 to 2015 at the Global, Regional, and National Level: Results from the Global Burden of Disease Study 2015. JAMA Oncol. 2017, 3, 1683–1691. [Google Scholar] [PubMed]
- Allen, A.M.; Therneau, T.M.; Larson, J.J.; Coward, A.; Somers, V.K.; Kamath, P.S. Nonalcoholic fatty liver disease incidence and impact on metabolic burden and death: A 20 year-community study. Hepatology 2017, 67, 1726–1736. [Google Scholar] [CrossRef] [PubMed]
- Siegel, A.B.; Zhu, A.X. Metabolic Syndrome and Hepatocellular Carcinoma: Two Growing Epidemics with a Potential Link. Cancer 2009, 115, 5651. [Google Scholar] [CrossRef] [PubMed]
- Chavez-Tapia, N.C.; Gall, S.M.-B.; Ordoñez-Vázquez, A.L.; Nuño-Lambarri, N.; Vidal-Cevallos, P.; Uribe, M. Understanding the Role of Metabolic Syndrome as a Risk Factor for Hepatocellular Carcinoma. J. Hepatocell Carcinoma 2022, 9, 583–593. [Google Scholar] [CrossRef]
- Tsoulfas, G. Hepatocellular carcinoma and metabolic syndrome: The times are changing and so should we. World J. Gastroenterol. 2019, 25, 3842. [Google Scholar] [CrossRef]
- Wu, W.K.K.; Zhang, L.; Chan, M.T.V. Autophagy, NAFLD and NAFLD-related HCC. Adv. Exp. Med. Biol. 2018, 1061, 127–138. [Google Scholar]
- Sanyal, A.; Poklepovic, A.; Moyneur, E.; Barghout, V. Population-based risk factors and resource utilization for HCC: US perspective. Curr. Med. Res. Opin. 2010, 26, 2183–2191. [Google Scholar] [CrossRef]
- Vieira Barbosa, J.; Lai, M. Nonalcoholic Fatty Liver Disease Screening in Type 2 Diabetes Mellitus Patients in the Primary Care Setting. Hepatol. Commun. 2021, 5, 158. [Google Scholar] [CrossRef]
- Singal, A.K.; Bataller, R.; Ahn, J.; Kamath, P.S.; Shah, V.H. ACG Clinical Guideline: Alcoholic Liver Disease. Am. J. Gastroenterol. 2018, 113, 175–194. [Google Scholar] [CrossRef]
- Nagaoki, Y.; Hyogo, H.; Ando, Y.; Kosaka, Y.; Uchikawa, S.; Nishida, Y.; Teraoka, Y.; Morio, K.; Fujino, H.; Ono, A.; et al. Increasing incidence of non-HBV- and non-HCV-related hepatocellular carcinoma: Single-institution 20-year study. BMC Gastroenterol. 2021, 21, 306. [Google Scholar] [CrossRef]
- Rinella, M.E. Nonalcoholic fatty liver disease: A systematic review. JAMA 2015, 313, 2263–2273. [Google Scholar] [CrossRef] [PubMed]
- Brunt, E.M.; Wong, V.W.S.; Nobili, V.; Day, C.P.; Sookoian, S.; Maher, J.J.; Bugianesi, E.; Sirlin, C.B.; Neuschwander-Tetri, B.A.; Rinella, M.E. Nonalcoholic fatty liver disease. Nat. Rev. Dis. Prim. 2015, 1, 15080. [Google Scholar] [CrossRef] [PubMed]
- Neuschwander-Tetri, B.A. Non-alcoholic fatty liver disease. BMC Med. 2017, 15, 45. [Google Scholar] [CrossRef] [PubMed]
- Billeter, A.T.; Müller, P.C.; Albrecht, T.; Roessler, S.; Löffler, M.; Lemekhova, A.; Mehrabi, A.; Müller-Stich, B.P.; Hoffmann, K. Impact of Type 2 Diabetes on Oncologic Outcomes of Hepatocellular Carcinomas in Non-Cirrhotic, Non-alcoholic Steatohepatitis: A Matched-Pair Analysis. J. Gastrointest. Surg. 2020, 25, 1193–1202. [Google Scholar] [CrossRef] [PubMed]
- Wong, R.J.; Aguilar, M.; Cheung, R.; Perumpail, R.B.; Harrison, S.A.; Younossi, Z.M.; Ahmed, A. Nonalcoholic Steatohepatitis Is the Second Leading Etiology of Liver Disease Among Adults Awaiting Liver Transplantation in the United States. Gastroenterology 2015, 148, 547–555. [Google Scholar] [CrossRef]
- Powell, E.E.; Wong, V.W.S.; Rinella, M. Non-alcoholic fatty liver disease. Lancet 2021, 397, 2212–2224. [Google Scholar] [CrossRef]
- Burra, P.; Becchetti, C.; Germani, G. NAFLD and liver transplantation: Disease burden, current management and future challenges. JHEP Rep. 2020, 2, 100192. [Google Scholar] [CrossRef]
- Paul, S.; Dhamija, E.; Kedia, S. Non-alcoholic fatty liver disease associated with hepatocellular carcinoma: An increasing concern. Indian J. Med. Res. 2019, 149, 9–17. [Google Scholar] [CrossRef]
- Mattos, Â.Z.; Debes, J.D.; Dhanasekaran, R.; Benhammou, J.N.; Arrese, M.; Patrício, A.L.V.; Zilio, A.C.; Mattos, A.A. Hepatocellular carcinoma in nonalcoholic fatty liver disease: A growing challenge. World J. Hepatol. 2021, 13, 1107–1121. [Google Scholar] [CrossRef]
- Pinyopornpanish, K.; Khoudari, G.; Saleh, M.A.; Angkurawaranon, C.; Pinyopornpanish, K.; Mansoor, E.; Dasarathy, S.; McCullough, A. Hepatocellular carcinoma in nonalcoholic fatty liver disease with or without cirrhosis: A population-based study. BMC Gastroenterol. 2021, 21, 394. [Google Scholar] [CrossRef]
- Frenette, C.T.; Isaacson, A.J.; Bargellini, I.; Saab, S.; Singal, A.G. A Practical Guideline for Hepatocellular Carcinoma Screening in Patients at Risk. Mayo Clin. Proc. Innov. Qual. Outcomes 2019, 3, 302–310. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Wang, W.P.; Lee, W.J.; Meloni, M.F.; Clevert, D.-A.; Chammas, M.C.; Tannapfel, A.; Forgione, A.; Piscaglia, F.; Dietrich, C.F. Hepatocellular carcinoma in the non-cirrhotic liver. Clin. Hemorheol. Microcirc. 2021, 80, 423–436. [Google Scholar] [CrossRef] [PubMed]
- Tarao, K.; Nozaki, A.; Ikeda, T.; Sato, A.; Komatsu, H.; Komatsu, T.; Taguri, M.; Tanaka, K. Real impact of liver cirrhosis on the development of hepatocellular carcinoma in various liver diseases—Meta-analytic assessment. Cancer Med. 2019, 8, 1054–1065. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Zhou, J.; Peng, Y.; He, W.; Huang, C. The role of long noncoding RNAs in hepatocellular carcinoma. Mol. Cancer 2020, 19, 77. [Google Scholar] [CrossRef]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2020, 22, 96–118. [Google Scholar] [CrossRef]
- Khalifa, O.; Errafii, K.; Al-Akl, N.S.; Arredouani, A. Noncoding RNAs in Nonalcoholic Fatty Liver Disease: Potential Diagnosis and Prognosis Biomarkers. Dis. Markers 2020, 2020, 8822859. [Google Scholar] [CrossRef]
- Pirlog, R.; Drula, R.; Nutu, A.; Calin, G.A.; Berindan-Neagoe, I. The Roles of the Colon Cancer Associated Transcript 2 (CCAT2) Long Non-Coding RNA in Cancer: A Comprehensive Characterization of the Tumorigenic and Molecular Functions. Int. J. Mol. Sci. 2021, 22, 12491. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef]
- Peng, Y.; Croce, C.M. The role of MicroRNAs in human cancer. Signal Transduct. Target Ther. 2016, 1, 1–9. [Google Scholar] [CrossRef]
- Morishita, A.; Oura, K.; Tadokoro, T.; Fujita, K.; Tani, J.; Masaki, T. MicroRNAs in the Pathogenesis of Hepatocellular Carcinoma: A Review. Cancers 2021, 13, 514. [Google Scholar] [CrossRef]
- Kim, Y.-A.; Park, K.-K.; Lee, S.-J. LncRNAs Act as a Link between Chronic Liver Disease and Hepatocellular Carcinoma. Int. J. Mol. Sci. 2020, 21, 2883. [Google Scholar] [CrossRef] [PubMed]
- Shabgah, A.G.; Norouzi, F.; Hedayati-Moghadam, M.; Soleimani, D.; Pahlavani, N.; Navashenaq, J.G. A comprehensive review of long non-coding RNAs in the pathogenesis and development of non-alcoholic fatty liver disease. Nutr. Metab. 2021, 18, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z.; Dou, G.; Wang, L. MicroRNAs in the Pathogenesis of Nonalcoholic Fatty Liver Disease. Int. J. Biol. Sci. 2021, 17, 1851–1863. [Google Scholar] [CrossRef] [PubMed]
