Drp1 Overexpression Decreases Insulin Content in Pancreatic MIN6 Cells
Abstract
1. Introduction
2. Results
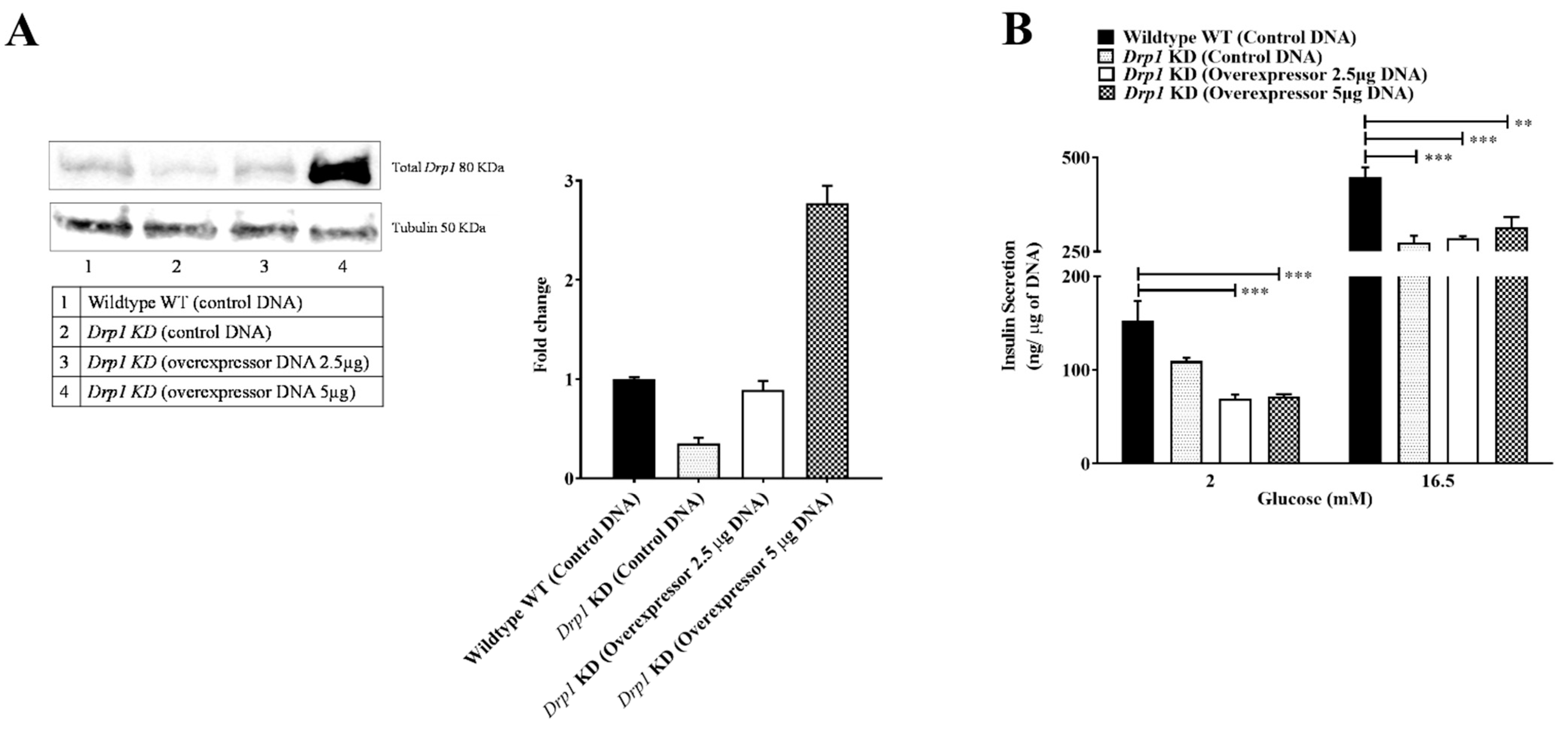
2.1. Drp1 Overexpression Does Not Rescue GSIS in Beta Cells
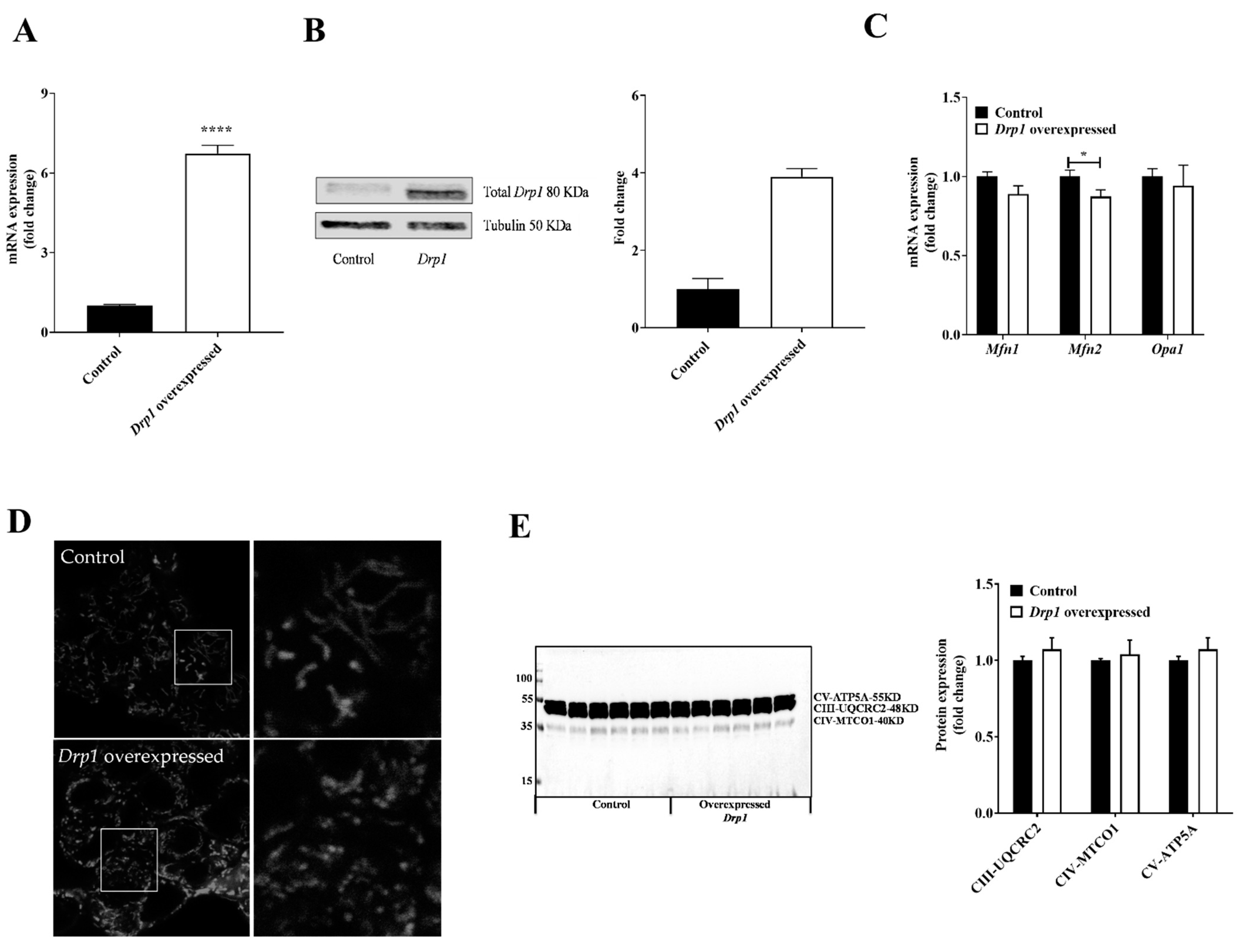
2.2. Drp1 Overexpression Altered Mitochondrial Morphology But Had No Effect on Mitochondrial Respiratory Complexes in MIN6 Cells
2.3. Reduced Insulin Content in Drp1-Overexpressing MIN6 Cells
2.4. Stress Pathway Activation upon Drp1-Overexpression in MIN6 Cells
3. Discussion
4. Materials and Methods
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Affourtit, C.; Alberts, B.; Barlow, J.; Carré, J.E.; Wynne, A.G. Control of pancreatic β-cell bioenergetics. Biochem. Soc. Trans. 2018, 46, 555–564. [Google Scholar] [CrossRef] [PubMed]
- Yapa, N.M.; Lisnyak, V.; Reljic, B.; Ryan, M.T. Mitochondrial dynamics in health and disease. FEBS Lett. 2021, 595, 1184–1204. [Google Scholar] [CrossRef] [PubMed]
- Wai, T.; Langer, T. Mitochondrial dynamics and metabolic regulation. Trends Endocrinol. Metab. 2016, 27, 105–117. [Google Scholar] [CrossRef] [PubMed]
- Anello, M.; Lupi, R.; Spampinato, D.; Piro, S.; Masini, M.; Boggi, U.; Marchetti, P. Functional and morphological alterations of mitochondria in pancreatic beta cells from type 2 diabetic patients. Diabetologia 2005, 48, 282–289. [Google Scholar] [CrossRef] [PubMed]
- Molina, A.J.; Wikstrom, J.D.; Stiles, L.; Las, G.; Mohamed, H.; Elorza, A.; Shirihai, O.S. Mitochondrial networking protects β-cells from nutrient-induced apoptosis. Diabetes 2009, 58, 2303–2315. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Wakabayashi, N.; Wakabayashi, J.; Tamura, Y.; Song, W.J.; Sereda, S.; Sesaki, H. The dynamin-related GTPase Opa1 is required for glucose-stimulated ATP production in pancreatic beta cells. Mol. Biol. Cell 2011, 22, 2235–2245. [Google Scholar] [CrossRef]
- Park, K.S.; Wiederkehr, A.; Kirkpatrick, C.; Mattenberger, Y.; Martinou, J.C.; Marchetti, P.; Wollheim, C.B. Selective Actions of Mitochondrial Fission/Fusion Genes on Metabolism-Secretion Coupling in Insulin-releasing Cells. J. Biol. Chem. 2008, 283, 33347–33356. [Google Scholar] [CrossRef]
