Rad17 Translocates to Nucleolus upon UV Irradiation through Nucleolar Localization Signal in the Central Basic Domain
Abstract
1. Introduction
2. Results and Discussion
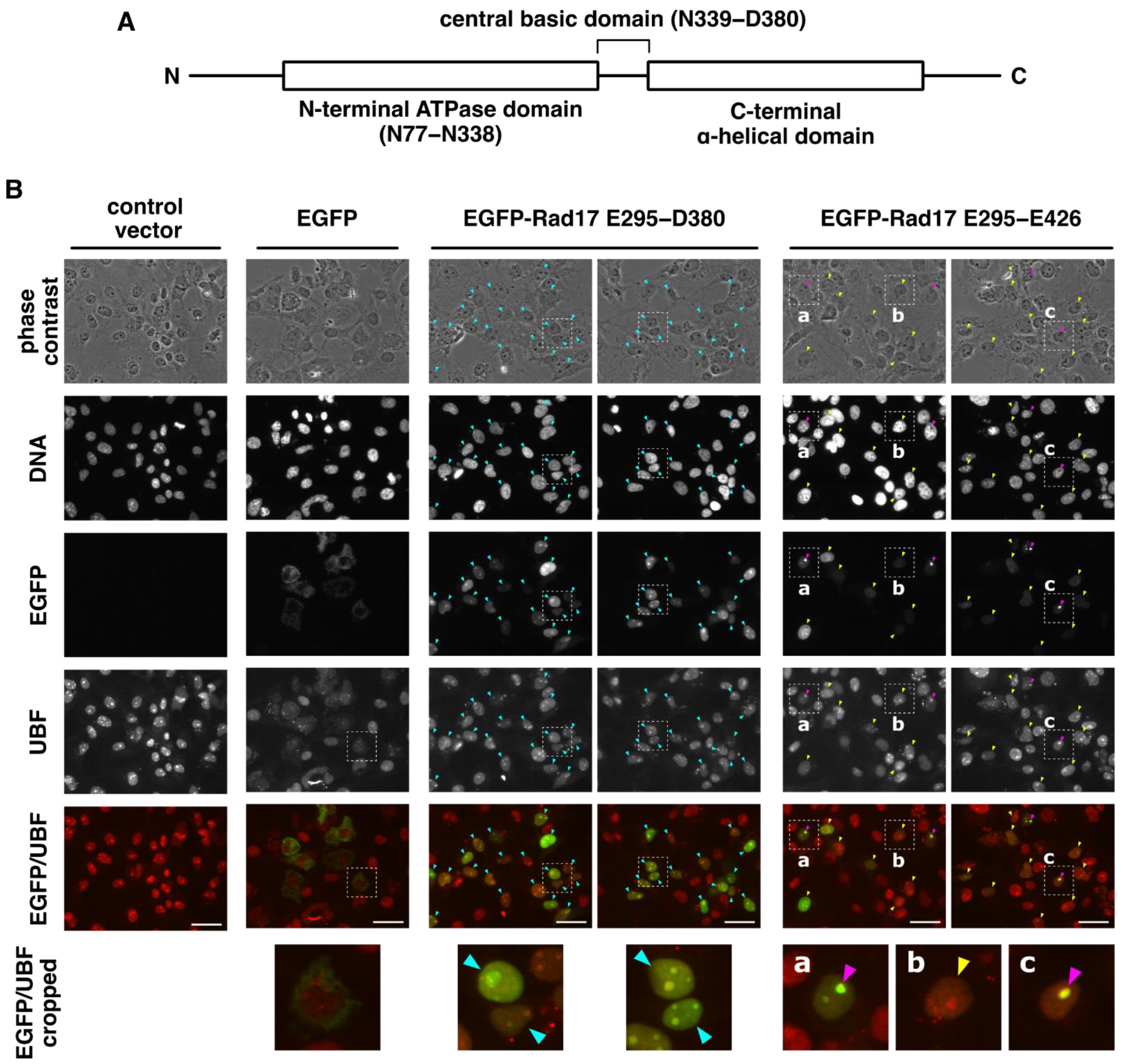
2.1. The Central Basic Domain of Rad17 Encodes a Nucleolar Localization Signal
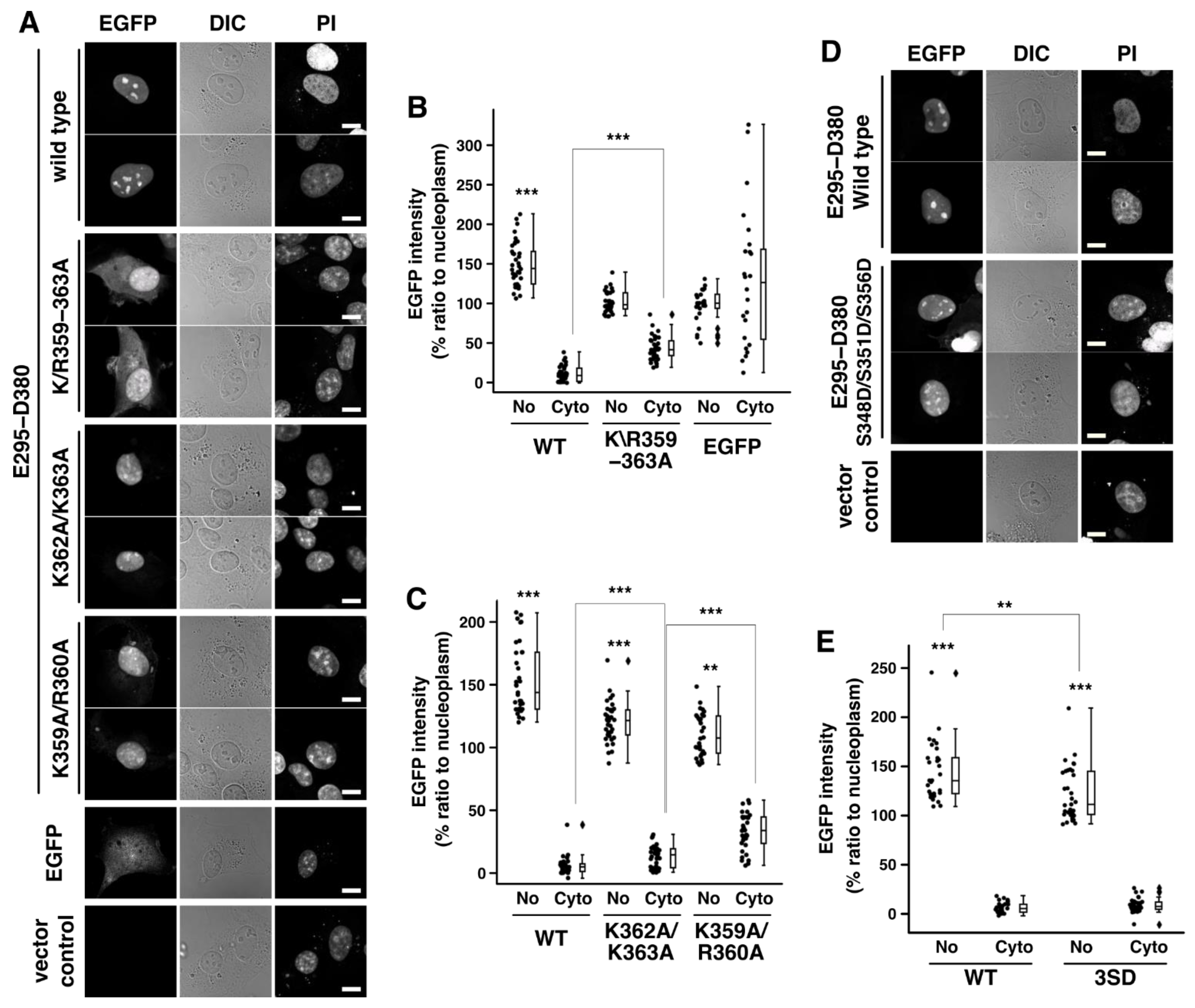
2.2. Rad17 K359–K363 Residues Encode the Nucleolar Localization Signal
2.3. Putative Phosphorylation Sites in the Central Basic Domain Regulate the Nucleolar Localization Signal of Rad17
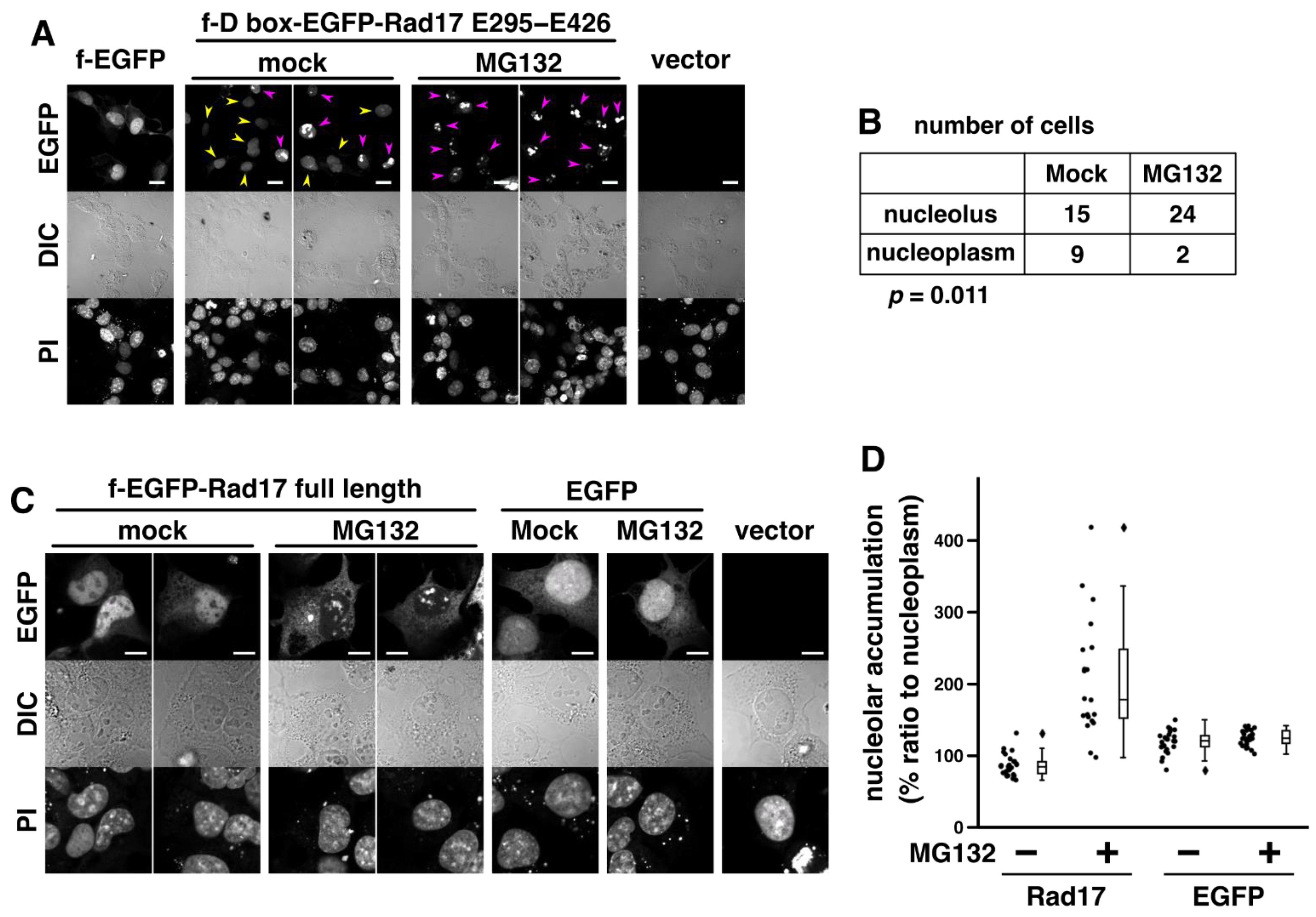
2.4. Proteasomal Degradation Negatively Regulates the Nucleolar Localization of Rad17
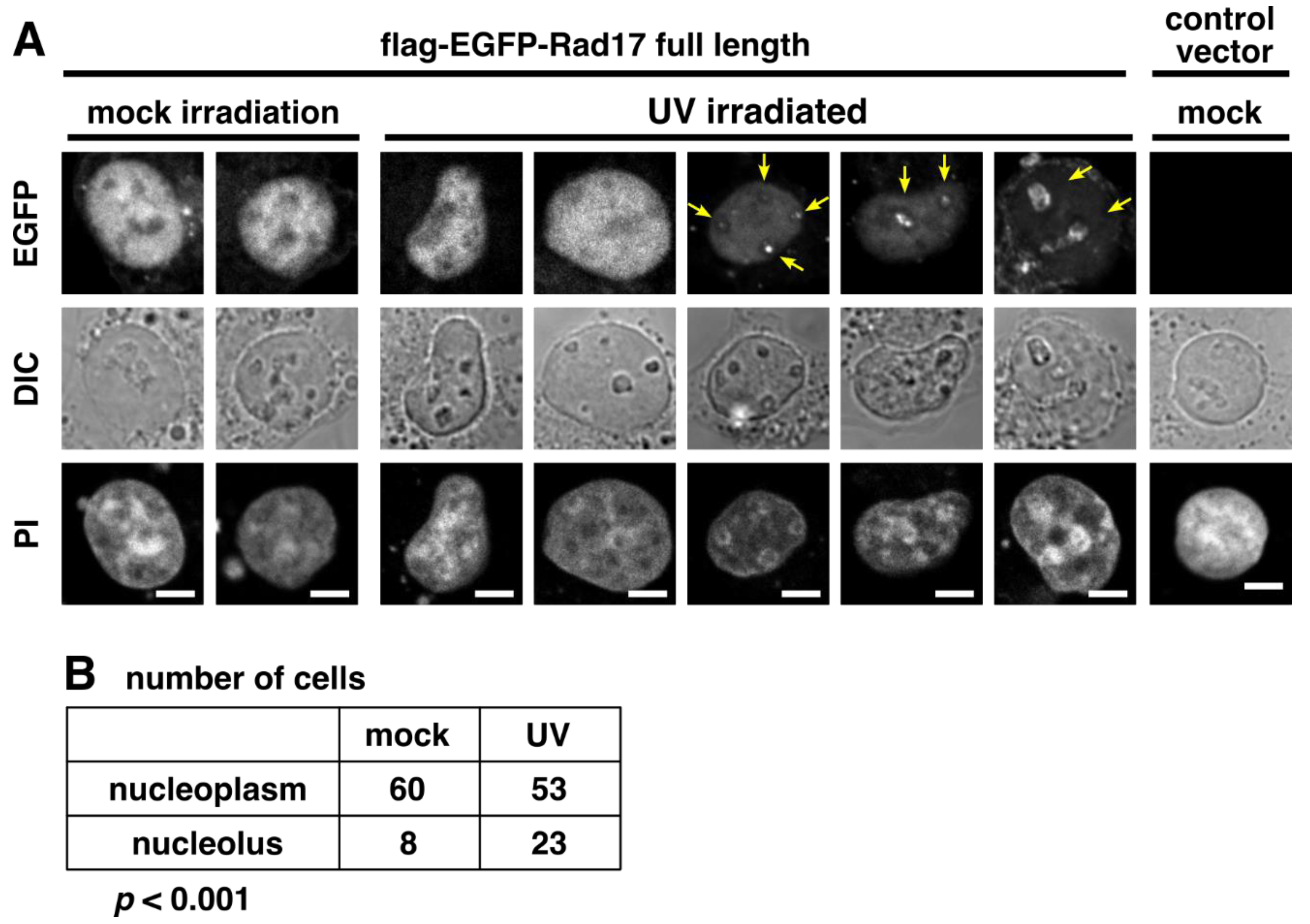
2.5. UV Irradiation Promotes the Nucleolar Localization of Rad17
3. Materials and Methods
3.1. Antibodies
3.2. Plasmids
3.3. Fluorescence Microscopy
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Lindstrom, M.S.; Jurada, D.; Bursac, S.; Orsolic, I.; Bartek, J.; Volarevic, S. Nucleolus as an emerging hub in maintenance of genome stability and cancer pathogenesis. Oncogene 2018, 37, 2351–2366. [Google Scholar] [CrossRef] [PubMed]
- Larsen, D.H.; Stucki, M. Nucleolar responses to DNA double-strand breaks. Nucleic Acids Res. 2016, 44, 538–544. [Google Scholar] [CrossRef] [PubMed]
- Saldivar, J.C.; Cortez, D.; Cimprich, K.A. The essential kinase ATR: Ensuring faithful duplication of a challenging genome. Nat. Rev. Mol. Cell. Biol. 2017, 18, 622–636. [Google Scholar] [CrossRef] [PubMed]