- Mahpour, A.; Mullen, A.C. Our emerging understanding of the roles of long non-coding RNAs in normal liver function, disease, and malignancy. JHEP Rep. 2020, 3, 100177. [Google Scholar] [CrossRef]
- Xu, X.; Tao, Y.; Shan, L.; Chen, R.; Jiang, H.; Qian, Z.; Cai, F.; Ma, L.; Yu, Y. The Role of MicroRNAs in Hepatocellular Carcinoma. J. Cancer 2018, 9, 3557–3569. [Google Scholar] [CrossRef]
- Ghafouri-Fard, S.; Gholipour, M.; Hussen, B.M.; Taheri, M. The Impact of Long Non-Coding RNAs in the Pathogenesis of Hepatocellular Carcinoma. Front. Oncol. 2021, 11, 1150. [Google Scholar] [CrossRef]
- Michelotti, G.A.; Machado, M.V.; Diehl, A.M. NAFLD, NASH and liver cancer. Nat. Rev. Gastroenterol. Hepatol. 2013, 10, 656–665. [Google Scholar] [CrossRef]
- White, D.L.; Kanwal, F.; El-Serag, H.B. Non-Alcoholic Fatty Liver Disease and Hepatocellular Cancer: A Systematic Review. Clin. Gastroenterol. Hepatol. 2012, 10, 1342. [Google Scholar] [CrossRef]
- Younes, R.; Bugianesi, E. Should we undertake surveillance for HCC in patients with NAFLD? J. Hepatol. 2018, 68, 326–334. [Google Scholar] [CrossRef]
- Sumida, Y.; Yoneda, M.; Seko, Y.; Ishiba, H.; Hara, T.; Toyoda, H.; Yasuda, S.; Kumada, T.; Hayashi, H.; Kobayashi, T.; et al. Surveillance of Hepatocellular Carcinoma in Nonalcoholic Fatty Liver Disease. Diagnostics 2020, 10, 579. [Google Scholar] [CrossRef]
- Peng, C.; Stewart, A.G.; Woodman, O.L.; Ritchie, R.H.; Qin, C.X. Non-Alcoholic Steatohepatitis: A Review of Its Mechanism, Models and Medical Treatments. Front. Pharmacol. 2020, 11, 1864. [Google Scholar] [CrossRef] [PubMed]
- Pouwels, S.; Sakran, N.; Graham, Y.; Leal, A.; Pintar, T.; Yang, W.; Kassir, R.; Singhal, R.; Mahawar, K.; Ramnarain, D. Non-alcoholic fatty liver disease (NAFLD): A review of pathophysiology, clinical management and effects of weight loss. BMC Endocr. Disord. 2022, 22, 63. [Google Scholar] [CrossRef] [PubMed]
- Foerster, F.; Gairing, S.J.; Müller, L.; Galle, P.R. NAFLD-driven HCC: Safety and efficacy of current and emerging treatment options. J. Hepatol. 2021, 76, 446–457. [Google Scholar] [CrossRef] [PubMed]
- Wainwright, P.; Byrne, C.D. Bidirectional Relationships and Disconnects between NAFLD and Features of the Metabolic Syndrome. Int. J. Mol. Sci. 2016, 17, 367. [Google Scholar] [CrossRef]
- Bertuccio, P.; Turati, F.; Carioli, G.; Rodriguez, T.; La Vecchia, C.; Malvezzi, M.; Negri, E. Global trends and predictions in hepatocellular carcinoma mortality. J. Hepatol. 2017, 67, 302–309. [Google Scholar] [CrossRef] [PubMed]
- Godoy-Matos, A.F.; Silva Júnior, W.S.; Valerio, C.M. NAFLD as a continuum: From obesity to metabolic syndrome and diabetes. Diabetol. Metab. Syndr. 2020, 12, 60. [Google Scholar] [CrossRef]
- Paik, J.M.; Golabi, P.; Younossi, Y.; Mishra, A.; Younossi, Z.M. Changes in the Global Burden of Chronic Liver Diseases From 2012 to 2017: The Growing Impact of NAFLD. Hepatology 2020, 72, 1605–1616. [Google Scholar] [CrossRef]
- Orci, L.A.; Sanduzzi-Zamparelli, M.; Caballol, B.; Sapena, V.; Colucci, N.; Torres, F.; Bruix, J.; Reig, M.; Toso, C. Incidence of Hepatocellular Carcinoma in Patients With Nonalcoholic Fatty Liver Disease: A Systematic Review, Meta-analysis, and Meta-regression. Clin. Gastroenterol. Hepatol. 2021, 20, 283–292.e10. [Google Scholar] [CrossRef]
- Farrell, A.; Ryan, M.; Howell, J. Epidemiology of non-alcoholic fatty liver disease-related hepatocellular carcinoma: A western perspective. Hepatoma Res. 2020, 6, 18. [Google Scholar] [CrossRef]
- Maurice, J.; Manousou, P. Non-alcoholic fatty liver disease. Clin. Med. 2018, 18, 245–250. [Google Scholar] [CrossRef]
- Younossi, Z.M.; Henry, L. Epidemiology of non-alcoholic fatty liver disease and hepatocellular carcinoma. JHEP Rep. 2021, 3, 100305. [Google Scholar] [CrossRef] [PubMed]
- Galle, P.R.; Forner, A.; Llovet, J.M.; Mazzaferro, V.; Piscaglia, F.; Raoul, J.L.; European Association for the Study of the Liver. EASL Clinical Practice Guidelines: Management of hepatocellular carcinoma. J. Hepatol. 2018, 69, 182–236. [Google Scholar] [CrossRef] [PubMed]
- Vogel, A.; Cervantes, A.; Chau, I.; Daniele, B.; Llovet, J.M.; Meyer, T.; Nault, J.-C.; Neumann, U.; Ricke, J.; Sangro, B. Correction to: “Hepatocellular carcinoma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up”. Ann. Oncol. 2019, 30, 871–873. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.T.; Tan, Y.J.; Oon, C.E. Molecular targeted therapy: Treating cancer with specificity. Eur. J. Pharmacol. 2018, 834, 188–196. [Google Scholar] [CrossRef]
- Bochis, O.V.; Irimie, A.; Pichler, M.; Berindan-Neagoe, I. The role of Skp2 and its substrate CDKN1B (p27) in colorectal cancer. J. Gastrointestin. Liver Dis. 2015, 24, 225–234. [Google Scholar] [CrossRef]
- Fujiwara, N.; Friedman, S.L.; Goossens, N.; Hoshida, Y. Risk factors and prevention of hepatocellular carcinoma in the era of precision medicine. J. Hepatol. 2018, 68, 526–549. [Google Scholar] [CrossRef]
- Loomba, R.; Lim, J.K.; Patton, H.; El-Serag, H.B. AGA Clinical Practice Update on Screening and Surveillance for Hepatocellular Carcinoma in Patients with Nonalcoholic Fatty Liver Disease: Expert Review. Gastroenterology 2020, 158, 1822–1830. [Google Scholar] [CrossRef]
- Friedman, S.L.; Neuschwander-Tetri, B.A.; Rinella, M.; Sanyal, A.J. Mechanisms of NAFLD development and therapeutic strategies. Nat. Med. 2018, 24, 908–922. [Google Scholar] [CrossRef]
- Stine, J.G.; Wentworth, B.J.; Zimmet, A.; Rinella, M.E.; Loomba, R.; Caldwell, S.H.; Argo, C.K. Systematic review with meta-analysis: Risk of hepatocellular carcinoma in non-alcoholic steatohepatitis without cirrhosis compared to other liver diseases. Aliment. Pharmacol. Ther. 2018, 48, 696–703. [Google Scholar] [CrossRef]
- Geh, D.; Anstee, Q.M.; Reeves, H.L. NAFLD-Associated HCC: Progress and Opportunities. J. Hepatocell. Carcinoma. 2021, 8, 223. [Google Scholar] [CrossRef]
- Friedman, S.L. Focus. J. Hepatol. 2014, 60, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Piscaglia, F.; Svegliati-Baroni, G.; Barchetti, A.; Pecorelli, A.; Marinelli, S.; Tiribelli, C.; Bellentani, S.; HCC-NAFLD Italian Study Group. Clinical patterns of hepatocellular carcinoma in nonalcoholic fatty liver disease: A multicenter prospective study. Hepatology 2016, 63, 827–838. [Google Scholar] [CrossRef]
- Mittal, S.; Sada, Y.H.; El-Serag, H.B.; Kanwal, F.; Duan, Z.; Temple, S.; May, S.B.; Kramer, J.R.; Richardson, P.A.; Davila, J.A. Temporal trends of nonalcoholic fatty liver disease-related hepatocellular carcinoma in the veteran affairs population. Clin. Gastroenterol. Hepatol. 2015, 13, 594–601.e1. [Google Scholar] [CrossRef] [PubMed]
- Weinmann, A.; Alt, Y.; Koch, S.; Nelles, C.; Düber, C.; Lang, H.; Otto, G.; Zimmermann, T.; Marquardt, J.U.; Galle, P.R.; et al. Treatment and survival of non-alcoholic steatohepatitis associated hepatocellular carcinoma. BMC Cancer 2015, 15, 210. [Google Scholar] [CrossRef]