- Jhun, B.S.; Lee, H.; Jin, Z.G.; Yoon, Y. Glucose stimulation induces dynamic change of mitochondrial morphology to promote insulin secretion in the insulinoma cell line INS-1E. PLoS ONE 2013, 8, e60810. [Google Scholar] [CrossRef]
- Kabra, U.D.; Pfuhlmann, K.; Migliorini, A.; Keipert, S.; Lamp, D.; Korsgren, O.; Jastroch, M. Direct Substrate Delivery Into Mitochondrial Fission–Deficient Pancreatic Islets Rescues Insulin Secretion. Diabetes 2017, 66, 1247–1257. [Google Scholar] [CrossRef]
- Kabra, U.D.; Affourtit, C.; Jastroch, M. Respiratory parameters for the classification of dysfunctional insulin secretion by pancreatic islets. Metabolites 2021, 11, 405. [Google Scholar] [CrossRef]
- Touvier, T.; De Palma, C.; Rigamonti, E.; Scagliola, A.; Incerti, E.; Mazelin, L.; Brunelli, S. Muscle-specific Drp1 overexpression impairs skeletal muscle growth via translational attenuation. Cell Death Dis. 2015, 6, e1663. [Google Scholar] [CrossRef] [PubMed]
- Vezza, T.; Díaz-Pozo, P.; Canet, F.; de Marañón, A.M.; Abad-Jiménez, Z.; García-Gargallo, C.; Víctor, V.M. The role of mitochondrial dynamic dysfunction in age-associated type 2 diabetes. World J. Men’s Health 2022, 40, 399. [Google Scholar] [CrossRef] [PubMed]
- Mulder, H.; Ling, C. Mitochondrial dysfunction in pancreatic β-cells in type 2 diabetes. Mol. Cell. Endocrinol. 2009, 297, 34–40. [Google Scholar] [CrossRef]
- Lo, M.C.; Chen, M.H.; Lee, W.S.; Lu, C.I.; Chang, C.R.; Kao, S.H.; Lee, H.M. N ε-(carboxymethyl) lysine-induced mitochondrial fission and mitophagy cause decreased insulin secretion from β-cells. Am. J. Physiol.-Endocrinol. Metab. 2015, 309, E829–E839. [Google Scholar] [CrossRef]
- Liu, J.; Chen, Z.; Zhang, Y.; Zhang, M.; Zhu, X.; Fan, Y.; Liu, Z. Rhein protects pancreatic β-cells from dynamin-related protein-1–mediated mitochondrial fission and cell apoptosis under hyperglycemia. Diabetes 2013, 62, 3927–3935. [Google Scholar] [CrossRef] [PubMed]
- Supale, S.; Li, N.; Brun, T.; Maechler, P. Mitochondrial dysfunction in pancreatic β cells. Trends Endocrinol. Metab. 2012, 23, 477–487. [Google Scholar] [CrossRef] [PubMed]