- Sokka, M.; Rilla, K.; Miinalainen, I.; Pospiech, H.; Syvaoja, J.E. High levels of TopBP1 induce ATR-dependent shut-down of rRNA transcription and nucleolar segregation. Nucleic Acids Res. 2015, 43, 4975–4989. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Goldstein, M.; Alexander, P.; Wakeman, T.P.; Sun, T.; Feng, J.; Lou, Z.; Kastan, M.B.; Wang, X.F. Rad17 recruits the MRE11-RAD50-NBS1 complex to regulate the cellular response to DNA double-strand breaks. EMBO J. 2014, 33, 862–877. [Google Scholar] [CrossRef]
- Tsao, C.C.; Geisen, C.; Abraham, R.T. Interaction between human MCM7 and Rad17 proteins is required for replication checkpoint signaling. EMBO J. 2004, 23, 4660–4669. [Google Scholar] [CrossRef]
- Mohni, K.N.; Dee, A.R.; Smith, S.; Schumacher, A.J.; Weller, K.S. Efficient herpes simplex virus 1 replication requires cellular ATR pathway proteins. J. Virol. 2013, 87, 531–542. [Google Scholar] [CrossRef]
- Zhang, L.; Park, C.H.; Wu, J.; Kim, H.; Liu, W.; Fujita, T.; Balasubramani, M.; Schreiber, E.M.; Wang, X.F.; Wan, Y. Proteolysis of Rad17 by Cdh1/APC regulates checkpoint termination and recovery from genotoxic stress. EMBO J. 2010, 29, 1726–1737. [Google Scholar] [CrossRef]
- Zou, L.; Cortez, D.; Elledge, S.J. Regulation of ATR substrate selection by Rad17-dependent loading of Rad9 complexes onto chromatin. Genes Dev. 2002, 16, 198–208. [Google Scholar] [CrossRef]