- Grgurevic, I.; Bozin, T.; Mikus, M.; Kukla, M.; O’Beirne, J. Hepatocellular Carcinoma in Non-Alcoholic Fatty Liver Disease: From Epidemiology to Diagnostic Approach. Cancers 2021, 13, 5844. [Google Scholar] [CrossRef]
- Masoodi, M.; Gastaldelli, A.; Hyötyläinen, T.; Arretxe, E.; Alonso, C.; Gaggini, M.; Brosnan, J.; Anstee, Q.M.; Millet, O.; Ortiz, P. Metabolomics and lipidomics in NAFLD: Biomarkers and non-invasive diagnostic tests. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 835–856. [Google Scholar] [CrossRef]
- Teng, Y.-X.; Xie, S.; Guo, P.-P.; Deng, Z.-J.; Zhang, Z.-Y.; Gao, W.; Zhang, W.-G.; Zhong, J.-H. Hepatocellular Carcinoma in Non-alcoholic Fatty Liver Disease: Current Progresses and Challenges. J. Clin. Transl. Hepatol. 2022. [Google Scholar] [CrossRef]
- Margini, C.; Dufour, J.F. The story of HCC in NAFLD: From epidemiology, across pathogenesis, to prevention and treatment. Liver Int. 2016, 36, 317–324. [Google Scholar] [CrossRef] [PubMed]
- Anstee, Q.M.; Reeves, H.L.; Kotsiliti, E.; Govaere, O.; Heikenwalder, M. From NASH to HCC: Current concepts and future challenges. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 411–428. [Google Scholar] [CrossRef]
- Calvisi, D.F.; Ladu, S.; Gorden, A.; Farina, M.; Conner, E.A.; Lee, J.S.; Factor, V.M.; Thorgeirsson, S.S. Ubiquitous Activation of Ras and Jak/Stat Pathways in Human HCC. Gastroenterology 2006, 130, 1117–1128. [Google Scholar] [CrossRef]
- He, G.; Yu, G.Y.; Temkin, V.; Ogata, H.; Kuntzen, C.; Sakurai, T.; Sieghart, W.; Peck-Radosavljevic, M.; Leffert, H.L.; Karin, M. Hepatocyte IKKβ/NF-κB inhibits tumor promotion and progression by preventing oxidative stress driven STAT3 activation. Cancer Cell 2010, 17, 286. [Google Scholar] [CrossRef] [PubMed]
- He, G.; Dhar, D.; Nakagawa, H.; Font-Burgada, J.; Ogata, H.; Jiang, Y.; Shalapour, S.; Seki, E.; Yost, S.E.; Jepsen, K.; et al. Identification of Liver Cancer Progenitors Whose Malignant Progression Depends on Autocrine IL-6 Signaling. Cell 2013, 155, 384–396. [Google Scholar] [CrossRef] [PubMed]
- Grohmann, M.; Wiede, F.; Dodd, G.T.; Gurzov, E.N.; Ooi, G.J.; Butt, T.; Rasmiena, A.A.; Kaur, S.; Gulati, T.; Goh, P.K.; et al. Obesity Drives STAT-1-Dependent NASH and STAT-3-Dependent HCC. Cell 2018, 175, 1289–1306.e20. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-L.; Patman, G.L.; Leathart, J.B.S.; Piguet, A.-C.; Burt, A.D.; Dufour, J.-F.; Day, C.P.; Daly, A.K.; Reeves, H.L.; Anstee, Q.M. Carriage of the PNPLA3 rs738409 C >G polymorphism confers an increased risk of non-alcoholic fatty liver disease associated hepatocellular carcinoma. J. Hepatol. 2014, 61, 75–81. [Google Scholar] [CrossRef]
- Liu, Y.-L.; Reeves, H.L.; Burt, A.D.; Tiniakos, D.; McPherson, S.; Leathart, J.B.S.; Allison, M.E.D.; Alexander, G.J.; Piguet, A.-C.; Anty, R.; et al. TM6SF2 rs58542926 influences hepatic fibrosis progression in patients with non-alcoholic fatty liver disease. Nat. Commun. 2014, 5, 4309. [Google Scholar] [CrossRef] [PubMed]
- Mancina, R.M.; Dongiovanni, P.; Petta, S.; Pingitore, P.; Meroni, M.; Rametta, R.; Borén, J.; Montalcini, T.; Pujia, A.; Wiklund, O.; et al. The MBOAT7-TMC4 Variant rs641738 Increases Risk of Nonalcoholic Fatty Liver Disease in Individuals of European Descent. Gastroenterology 2016, 150, 1219–1230.e6. [Google Scholar] [CrossRef]
- Seko, Y.; Sumida, Y.; Tanaka, S.; Mori, K.; Taketani, H.; Ishiba, H.; Hara, T.; Okajima, A.; Umemura, A.; Nishikawa, T.; et al. Development of hepatocellular carcinoma in Japanese patients with biopsy-proven non-alcoholic fatty liver disease: Association between PNPLA3 genotype and hepatocarcinogenesis/fibrosis progression. Hepatol. Res. 2016, 47, 1083–1092. [Google Scholar] [CrossRef]
- Tang, S.; Zhang, J.; Mei, T.-T.; Guo, H.-Q.; Wei, X.-H.; Zhang, W.-Y.; Liu, Y.-L.; Liang, S.; Fan, Z.-P.; Ma, L.-X.; et al. Association of TM6SF2 rs58542926 T/C gene polymorphism with hepatocellular carcinoma: A meta-analysis. BMC Cancer 2019, 19, 1128. [Google Scholar] [CrossRef]
- Manna, D.; Sarkar, D. Non-Coding RNAs: Regulating Disease Progression and Therapy Resistance in Hepatocellular Carcinoma. Cancers 2020, 12, 1243. [Google Scholar] [CrossRef]
- Sulaiman, S.A.; Muhsin, N.I.A.; Jamal, R. Regulatory Non-coding RNAs Network in Non-alcoholic Fatty Liver Disease. Front. Physiol. 2019, 10, 279. [Google Scholar] [CrossRef]
- Zhou, H.; Xu, Q.; Ni, C.; Ye, S.; Xu, X.; Hu, X.; Jiang, J.; Hong, Y.; Huang, D.; Yang, L. Prospects of Noncoding RNAs in Hepatocellular Carcinoma. BioMed Res. Int. 2018, 2018, 6579436. [Google Scholar] [CrossRef] [PubMed]
- Heo, M.J.; Yun, J.; Kim, S.G. Role of non-coding RNAs in liver disease progression to hepatocellular carcinoma. Arch. Pharmacal Res. 2019, 42, 48–62. [Google Scholar] [CrossRef] [PubMed]
- Evangelista, J.; Zaninotto, E.; Gaglio, A.; Ghidini, M.; Raimondi, L. MicroRNAs as Diagnostic Tools in Hepatocellular Carcinoma. Gastrointest. Disord. 2021, 3, 237–246. [Google Scholar] [CrossRef]
- Tessitore, A.; Cicciarelli, G.; Del Vecchio, F.; Gaggiano, A.; Verzella, D.; Fischietti, M.; Mastroiaco, V.; Vetuschi, A.; Sferra, R.; Barnabei, R.; et al. MicroRNA expression analysis in high fat diet-induced NAFLD-NASH-HCC progression: Study on C57BL/6J mice. BMC Cancer 2016, 16, 2860. [Google Scholar] [CrossRef]
- Guo, Y.; Xiong, Y.; Sheng, Q.; Zhao, S.; Wattacheril, J.; Flynn, C.R. A micro-RNA expression signature for human NAFLD progression. J. Gastroenterol. 2016, 51, 1022–1030. [Google Scholar] [CrossRef]
- Berindan-Neagoe, I.; Calin, G.A. Molecular Pathways: microRNAs, Cancer Cells, and Microenvironment. Clin. Cancer Res. 2014, 20, 6247. [Google Scholar] [CrossRef]
- Berindan-Neagoe, I.; Bs, P.D.C.M.; Pasculli, B.; Calin, G.A. MicroRNAome genome: A treasure for cancer diagnosis and therapy. CA: A Cancer J. Clin. 2014, 64, 311–336. [Google Scholar] [CrossRef]
- Annese, T.; Tamma, R.; De Giorgis, M.; Ribatti, D. microRNAs Biogenesis, Functions and Role in Tumor Angiogenesis. Front. Oncol. 2020, 10, 2610. [Google Scholar] [CrossRef]
- Szabo, G.; Bala, S. MicroRNAs in liver disease. Nat. Rev. Gastroenterol. Hepatol. 2013, 10, 542. [Google Scholar] [CrossRef]
- Yin, S.; Yu, Y.; Reed, R. Primary microRNA processing is functionally coupled to RNAP II transcription in vitro. Sci. Rep. 2015, 5, 11992. [Google Scholar] [CrossRef]
- Graves, P.; Zeng, Y. Biogenesis of Mammalian MicroRNAs: A Global View. Genom. Proteom. Bioinform. 2012, 10, 239–245. [Google Scholar] [CrossRef]
- Okada, C.; Yamashita, E.; Lee, S.J.; Shibata, S.; Katahira, J.; Nakagawa, A.; Yoneda, Y.; Tsukihara, T. A high-resolution structure of the pre-microRNA nuclear export machinery. Science 2009, 326, 1275–1279. [Google Scholar] [CrossRef]
- Chendrimada, T.P.; Gregory, R.I.; Kumaraswamy, E.; Norman, J.; Cooch, N.; Nishikura, K.; Shiekhattar, R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 2005, 436, 740–744. [Google Scholar] [CrossRef]