- Wiederkehr, A.; Wollheim, C.B. Mitochondrial signals drive insulin secretion in the pancreatic β-cell. Mol. Cell. Endocrinol. 2012, 353, 128–137. [Google Scholar] [CrossRef] [PubMed]
- Haythorne, E.; Rohm, M.; van de Bunt, M.; Brereton, M.F.; Tarasov, A.I.; Blacker, T.S.; Ashcroft, F.M. Diabetes causes marked inhibition of mitochondrial metabolism in pancreatic β-cells. Nat. Commun. 2019, 10, 2474. [Google Scholar] [CrossRef] [PubMed]
- Brand, M.D.; Parker, N.; Affourtit, C.; Mookerjee, S.A.; Azzu, V. Mitochondrial uncoupling protein 2 in pancreatic β-cells. Diabetes Obes. Metab. 2010, 12, 134–140. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, S.G.; Gromada, J.; Urano, F. Endoplasmic reticulum stress and pancreatic β-cell death. Trends Endocrinol. Metab. 2011, 22, 266–274. [Google Scholar] [CrossRef] [PubMed]
- Back, S.H.; Kaufman, R.J. Endoplasmic reticulum stress and type 2 diabetes. Annu. Rev. Biochem. 2012, 81, 767. [Google Scholar] [CrossRef] [PubMed]
- Kaufman, R.J.; Back, S.H.; Song, B.; Han, J.; Hassler, J. The unfolded protein response is required to maintain the integrity of the endoplasmic reticulum, prevent oxidative stress and preserve differentiation in β-cells. Diabetes Obes. Metab. 2010, 12, 99–107. [Google Scholar] [CrossRef]
- Huang, C.J.; Haataja, L.; Gurlo, T.; Butler, A.E.; Wu, X.; Soeller, W.C.; Butler, P.C. Induction of endoplasmic reticulum stress-induced β-cell apoptosis and accumulation of polyubiquitinated proteins by human islet amyloid polypeptide. Am. J. Physiol. -Endocrinol. Metab. 2007, 293, E1656–E1662. [Google Scholar] [CrossRef]
- Volchuk, A.; Ron, D. The endoplasmic reticulum stress response in the pancreatic β-cell. Diabetes Obes. Metab. 2010, 12, 48–57. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, S.G.; Lipson, K.L.; Urano, F. Endoplasmic reticulum stress signaling in pancreatic β-cells. Antioxid. Redox Signal. 2007, 9, 2335–2344. [Google Scholar] [CrossRef] [PubMed]
- Oyadomari, S.; Araki, E.; Mori, M. Endoplasmic reticulum stress-mediated apoptosis in pancreatic β-cells. Apoptosis 2002, 7, 335–345. [Google Scholar] [CrossRef] [PubMed]