- Post, S.M.; Tomkinson, A.E.; Lee, E.Y. The human checkpoint Rad protein Rad17 is chromatin-associated throughout the cell cycle, localizes to DNA replication sites, and interacts with DNA polymerase epsilon. Nucleic Acids Res. 2003, 31, 5568–5575. [Google Scholar] [CrossRef]
- Chang, M.S.; Sasaki, H.; Campbell, M.S.; Kraeft, S.K.; Sutherland, R.; Yang, C.Y.; Liu, Y.; Auclair, D.; Hao, L.; Sonoda, H.; et al. HRad17 colocalizes with NHP2L1 in the nucleolus and redistributes after UV irradiation. J. Biol. Chem. 1999, 274, 36544–36549. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bao, S.; Shen, X.; Shen, K.; Liu, Y.; Wang, X.F. The mammalian Rad24 homologous to yeast Saccharomyces cerevisiae Rad24 and Schizosaccharomyces pombe Rad17 is involved in DNA damage checkpoint. Cell Growth Differ. 1998, 9, 961–967. [Google Scholar] [PubMed]
- Perez-Castro, A.J.; Freire, R. Rad9B responds to nucleolar stress through ATR and JNK signalling, and delays the G1-S transition. J. Cell Sci. 2012, 125, 1152–1164. [Google Scholar] [CrossRef] [PubMed]
- Fukumoto, Y.; Ikeuchi, M.; Qu, L.; Hoshino, T.; Yamaguchi, N.; Nakayama, Y.; Ogra, Y. Nuclear translocation promotes proteasomal degradation of human Rad17 protein through the N-terminal destruction boxes. J. Biol. Chem. 2021, 297, 100831. [Google Scholar] [CrossRef] [PubMed]