- Kim, Y.; Kim, V.N. MicroRNA Factory: RISC Assembly from Precursor MicroRNAs. Mol. Cell 2012, 46, 384–386. [Google Scholar] [CrossRef]
- Ezaz, G.; Trivedi, H.D.; Connelly, M.A.; Filozof, C.; Howard, K.L.; Parrish, M.; Kim, M.; Herman, M.A.; Nasser, I.; Afdhal, N.H.; et al. Differential Associations of Circulating MicroRNAs With Pathogenic Factors in NAFLD. Hepatol. Commun. 2020, 4, 670–680. [Google Scholar] [CrossRef]
- Hon, C.-C.; Ramilowski, J.A.; Harshbarger, J.; Bertin, N.; Rackham, O.J.L.; Gough, J.; Denisenko, E.; Schmeier, S.; Poulsen, T.M.; Severin, J.; et al. An atlas of human long non-coding RNAs with accurate 5′ ends. Nature 2017, 543, 199–204. [Google Scholar] [CrossRef]
- Huarte, M. The emerging role of lncRNAs in cancer. Nat. Med. 2015, 21, 1253–1261. [Google Scholar] [CrossRef]
- Kopp, F.; Mendell, J.T. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 2018, 172, 393–407. [Google Scholar] [CrossRef]
- Wang, K.C.; Chang, H.Y. Molecular Mechanisms of Long Noncoding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, W.; Zhu, W.; Dong, J.; Cheng, Y.; Yin, Z.; Shen, F. Mechanisms and Functions of Long Non-Coding RNAs at Multiple Regulatory Levels. Int. J. Mol. Sci. 2019, 20, 5573. [Google Scholar] [CrossRef]
- Fang, Y.; Fullwood, M.J. Roles, Functions, and Mechanisms of Long Non-coding RNAs in Cancer. Genom. Proteom. Bioinform. 2016, 14, 42–54. [Google Scholar] [CrossRef]
- Gao, N.; Li, Y.; Li, J.; Gao, Z.; Yang, Z.; Li, Y.; Liu, H.; Fan, T. Long Non-Coding RNAs: The Regulatory Mechanisms, Research Strategies, and Future Directions in Cancers. Front. Oncol. 2020, 10, 2903. [Google Scholar] [CrossRef]
- Guh, C.-Y.; Hsieh, Y.-H.; Chu, H.-P. Functions and properties of nuclear lncRNAs-from systematically mapping the interactomes of lncRNAs. J. Biomed. Sci. 2020, 27, 1–14. [Google Scholar] [CrossRef]
- Ye, D.; Zhang, T.; Lou, G.; Liu, Y. Role of miR-223 in the pathophysiology of liver diseases. Exp. Mol. Med. 2018, 50, 1–12. [Google Scholar] [CrossRef]
- Cui, J.; Wang, Y.; Xue, H. Long non-coding RNA GAS5 contributes to the progression of nonalcoholic fatty liver disease by targeting the microRNA-29a-3p/NOTCH2 axis. Bioengineered 2022, 13, 8370–8381. [Google Scholar] [CrossRef]
- Qian, G.; Morral, N. Role of non-coding RNAs on liver metabolism and NAFLD pathogenesis. Hum. Mol. Genet. 2022, 1–18. [Google Scholar] [CrossRef]
- López-Sánchez, G.N.; Dóminguez-Pérez, M.; Uribe, M.; Chávez-Tapia, N.C.; Nuño-Lámbarri, N. Non-alcoholic fatty liver disease and microRNAs expression, how it affects the development and progression of the disease. Ann. Hepatol. 2021, 21, 100212. [Google Scholar] [CrossRef]
- Agbu, P.; Carthew, R.W. MicroRNA-mediated regulation of glucose and lipid metabolism. Nat. Rev. Mol. Cell. Biol. 2021, 22, 425–438. [Google Scholar] [CrossRef]
- Degasperi, E.; Colombo, M. Distinctive features of hepatocellular carcinoma in non-alcoholic fatty liver disease. Lancet Gastroenterol. Hepatol. 2016, 1, 156–164. [Google Scholar] [CrossRef]
- Cheung, O.; Puri, P.; Eicken, C.; Contos, M.J.; Mirshahi, F.; Maher, J.W.; Kellum, J.M.; Min, H.; Luketic, V.A.; Sanyal, A.J. Nonalcoholic steatohepatitis is associated with altered hepatic MicroRNA expression. Hepatology 2008, 48, 1810–1820. [Google Scholar] [CrossRef]
- Ambade, A.; Satishchandran, A.; Szabo, G. Alcoholic hepatitis accelerates early hepatobiliary cancer by increasing stemness and miR-122-mediated HIF-1α activation. Sci. Rep. 2016, 6, 21340. [Google Scholar] [CrossRef]
- Gramantieri, L.; Ferracin, M.; Fornari, F.; Veronese, A.; Sabbioni, S.; Liu, C.-G.; Calin, G.A.; Giovannini, C.; Ferrazzi, E.; Grazi, G.L.; et al. Cyclin G1 Is a Target of miR-122a, a MicroRNA Frequently Down-regulated in Human Hepatocellular Carcinoma. Cancer Res. 2007, 67, 6092–6099. [Google Scholar] [CrossRef]
- Kutay, H.; Bai, S.; Datta, J.; Motiwala, T.; Pogribny, I.; Frankel, W.; Jacob, S.T.; Ghoshal, K. Downregulation of miR-122 in the rodent and human hepatocellular carcinomas. J. Cell Biochem. 2006, 99, 671–678. [Google Scholar] [CrossRef]
- Coulouarn, C.; Factor, V.M.; Andersen, J.B.; Durkin, M.E.; Thorgeirsson, S.S. Loss of miR-122 expression in liver cancer correlates with suppression of the hepatic phenotype and gain of metastatic properties. Oncogene 2009, 28, 3526–3536. [Google Scholar] [CrossRef]
- Tsai, W.-C.; Hsu, P.W.-C.; Lai, T.-C.; Chau, G.-Y.; Lin, C.-W.; Chen, C.-M.; Lin, C.-D.; Liao, Y.-L.; Wang, J.-L.; Chau, Y.-P.; et al. MicroRNA-122, a tumor suppressor microRNA that regulates intrahepatic metastasis of hepatocellular carcinoma. Hepatology 2009, 49, 1571–1582. [Google Scholar] [CrossRef]
- Kojima, K.; Takata, A.; Vadnais, C.; Otsuka, M.; Yoshikawa, T.; Akanuma, M.; Kondo, Y.; Kang, Y.J.; Kishikawa, T.; Kato, N.; et al. MicroRNA122 is a key regulator of α-fetoprotein expression and influences the aggressiveness of hepatocellular carcinoma. Nat. Commun. 2011, 2, 338. [Google Scholar] [CrossRef]
- Wu, F.-L.; Jin, W.-B.; Li, J.-H.; Guo, A.-G. Targets for human encoded microRNAs in HBV genes. Virus Genes 2010, 42, 157–161. [Google Scholar] [CrossRef]
- Akuta, N.; Kawamura, Y.; Suzuki, F.; Saitoh, S.; Arase, Y.; Kunimoto, H.; Sorin, Y.; Fujiyama, S.; Sezaki, H.; Hosaka, T.; et al. Impact of circulating miR-122 for histological features and hepatocellular carcinoma of nonalcoholic fatty liver disease in Japan. Hepatol. Int. 2016, 10, 647–656. [Google Scholar] [CrossRef]
- Tsai, W.-C.; Hsu, S.-D.; Hsu, C.-S.; Lai, T.-C.; Chen, S.-J.; Shen, R.; Huang, Y.; Chen, H.-C.; Lee, C.-H.; Tsai, T.-F.; et al. MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J. Clin. Investig. 2012, 122, 2884–2897. [Google Scholar] [CrossRef]
- Jin, K.; Li, T.; Sánchez-Duffhues, G.; Zhou, F.; Zhang, L. Involvement of inflammation and its related microRNAs in hepatocellular carcinoma. Oncotarget 2016, 8, 22145–22165. [Google Scholar] [CrossRef]
- Franck, M.; Schütte, K.; Malfertheiner, P.; Link, A. Prognostic value of serum microRNA-122 in hepatocellular carcinoma is dependent on coexisting clinical and laboratory factors. World J. Gastroenterol. 2020, 26, 86. [Google Scholar] [CrossRef]
- Pop-Bica, C.; Pintea, S.; Magdo, L.; Cojocneanu, R.; Gulei, D.; Ferracin, M.; Berindan-Neagoe, I. The Clinical Utility of miR-21 and let-7 in Non-small Cell Lung Cancer (NSCLC). A Systematic Review and Meta-Analysis. Front. Oncol. 2020, 10, 516850. [Google Scholar] [CrossRef]
- Loyer, X.; Paradis, V.; Hénique, C.; Vion, A.-C.; Colnot, N.; Guerin, C.L.; Devue, C.; On, S.; Scetbun, J.; Romain, M.; et al. Liver microRNA-21 is overexpressed in non-alcoholic steatohepatitis and contributes to the disease in experimental models by inhibiting PPARα expression. Gut 2015, 65, 1882–1894. [Google Scholar] [CrossRef]
- Liu, J.; Xiao, Y.; Wu, X.; Jiang, L.; Yang, S.; Ding, Z.; Fang, Z.; Hua, H.; Kirby, M.S.; Shou, J. A circulating microRNA signature as noninvasive diagnostic and prognostic biomarkers for nonalcoholic steatohepatitis. BMC Genom. 2018, 19, 188. [Google Scholar] [CrossRef]