- Song, B.; Scheuner, D.; Ron, D.; Pennathur, S.; Kaufman, R.J. Chop deletion reduces oxidative stress, improves β cell function, and promotes cell survival in multiple mouse models of diabetes. J. Clin. Investig. 2008, 118, 3378–3389. [Google Scholar] [CrossRef] [PubMed]
- Engin, F.; Nguyen, T.; Yermalovich, A.; Hotamisligil, G.S. Aberrant islet unfolded protein response in type 2 diabetes. Sci. Rep. 2014, 4, 4054. [Google Scholar] [CrossRef] [PubMed]
- Ježek, P.; Dlasková, A. Dynamic of mitochondrial network, cristae, and mitochondrial nucleoids in pancreatic β-cells. Mitochondrion 2019, 49, 245–258. [Google Scholar] [CrossRef]
- Wikstrom, J.D.; Israeli, T.; Bachar-Wikstrom, E.; Swisa, A.; Ariav, Y.; Waiss, M.; Leibowitz, G. AMPK regulates ER morphology and function in stressed pancreatic β-cells via phosphorylation of DRP1. Mol. Endocrinol. 2013, 27, 1706–1723. [Google Scholar] [CrossRef]
- Bachar, E.; Ariav, Y.; Ketzinel-Gilad, M.; Cerasi, E.; Kaiser, N.; Leibowitz, G. Glucose amplifies fatty acid-induced endoplasmic reticulum stress in pancreatic β-cells via activation of mTORC1. PLoS ONE 2009, 4, e4954. [Google Scholar] [CrossRef]
- Divakaruni, A.S.; Jastroch, M. A practical guide for the analysis, standardization and interpretation of oxygen consumption measurements. Nat. Metab. 2022, 4, 978–994. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kabra, U.D.; Moruzzi, N.; Berggren, P.-O.; Jastroch, M. Drp1 Overexpression Decreases Insulin Content in Pancreatic MIN6 Cells. Int. J. Mol. Sci. 2022, 23, 12338. https://doi.org/10.3390/ijms232012338
Kabra UD, Moruzzi N, Berggren P-O, Jastroch M. Drp1 Overexpression Decreases Insulin Content in Pancreatic MIN6 Cells. International Journal of Molecular Sciences. 2022; 23(20):12338. https://doi.org/10.3390/ijms232012338
Chicago/Turabian StyleKabra, Uma D., Noah Moruzzi, Per-Olof Berggren, and Martin Jastroch. 2022. "Drp1 Overexpression Decreases Insulin Content in Pancreatic MIN6 Cells" International Journal of Molecular Sciences 23, no. 20: 12338. https://doi.org/10.3390/ijms232012338
APA StyleKabra, U. D., Moruzzi, N., Berggren, P.-O., & Jastroch, M. (2022). Drp1 Overexpression Decreases Insulin Content in Pancreatic MIN6 Cells. International Journal of Molecular Sciences, 23(20), 12338. https://doi.org/10.3390/ijms232012338