- Greig, J.A.; Nguyen, T.A.; Lee, M.; Holehouse, A.S.; Posey, A.E.; Pappu, R.V.; Jedd, G. Arginine-Enriched Mixed-Charge Domains Provide Cohesion for Nuclear Speckle Condensation. Mol. Cell 2020, 77, 1237–1250.e4. [Google Scholar] [CrossRef] [PubMed]
- Sztacho, M.; Salovska, B.; Cervenka, J.; Balaban, C.; Hoboth, P.; Hozak, P. Limited Proteolysis-Coupled Mass Spectrometry Identifies Phosphatidylinositol 4,5-Bisphosphate Effectors in Human Nuclear Proteome. Cells 2021, 10, 68. [Google Scholar] [CrossRef] [PubMed]
- Hoboth, P.; Sztacho, M.; Sebesta, O.; Schatz, M.; Castano, E.; Hozak, P. Nanoscale mapping of nuclear phosphatidylinositol phosphate landscape by dual-color dSTORM. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2021, 1866, 158890. [Google Scholar] [CrossRef] [PubMed]
- Emmott, E.; Hiscox, J.A. Nucleolar targeting: The hub of the matter. EMBO Rep. 2009, 10, 231–238. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Ching, Y.P.; Chun, A.C.; Jin, D.Y. Nuclear localization of the cell cycle regulator CDH1 and its regulation by phosphorylation. J. Biol. Chem. 2003, 278, 12530–12536. [Google Scholar] [CrossRef] [PubMed]
- Latonen, L.; Moore, H.M.; Bai, B.; Jaamaa, S.; Laiho, M. Proteasome inhibitors induce nucleolar aggregation of proteasome target proteins and polyadenylated RNA by altering ubiquitin availability. Oncogene 2011, 30, 790–805. [Google Scholar] [CrossRef]