- Meng, F.; Henson, R.; Wehbe–Janek, H.; Ghoshal, K.; Jacob, S.T.; Patel, T. MicroRNA-21 Regulates Expression of the PTEN Tumor Suppressor Gene in Human Hepatocellular Cancer. Gastroenterology 2007, 133, 647–658. [Google Scholar] [CrossRef]
- Wagenaar, T.R.; Zabludoff, S.; Ahn, S.M.; Allerson, C.; Arlt, H.; Baffa, R.; Cao, H.; Davis, S.; Garcia-Echeverria, C.; Gaur, R.; et al. Anti-miR-21 Suppresses Hepatocellular Carcinoma Growth via Broad Transcriptional Network Deregulation. Mol. Cancer Res. 2015, 13, 1009–1021. [Google Scholar] [CrossRef]
- Sun, C.; Huang, F.; Liu, X.; Xiao, X.; Yang, M.; Hu, G.; Liu, H.; Liao, L. miR-21 regulates triglyceride and cholesterol metabolism in non-alcoholic fatty liver disease by targeting HMGCR. Int. J. Mol. Med. 2015, 35, 847–853. [Google Scholar] [CrossRef]
- Yamada, H.; Suzuki, K.; Ichino, N.; Ando, Y.; Sawada, A.; Osakabe, K.; Sugimoto, K.; Ohashi, K.; Teradaira, R.; Inoue, T.; et al. Associations between circulating microRNAs (miR-21, miR-34a, miR-122 and miR-451) and non-alcoholic fatty liver. Clin. Chim. Acta 2013, 424, 99–103. [Google Scholar] [CrossRef]
- Gramantieri, L.; Fornari, F.; Callegari, E.; Sabbioni, S.; Lanza, G.; Croce, C.M.; Bolondi, L.; Negrini, M. MicroRNA involvement in hepatocellular carcinoma. J. Cell Mol. Med. 2008, 12, 2189–2204. [Google Scholar] [CrossRef]
- Calo, N.; Ramadori, P.; Sobolewski, C.; Romero, Y.; Maeder, C.; Fournier, M.; Rantakari, P.; Zhang, F.-P.; Poutanen, M.; Dufour, J.-F.; et al. Stress-activated miR-21/miR-21* in hepatocytes promotes lipid and glucose metabolic disorders associated with high-fat diet consumption. Gut 2016, 65, 1871–1881. [Google Scholar] [CrossRef]
- Wu, H.; Ng, R.; Chen, X.; Steer, C.J.; Song, G. MicroRNA-21 is a potential link between non-alcoholic fatty liver disease and hepatocellular carcinoma via modulation of the HBP1-p53-Srebp1c pathway. Gut 2016, 65, 1850–1860. [Google Scholar] [CrossRef]
- Wei, J.; Feng, L.; Li, Z.; Xu, G.; Fan, X. MicroRNA-21 activates hepatic stellate cells via PTEN/Akt signaling. Biomed. Pharmacother. 2013, 67, 387–392. [Google Scholar] [CrossRef]
- Zhao, J.; Tang, N.; Wu, K.; Dai, W.; Ye, C.; Shi, J.; Zhang, J.; Ning, B.; Zeng, X.; Lin, Y. MiR-21 Simultaneously Regulates ERK1 Signaling in HSC Activation and Hepatocyte EMT in Hepatic Fibrosis. PLoS ONE 2014, 9, e108005. [Google Scholar] [CrossRef]
- Kitano, M.; Bloomston, P.M. Hepatic Stellate Cells and microRNAs in Pathogenesis of Liver Fibrosis. J. Clin. Med. 2016, 5, 38. [Google Scholar] [CrossRef]
- Ning, Z.W.; Luo, X.Y.; Wang, G.Z.; Li, Y.; Pan, M.X.; Yang, R.Q.; Ling, X.-G.; Huang, S.; Ma, X.X.; Jin, S.Y.; et al. MicroRNA-21 Mediates Angiotensin II-Induced Liver Fibrosis by Activating NLRP3 Inflammasome/IL-1β Axis via Targeting Smad7 and Spry1. Antioxid. Redox Signal. 2017, 27, 1. [Google Scholar] [CrossRef]
- Rodrigues, P.M.; Afonso, M.B.; Simaõ, A.L.; Carvalho, C.C.; Trindade, A.; Duarte, A.; Borralho, P.M.; Machado, M.V.; Cortez-Pinto, H.; Rodrigues, C.M.P.; et al. miR-21 ablation and obeticholic acid ameliorate nonalcoholic steatohepatitis in mice. Cell Death Dis. 2017, 8, e2748. [Google Scholar]
- Kennedy, L.L.; Meng, F.; Venter, J.K.; Zhou, T.; Karstens, W.A.; Hargrove, L.A.; Wu, N.; Kyritsi, K.; Greene, J.; Invernizzi, P.; et al. Knockout of microRNA-21 reduces biliary hyperplasia and liver fibrosis in cholestatic bile duct ligated mice. Lab. Investig. 2016, 96, 1256–1267. [Google Scholar]
- Wang, J.; Chu, Y.; Xu, M.; Zhang, X.; Zhou, Y. miR-21 promotes cell migration and invasion of hepatocellular carcinoma by targeting KLF5. Oncol. Lett. 2018, 17, 2221–2227. [Google Scholar] [CrossRef]
- Zhou, Y.; Ren, H.; Dai, B.; Li, J.; Shang, L.; Huang, J.; Shi, X. Hepatocellular carcinoma-derived exosomal miRNA-21 contributes to tumor progression by converting hepatocyte stellate cells to cancer-associated fibroblasts. J. Exp. Clin. Cancer Res. 2018, 37, 324. [Google Scholar] [CrossRef]
- Karakatsanis, A.; Papaconstantinou, I.; Gazouli, M.; Lyberopoulou, A.; Polymeneas, G.; Voros, D. Expression of microRNAs, miR-21, miR-31, miR-122, miR-145, miR-146a, miR-200c, miR-221, miR-222, and miR-223 in patients with hepatocellular carcinoma or intrahepatic cholangiocarcinoma and its prognostic significance. Mol. Carcinog. 2011, 52, 297–303. [Google Scholar] [CrossRef]
- Guo, X.; Lv, X.; Lv, X.; Ma, Y.; Chen, L.; Chen, Y. Circulating miR-21 serves as a serum biomarker for hepatocellular carcinoma and correlated with distant metastasis. Oncotarget 2017, 8, 44050–44058. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.S.; Kim, G.; Lee, Y.R.; Park, S.Y.; Tak, W.Y.; Kweon, Y.O.; Park, J.G.; Han, Y.S.; Ha, H.T.; Chun, J.M.; et al. Clinical significance of microRNA-21 expression in disease progression of patients with hepatocellular carcinoma. Biomark Med. 2018, 12, 1105–1114. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.-S.; Yu, W.; Cui, H.; Wang, Y.-J.; Zhang, L.; Han, F.; Huang, T. Increased expression of miR-21 predicts poor prognosis in patients with hepatocellular carcinoma. Int. J. Clin. Exp. Pathol. 2015, 8, 7234. [Google Scholar] [PubMed]
- Rodrigues, P.M.; Afonso, M.B.; Simão, A.; Islam, T.; Gaspar, M.M.; O Rourke, C.; Andersen, J.; Santos-Laso, A.; Jimenez-Aguero, R.; Eizaguirre, E.; et al. mIR-21 is increased in patients with NASH-associated HCC and contributes to hepatocarcinogenesis in mice with NAFLD. J. Hepatol. 2020, 73, S677–S678. [Google Scholar] [CrossRef]
- Pirola, C.J.; Fernandez Gianotti, T.; Castano, G.O.; Mallardi, P.; San Martino, J.; Mora Gonzalez Lopez Ledesma, M.; Flichman, D.; Mirshahi, F.; Sanyal, A.J.; Sookoian, S. Circulating microRNA signature in non-alcoholic fatty liver disease: From serum non-coding RNAs to liver histology and disease pathogenesis. Gut 2015, 64, 800–812. [Google Scholar] [CrossRef] [PubMed]
- Lei, L.; Zhou, C.; Yang, X.; Li, L. Down-regulation of microRNA-375 regulates adipokines and inhibits inflammatory cytokines by targeting AdipoR2 in non-alcoholic fatty liver disease. Clin. Exp. Pharmacol. Physiol. 2018, 45, 819–831. [Google Scholar] [CrossRef]
- Li, L.; Jia, L.; Ding, Y. Upregulation of miR-375 inhibits human liver cancer cell growth by modulating cell proliferation and apoptosis via targeting ErbB2. Oncol. Lett. 2018, 16, 3319–3326. [Google Scholar] [CrossRef]
- He, X.X.; Chang, Y.; Meng, F.Y.; Wang, M.Y.; Xie, Q.H.; Tang, F.; Li, P.-Y.; Song, Y.-H.; Lin, J.-S. MicroRNA-375 targets AEG-1 in hepatocellular carcinoma and suppresses liver cancer cell growth in vitro and in vivo. Oncogene 2012, 31, 3357–3369. [Google Scholar] [CrossRef]
- Liu, A.M.; Poon, R.T.; Luk, J.M. MicroRNA-375 targets Hippo-signaling effector YAP in liver cancer and inhibits tumor properties. Biochem. Biophys. Res. Commun. 2010, 394, 623–627. [Google Scholar] [CrossRef]
- Zheng, L.; Lv, G.C.; Sheng, J.; DaYang, Y. Effect of miRNA-10b in regulating cellular steatosis level by targeting PPAR-alpha expression, a novel mechanism for the pathogenesis of NAFLD. J. Gastroenterol. Hepatol. 2010, 25, 156–163. [Google Scholar] [CrossRef]
- Celikbilek, M.; Baskol, M.; Taheri, S.; Deniz, K.; Dogan, S.; Zararsiz, G.; Gursoy, S.; Guven, K.; Ozbakır, O.; Dundar, M.; et al. Circulating microRNAs in patients with non-alcoholic fatty liver disease. World J. Hepatol. 2014, 6, 613–620. [Google Scholar] [CrossRef] [PubMed]
- Liao, C.; Kong, L.; Zhou, P.; Yang, X.; Huang, J.; Zhang, H.; Lu, N. miR-10b is overexpressed in hepatocellular carcinoma and promotes cell proliferation, migration and invasion through RhoC, uPAR and MMPs. J. Transl. Med. 2014, 12, 234. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Cheng, Q.; Zhang, B.H.; Zhang, M.Z. Circulating microRNAs as biomarkers in hepatocellular carcinoma screening: A validation set from China. Medicine 2015, 94, e603. [Google Scholar] [CrossRef] [PubMed]
- Castro, R.E.; Ferreira, D.M.S.; Afonso, M.B.; Borralho, P.M.; MacHado, M.V.; Cortez-Pinto, H.; Rodrigues, C.M. miR-34a/SIRT1/p53 is suppressed by ursodeoxycholic acid in the rat liver and activated by disease severity in human non-alcoholic fatty liver disease. J. Hepatol. 2013, 58, 119–125. [Google Scholar] [CrossRef]
- Xu, Y.; Zhu, Y.; Hu, S.; Pan, X.; Bawa, F.C.; Wang, H.H.; Wang, D.Q.; Yin, L.; Zhang, Y. Hepatocyte miR-34a is a key regulator in the development and progression of non-alcoholic fatty liver disease. Mol. Metab. 2021, 51, 101244. [Google Scholar] [CrossRef] [PubMed]
- Budhu, A.; Jia, H.L.; Forgues, M.; Liu, C.G.; Goldstein, D.; Lam, A.; Zanetti, K.A.; Ye, Q.H.; Qin, L.X.; Croce, C.M.; et al. Identification of metastasis-related microRNAs in hepatocellular carcinoma. Hepatology 2008, 47, 897–907. [Google Scholar] [CrossRef]
- Sun, T.-Y.; Xie, H.-J.; Li, Z.; Kong, L.-F.; Gou, X.-N.; Li, D.-J.; Shi, Y.-J.; Ding, Y.-Z. miR-34a regulates HDAC1 expression to affect the proliferation and apoptosis of hepatocellular carcinoma. Am. J. Transl. Res. 2017, 9, 103–114. [Google Scholar]
- Freese, K.; Seitz, T.; Dietrich, P.; Lee, S.M.L.; Thasler, W.E.; Bosserhoff, A.; Hellerbrand, C. Histone Deacetylase Expressions in Hepatocellular Carcinoma and Functional Effects of Histone Deacetylase Inhibitors on Liver Cancer Cells In Vitro. Cancers 2019, 11, 1587. [Google Scholar] [CrossRef]
- Ler, S.Y.; Leung, C.H.W.; Khin, L.W.; Lu, G.-D.; Salto-Tellez, M.; Hartman, M.; Iau, P.T.C.; Yap, C.T.; Hooi, S.C. HDAC1 and HDAC2 independently predict mortality in hepatocellular carcinoma by a competing risk regression model in a Southeast Asian population. Oncol. Rep. 2015, 34, 2238–2250. [Google Scholar] [CrossRef]
- Miller, A.M.; Gilchrist, D.S.; Nijjar, J.; Araldi, E.; Ramírez, C.M.; Lavery, C.A.; Fernandez-Hernando, C.; McInnes, I.B.; Kurowska-Stolarska, M. MiR-155 Has a Protective Role in the Development of Non-Alcoholic Hepatosteatosis in Mice. PLoS ONE 2013, 8, e72324. [Google Scholar] [CrossRef]
- Lin, X.; Jia, J.; Du, T.; Li, W.; Wang, X.; Wei, J.; Lin, X.; Zeng, H.; Yao, L.; Chen, X.; et al. Overexpression of miR-155 in the Liver of Transgenic Mice Alters the Expression Profiling of Hepatic Genes Associated with Lipid Metabolism. PLoS ONE 2015, 10, e0118417. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Majumder, S.; Nuovo, G.; Kutay, H.; Volinia, S.; Patel, T.; Schmittgen, T.D.; Croce, C.; Ghoshal, K.; Jacob, S.T.; et al. Role of miR-155 at early stages of hepatocarcinogenesis induced by choline-deficient and amino acid defined diet in C57BL/6 mice. Hepatology 2009, 50, 1152. [Google Scholar] [CrossRef] [PubMed]
- Susman, S.; Pîrlog, R.; Leucuța, D.; Mitre, A.O.; Padurean, V.A.; Melincovici, C.; Moldovan, I.; Crișan, D.; Florian, S.I. The role of p-Stat3 Y705 immunohistochemistry in glioblastoma prognosis. Diagn. Pathol. 2019, 14, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.-L.; Jia, Y.-L.; Chen, L.; Zeng, Q.; Zhou, J.-N.; Fu, C.-J.; Chen, H.-X.; Yuan, H.-F.; Li, Z.-W.; Shi, L.; et al. Hepatocellular carcinoma-associated mesenchymal stem cells promote hepatocarcinoma progression: Role of the S100A4-miR155-SOCS1-MMP9 axis. Hepatology 2013, 57, 2274–2286. [Google Scholar] [CrossRef] [PubMed]
- Zanoaga, O.; Braicu, C.; Chiroi, P.; Andreea, N.; Hajjar, N.; Mărgărit, S.; Korban, S.; Berindan-Neagoe, I. The Role of miR-155 in Nutrition: Modulating Cancer-Associated Inflammation. Nutrients 2021, 13, 2245. [Google Scholar] [CrossRef]
- Bala, S.; Csak, T.; Momen-Heravi, F.; Lippai, D.; Kodys, K.; Catalano, D.; Satishchandran, A.; Ambros, V.; Szabo, G. Biodistribution and function of extracellular miRNA-155 in mice. Sci. Rep. 2015, 5, 10721. [Google Scholar] [CrossRef]
- Pillai, S.S.; Lakhani, H.V.; Zehra, M.; Wang, J.; Dilip, A.; Puri, N.; O’Hanlon, K.; Sodhi, K. Predicting Nonalcoholic Fatty Liver Disease through a Panel of Plasma Biomarkers and MicroRNAs in Female West Virginia Population. Int. J. Mol. Sci. 2020, 21, 6698. [Google Scholar] [CrossRef]
- Cermelli, S.; Ruggieri, A.; Marrero, J.A.; Ioannou, G.N.; Beretta, L. Circulating MicroRNAs in Patients with Chronic Hepatitis C and Non-Alcoholic Fatty Liver Disease. PLoS ONE 2011, 6, e23937. [Google Scholar] [CrossRef]
- Dongiovanni, P.; Meroni, M.; Longo, M.; Fargion, S.; Fracanzani, A.L. miRNA Signature in NAFLD: A Turning Point for a Non-Invasive Diagnosis. Int. J. Mol. Sci. 2018, 19, 3966. [Google Scholar] [CrossRef]
- Ge, W.; Yu, D.C.; Li, Q.G.; Chen, X.; Zhang, C.Y.; Ding, Y.T. Expression of serum miR-16, let-7f, and miR-21 in patients with hepatocellular carcinoma and their clinical significances. Clin. Lab. 2014, 60, 427–434. [Google Scholar] [CrossRef]
- Qu, K.Z.; Zhang, K.; Li, H.; Afdhal, N.; Albitar, M. Circulating MicroRNAs as Biomarkers for Hepatocellular Carcinoma. J. Clin. Gastroenterol. 2011, 45, 355–360. [Google Scholar] [CrossRef] [PubMed]
- Cheng, B.; Ding, F.; Huang, C.Y.; Xiao, H.; Fei, F.Y.; Li, J. Role of miR-16-5p in the proliferation and metastasis of hepatocellular carcinoma. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 137–145. [Google Scholar] [PubMed]
- Wang, B.; Hsu, S.H.; Frankel, W.; Ghoshal, K.; Jacob, S.T. Stat3-mediated activation of miR-23a suppresses gluconeogenesis in hepatocellular carcinoma by downregulating G6PC and PGC-1α. Hepatology 2012, 56, 186. [Google Scholar] [CrossRef]
- Pan, J.H.; Cha, H.; Tang, J.; Lee, S.; Lee, S.H.; Le, B.; Redding, M.C.; Kim, S.; Batish, M.; Kong, B.C.; et al. The role of microRNA-33 as a key regulator in hepatic lipogenesis signaling and a potential serological biomarker for NAFLD with excessive dietary fructose consumption in C57BL/6N mice. Food Funct. 2020, 12, 656–667. [Google Scholar] [CrossRef] [PubMed]
- Erhartova, D.; Cahova, M.; Dankova, H.; Heczkova, M.; Mikova, I.; Sticova, E.; Spicak, J.; Seda, O.; Trunecka, P. Serum miR-33a is associated with steatosis and inflammation in patients with non-alcoholic fatty liver disease after liver transplantation. PLoS ONE 2019, 14, e0224820. [Google Scholar]