- Fukumoto, Y.; Ikeuchi, M.; Nakayama, Y.; Yamaguchi, N. The KYxxL motif in Rad17 protein is essential for the interaction with the 9-1-1 complex. Biochem. Biophys. Res. Commun. 2016, 477, 982–987. [Google Scholar] [CrossRef] [PubMed]
- Caspari, T.; Dahlen, M.; Kanter-Smoler, G.; Lindsay, H.D.; Hofmann, K.; Papadimitriou, K.; Sunnerhagen, P.; Carr, A.M. Characterization of Schizosaccharomyces pombe Hus1: A PCNA-related protein that associates with Rad1 and Rad9. Mol. Cell Biol. 2000, 20, 1254–1262. [Google Scholar] [CrossRef] [PubMed]
- Larsen, D.H.; Hari, F.; Clapperton, J.A.; Gwerder, M.; Gutsche, K.; Altmeyer, M.; Jungmichel, S.; Toledo, L.I.; Fink, D.; Rask, M.B.; et al. The NBS1-Treacle complex controls ribosomal RNA transcription in response to DNA damage. Nat. Cell. Biol. 2014, 16, 792–803. [Google Scholar] [CrossRef] [PubMed]
- Ciccia, A.; Huang, J.W.; Izhar, L.; Sowa, M.E.; Harper, J.W.; Elledge, S.J. Treacher Collins syndrome TCOF1 protein cooperates with NBS1 in the DNA damage response. Proc. Natl. Acad. Sci. USA 2014, 111, 18631–18636. [Google Scholar] [CrossRef] [PubMed]
- Ikeuchi, M.; Fukumoto, Y.; Honda, T.; Kuga, T.; Saito, Y.; Yamaguchi, N.; Nakayama, Y. v-Src Causes Chromosome Bridges in a Caffeine-Sensitive Manner by Generating DNA Damage. Int. J. Mol. Sci. 2016, 17, 871. [Google Scholar] [CrossRef] [PubMed]