- Chang, W.; Zhang, L.; Xian, Y.; Yu, Z. MicroRNA-33a promotes cell proliferation and inhibits apoptosis by targeting PPARα in human hepatocellular carcinoma. Exp. Ther. Med. 2017, 13, 2507–2514. [Google Scholar] [CrossRef]
- Zhang, Y.; Cheng, X.; Lu, Z.; Wang, J.; Chen, H.; Fan, W.; Gao, X.; Lu, D. Upregulation of miR-15b in NAFLD models and in the serum of patients with fatty liver disease. Diabetes Res. Clin. Pr. 2013, 99, 327–334. [Google Scholar] [CrossRef]
- Xia, H.; Qi, Y.; Ng, S.S.; Chen, X.; Chen, S.; Fang, M.; Li, D.; Zhao, Y.; Ge, R.; Li, G.; et al. MicroRNA-15b regulates cell cycle progression by targeting cyclins in glioma cells. Biochem. Biophys. Res. Commun. 2009, 380, 205–210. [Google Scholar] [CrossRef]
- Cimmino, A.; Calin, G.A.; Fabbri, M.; Iorio, M.V.; Ferracin, M.; Shimizu, M.; Wojcik, S.E.; Aqeilan, R.I.; Zupo, S.; Dono, M.; et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. USA 2005, 102, 13944–13949. [Google Scholar] [CrossRef]
- Jiang, M.; Zhang, Y.; Liu, M.; Lan, M.S.; Fei, J.; Fan, W.; Gao, X.; Lu, D. Hypermethylation of Hepatic Glucokinase and L-Type Pyruvate Kinase Promoters in High-Fat Diet–Induced Obese Rats. Endocrinology 2011, 152, 1284–1289. [Google Scholar] [CrossRef]
- Finck, B.N.; Kelly, D.P. PGC-1 coactivators: Inducible regulators of energy metabolism in health and disease. J. Clin. Invest. 2006, 116, 615–622. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.-M.; Jeong, H.-J.; Park, S.-W.; Lee, W. Obesity-induced miR-15b is linked causally to the development of insulin resistance through the repression of the insulin receptor in hepatocytes. Mol. Nutr. Food Res. 2015, 59, 2303–2314. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.-H.; Chung, G.E.; Myung, S.J.; Lee, J.-H.; Lee, S.-H.; Lee, S.-M.; Kim, S.-J.; Hwang, S.Y.; Lee, H.-S.; Kim, C.Y. High expression of microRNA-15b predicts a low risk of tumor recurrence following curative resection of hepatocellular carcinoma. Oncol. Rep. 2009, 23, 113–119. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, J.; Liu, Y.; Li, S.; Huang, P. Plasma miR-15b-5p, miR-338-5p, and miR-764 as Biomarkers for Hepatocellular Carcinoma. Med. Sci. Monit. 2015, 21, 1864. [Google Scholar] [CrossRef]
- de Conti, A.; Ortega, J.F.; Tryndyak, V.; Dreval, K.; Moreno, F.S.; Rusyn, I.; Beland, F.A.; Pogribny, I.P. MicroRNA deregulation in nonalcoholic steatohepatitis-associated liver carcinogenesis. Oncotarget 2017, 8, 88517. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhu, K.; Yu, W.; Wang, H.; Liu, L.; Wu, Q.; Li, S.; Guo, J. MiR-181b regulates steatosis in nonalcoholic fatty liver disease via targeting SIRT1. Biochem. Biophys. Res. Commun. 2017, 493, 227–232. [Google Scholar] [CrossRef]
- Wang, B.; Hsu, S.-H.; Majumder, S.; Kutay, H.; Huang, W.; Jacob, S.T.; Ghoshal, K. TGFβ-mediated upregulation of hepatic miR-181b promotes hepatocarcinogenesis by targeting TIMP3. Oncogene 2010, 29, 1787–1797. [Google Scholar] [CrossRef]
- Esau, C.; Kang, X.; Peralta, E.; Hanson, E.; Marcusson, E.G.; Ravichandran, L.V.; Sun, Y.; Koo, S.; Perera, R.J.; Jain, R.; et al. MicroRNA-143 regulates adipocyte differentiation. J. Biol. Chem. 2004, 279, 52361–52365. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, S.; Hu, T.; Liu, S.; He, Y.; Sun, S. Up-regulated microRNA-143 transcribed by nuclear factor kappa B enhances hepatocarcinoma metastasis by repressing fibronectin expression. Hepatology 2009, 50, 490–499. [Google Scholar] [CrossRef]
- Liu, X.-L.; Cao, H.-X.; Wang, B.-C.; Xin, F.-Z.; Zhang, R.-N.; Zhou, D.; Yang, R.-X.; Zhao, Z.-H.; Pan, Q.; Fan, J.-G. miR-192-5p regulates lipid synthesis in non-alcoholic fatty liver disease through SCD-1. World J. Gastroenterol. 2017, 23, 8140–8151. [Google Scholar] [CrossRef]
- Lian, J.; Jing, Y.; Dong, Q.; Huan, L.; Chen, D.; Bao, C.; Wang, Q.; Zhao, F.; Li, J.; Yao, M.; et al. miR-192, a prognostic indicator, targets the SLC39A6/SNAIL pathway to reduce tumor metastasis in human hepatocellular carcinoma. Oncotarget 2016, 7, 2672. [Google Scholar] [CrossRef] [PubMed]
- Nie, H.; Song, C.; Wang, D.; Cui, S.; Ren, T.; Cao, Z.; Liu, Q.; Chen, Z.; Chen, X.; Zhou, Y. MicroRNA-194 inhibition improves dietary-induced non-alcoholic fatty liver disease in mice through targeting on FXR. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2017, 1863, 3087–3094. [Google Scholar] [CrossRef] [PubMed]
- Niu, X.; Nong, S.; Gong, J.; Zhang, X.; Tang, H.; Zhou, T.; Li, W. MiR-194 promotes hepatocellular carcinoma through negative regulation of CADM1. Int. J. Clin. Exp. Pathol. 2020, 13, 1518. [Google Scholar] [PubMed]
- Hanson, A.; Wilhelmsen, D.; DiStefano, J.K. The Role of Long Non-Coding RNAs (lncRNAs) in the Development and Progression of Fibrosis Associated with Nonalcoholic Fatty Liver Disease (NAFLD). Non-Coding RNA 2018, 4, 18. [Google Scholar] [CrossRef] [PubMed]
- Wang, X. Down-regulation of lncRNA-NEAT1 alleviated the non-alcoholic fatty liver disease via mTOR/S6K1 signaling pathway. J. Cell Biochem. 2017, 119, 1567–1574. [Google Scholar] [CrossRef]
- Sun, Y.; Song, Y.; Liu, C.; Geng, J. LncRNA NEAT1-MicroRNA-140 axis exacerbates nonalcoholic fatty liver through interrupting AMPK/SREBP-1 signaling. Biochem. Biophys. Res. Commun. 2019, 516, 584–590. [Google Scholar] [CrossRef]
- Chen, X.; Tan, X.-R.; Li, S.-J.; Zhang, X.-X. LncRNA NEAT1 promotes hepatic lipid accumulation via regulating miR-146a-5p/ROCK1 in nonalcoholic fatty liver disease. Life Sci. 2019, 235, 116829. [Google Scholar] [CrossRef]
- Jin, S.S.; Lin, X.F.; Zheng, J.Z.; Wang, Q.; Guan, H.Q. lncRNA NEAT1 regulates fibrosis and inflammatory response induced by nonalcoholic fatty liver by regulating miR-506/GLI3. Eur. Cytokine Netw. 2019, 30, 98–106. [Google Scholar]
- Guo, S.; Chen, W.; Luo, Y.; Ren, F.; Zhong, T.; Rong, M.; Dang, Y.; Feng, Z.; Chen, G. Clinical implication of long non-coding RNA NEAT1 expression in hepatocellular carcinoma patients. Int. J. Clin. Exp. Pathol. 2015, 8, 5395. [Google Scholar]
- Fang, L.; Sun, J.; Pan, Z.; Song, Y.; Zhong, L.; Zhang, Y.; Liu, Y.; Zheng, X.; Huang, P. Long non-coding RNA NEAT1 promotes hepatocellular carcinoma cell proliferation through the regulation of miR-129-5p-VCP-IκB. Am. J. Physiol. Gastrointest. Liver Physiol. 2017, 313, G150–G156. [Google Scholar] [CrossRef]
- Zhang, Y.; Huang, C.; Zhu, Z.; Hou, Y.; Sun, C.; Tang, Y.; Zhang, Z.; Chen, H.; Ju, W.; Qiao, X.; et al. lncRNA NEAT1 regulates the proliferation and migration of hepatocellular carcinoma cells by acting as a miR-320a molecular sponge and targeting L antigen family member 3. Int. J. Oncol. 2020, 57, 1001–1012. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Xiao, T.; Xiao, Y.; Li, Y. Silencing of long non-coding RNA NEAT1 inhibits hepatocellular carcinoma progression by downregulating SMO by sponging microRNA-503. Mol. Med. Rep. 2020, 23, 1. [Google Scholar] [CrossRef] [PubMed]
- Yeermaike, A.; Gu, P.; Liu, D.; Nadire, T. LncRNA NEAT1 sponges miR-214 to promoted tumor growth in hepatocellular carcinoma. Mamm. Genome 2022, 33, 525–533. [Google Scholar] [CrossRef]
- Sakaguchi, H.; Tsuchiya, H.; Kitagawa, Y.; Tanino, T.; Yoshida, K.; Uchida, N.; Shiota, G. NEAT1 Confers Radioresistance to Hepatocellular Carcinoma Cells by Inducing Autophagy through GABARAP. Int. J. Mol. Sci. 2022, 23, 711. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Guo, H.; Xu, J.; Wang, J. Inhibition of lncRNA HULC improves hepatic fibrosis and hepatocyte apoptosis by inhibiting the MAPK signaling pathway in rats with nonalcoholic fatty liver disease. J. Cell Physiol. 2019, 234, 18169–18179. [Google Scholar] [CrossRef] [PubMed]
- Li, S.-P.; Xu, H.-X.; Yu, Y.; He, J.-D.; Wang, Z.; Xu, Y.-J.; Wang, C.-Y.; Zhang, H.-M.; Zhang, R.-X.; Zhang, J.-J.; et al. LncRNA HULC enhances epithelial-mesenchymal transition to promote tumorigenesis and metastasis of hepatocellular carcinoma via the miR-200a-3p/ZEB1 signaling pathway. Oncotarget 2016, 7, 42431–42446. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Li, X.; Hu, W.; Zhou, Y.; Din, Y. Silencing of lncRNA SNHG20 delays the progression of nonalcoholic fatty liver disease to hepatocellular carcinoma via regulating liver Kupffer cells polarization. IUBMB Life 2019, 71, 1952–1961. [Google Scholar] [CrossRef]
- Bechmann, L.P.; Hannivoort, R.A.; Gerken, G.; Hotamisligil, G.S.; Trauner, M.; Canbay, A. The interaction of hepatic lipid and glucose metabolism in liver diseases. J. Hepatol. 2012, 56, 952–964. [Google Scholar] [CrossRef]
- Ray, K. NAFLD-HCC: Target cholesterol. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 390. [Google Scholar] [CrossRef]
- Zhang, X.; Coker, O.O.; Chu, E.S.; Fu, K.; Lau, H.C.H.; Wang, Y.-X.; Chan, A.W.H.; Wei, H.; Yang, X.; Sung, J.J.Y.; et al. Dietary cholesterol drives fatty liver-associated liver cancer by modulating gut microbiota and metabolites. Gut 2020, 70, 761–774. [Google Scholar] [CrossRef]
- Liu, F.; Tian, T.; Zhang, Z.; Xie, S.; Yang, J.; Zhu, L.; Wang, W.; Shi, C.; Sang, L.; Guo, K.; et al. Long non-coding RNA SNHG6 couples cholesterol sensing with mTORC1 activation in hepatocellular carcinoma. Nat. Metab. 2022, 4, 1022–1240. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Ke, S.; Li, M.; Lin, C.; Liu, X.; Pan, Q. Downregulation of LncRNA GAS5 promotes liver cancer proliferation and drug resistance by decreasing PTEN expression. Mol. Genet. Genom. 2019, 295, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Zhong, Z.; Wang, A.; Yuan, C.; Ning, K.; Hu, H.; Wang, C.; Yin, X. LncRNA FTX represses the progression of non-alcoholic fatty liver disease to hepatocellular carcinoma via regulating the M1/M2 polarization of Kupffer cells. Cancer Cell Int. 2020, 20, 266. [Google Scholar] [CrossRef]
- Pirlog, R.; Cismaru, A.; Nutu, A.; Berindan-Neagoe, I. Field Cancerization in NSCLC: A New Perspective on MicroRNAs in Macrophage Polarization. Int. J. Mol. Sci. 2021, 22, 746. [Google Scholar] [CrossRef] [PubMed]
- Chi, Y.; Gong, Z.; Xin, H.; Wang, Z.; Liu, Z. Long noncoding RNA lncARSR promotes nonalcoholic fatty liver disease and hepatocellular carcinoma by promoting YAP1 and activating the IRS2/AKT pathway. J. Transl. Med. 2020, 18, 126. [Google Scholar] [CrossRef] [PubMed]
- Xiang, J.; Deng, Y.-Y.; Liu, H.-X.; Pu, Y. LncRNA MALAT1 Promotes PPARα/CD36-Mediated Hepatic Lipogenesis in Nonalcoholic Fatty Liver Disease by Modulating miR-206/ARNT Axis. Front. Bioeng. Biotechnol. 2022, 10, 858558. [Google Scholar] [CrossRef]
- Wu, Y.; Liu, X.; Zhou, Q.; Huang, C.; Meng, X.; Xu, F.; Li, J. Silent information regulator 1 (SIRT1) ameliorates liver fibrosis via promoting activated stellate cell apoptosis and reversion. Toxicol. Appl. Pharmacol. 2015, 289, 163–176. [Google Scholar] [CrossRef]
- Malakar, P.; Shilo, A.; Mogilevsky, A.; Stein, I.; Pikarsky, E.; Nevo, Y.; Benyamini, H.; Elgavish, S.; Zong, X.; Prasanth, K.V.; et al. Long noncoding RNA MALAT1 promotes hepatocellular carcinoma development by SRSF1 up-regulation and mTOR activation. Cancer Res. 2017, 77, 1155. [Google Scholar] [CrossRef]
- Liu, J.; Tang, T.; Wang, G.D.; Liu, B. LncRNA-H19 promotes hepatic lipogenesis by directly regulating miR-130a/PPARγ axis in non-alcoholic fatty liver disease. Biosci. Rep. 2019, 39, BSR20181722. [Google Scholar] [CrossRef]
- Zhou, Y.; Fan, R.-G.; Qin, C.-L.; Jia, J.; Wu, X.-D.; Zha, W.-Z. LncRNA-H19 activates CDC42/PAK1 pathway to promote cell proliferation, migration and invasion by targeting miR-15b in hepatocellular carcinoma. Genomics 2018, 111, 1862–1872. [Google Scholar] [CrossRef]
- Guo, B.; Cheng, Y.; Yao, L.; Zhang, J.; Lu, J.; Qi, H.; Chen, H. LncRNA HOTAIR regulates the lipid accumulation in non-alcoholic fatty liver disease via miR-130b-3p/ROCK1 axis. Cell Signal. 2021, 90, 110190. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Liu, B.; Huang, T.; Qi, X.; Li, S. HOTAIR modulates hepatocellular carcinoma progression by activating FUT8/core-fucosylated Hsp90/MUC1/STAT3 feedback loop via JAK1/STAT3 cascade. Dig. Liver Dis. 2022. [Google Scholar] [CrossRef] [PubMed]
- Zou, D.; Liu, L.; Zeng, Y.; Wang, H.; Dai, D.; Xu, M. LncRNA MEG3 up-regulates SIRT6 by ubiquitinating EZH2 and alleviates nonalcoholic fatty liver disease. Cell Death Discov. 2022, 8, 103. [Google Scholar] [CrossRef]
- Liu, Z.; Chen, J.Y.; Zhong, Y.; Xie, L.; Li, J.S. lncRNA MEG3 inhibits the growth of hepatocellular carcinoma cells by sponging miR-9-5p to upregulate SOX11. Braz. J. Med Biol. Res. 2019, 52, e8631. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Chen, X.; Zhang, F.; Zhao, M. RMRP inhibition prevents NAFLD progression in rats via regulating miR-206/PTPN1 axis. Mamm. Genome 2022, 33, 480–489. [Google Scholar] [CrossRef]
- Yukimoto, A.; Watanabe, T.; Sunago, K.; Nakamura, Y.; Tanaka, T.; Koizumi, Y.; Yoshida, O.; Tokumoto, Y.; Hirooka, M.; Abe, M.; et al. The long noncoding RNA of RMRP is downregulated by PERK, which induces apoptosis in hepatocellular carcinoma cells. Sci. Rep. 2021, 11, 7926. [Google Scholar] [CrossRef]
- Xia, Y.; Zhang, Y.; Wang, H. Upregulated lncRNA HCG18 in Patients with Non-Alcoholic Fatty Liver Disease and Its Regulatory Effect on Insulin Resistance. Diabetes, Metab. Syndr. Obesity Targets Ther. 2021, 14, 4747–4756. [Google Scholar] [CrossRef]
- Zou, Y.; Sun, Z.; Sun, S. LncRNA HCG18 contributes to the progression of hepatocellular carcinoma via miR-214-3p/CENPM axis. J. Biochem. 2020, 168, 535–546. [Google Scholar] [CrossRef]
- Bao, M.H.R.; Wong, C.C.L. Hypoxia, Metabolic Reprogramming, and Drug Resistance in Liver Cancer. Cells 2021, 10, 1715. [Google Scholar] [CrossRef]
- Razavi, Z.S.; Asgarpour, K.; Mahjoubin-Tehran, M.; Rasouli, S.; Khan, H.; Shahrzad, M.K.; Hamblin, M.R.; Mirzaei, H. Angiogenesis-related non-coding RNAs and gastrointestinal cancer. Mol. Ther. -Oncolytics 2021, 21, 220–241. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).