- Fukumoto, Y.; Obata, Y.; Ishibashi, K.; Tamura, N.; Kikuchi, I.; Aoyama, K.; Hattori, Y.; Tsuda, K.; Nakayama, Y.; Yamaguchi, N. Cost-effective gene transfection by DNA compaction at pH 4.0 using acidified, long shelf-life polyethylenimine. Cytotechnology 2010, 62, 73–82. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fukumoto, Y.; Ikeuchi, M.; Nakayama, Y.; Ogra, Y. Rad17 Translocates to Nucleolus upon UV Irradiation through Nucleolar Localization Signal in the Central Basic Domain. Int. J. Mol. Sci. 2022, 23, 12300. https://doi.org/10.3390/ijms232012300
Fukumoto Y, Ikeuchi M, Nakayama Y, Ogra Y. Rad17 Translocates to Nucleolus upon UV Irradiation through Nucleolar Localization Signal in the Central Basic Domain. International Journal of Molecular Sciences. 2022; 23(20):12300. https://doi.org/10.3390/ijms232012300
Chicago/Turabian StyleFukumoto, Yasunori, Masayoshi Ikeuchi, Yuji Nakayama, and Yasumitsu Ogra. 2022. "Rad17 Translocates to Nucleolus upon UV Irradiation through Nucleolar Localization Signal in the Central Basic Domain" International Journal of Molecular Sciences 23, no. 20: 12300. https://doi.org/10.3390/ijms232012300
APA StyleFukumoto, Y., Ikeuchi, M., Nakayama, Y., & Ogra, Y. (2022). Rad17 Translocates to Nucleolus upon UV Irradiation through Nucleolar Localization Signal in the Central Basic Domain. International Journal of Molecular Sciences, 23(20), 12300. https://doi.org/10.3390/ijms232012300






