Abstract
Pesticide resistance in insects is an example of adaptive evolution occurring in pest species and is driven by the artificial introduction of pesticides. The diamondback moth (DBM), Plutella xylostella (Lepidoptera: Plutellidae), has evolved resistance to various insecticides. Understanding the genetic changes underpinning the resistance to pesticides is necessary for the implementation of pest control measures. We sequenced the genome of six resistant and six susceptible DBM individuals separately and inferred the genomic regions of greatest divergence between strains using FST and θπ. Among several genomic regions potentially related to insecticide resistance, CYP6B6-like was observed with significant divergence between the resistant and susceptible strains, with a missense mutation located near the substrate recognition site (SRS) and four SNPs in the promoter. To characterize the relative effects of directional selection via insecticide tolerance (‘strain’) as compared to acute exposure to insecticide (‘treatment’), four pairwise comparisons were carried out between libraries to determine the differentially expressed genes. Most resistance-related differentially expressed genes were identified from the comparison of the strains and enriched in pathways for exogenous detoxification including cytochrome P450 and the ABC transporter. Further confirmation came from the weighted gene co-expression network analysis, which indicated that genes in the significant module associated with chlorantraniliprole resistance were enriched in pathways for exogenous detoxification, and that CYP6B6-like represented a hub gene in the “darkred” module. Furthermore, RNAi knock-down of CYP6B6-like increases P. xylostella sensitivity to chlorantraniliprole. Our study thus provides a genetic foundation underlying selection for pesticide resistance and plausible mechanisms to explain fast evolved adaptation through genomic divergence and altered gene expression in insects.
1. Introduction
Pesticide resistance in insects represents an excellent genetic model to explain how insects adapt to the relatively short-term and strong selection pressure exerted by non-native chemicals. Understanding the genetic mechanisms underlying pesticide resistance is thus advantageous for pest control management. As a major pest of Brassica vegetable and oilseed crops throughout the world, the diamondback moth (DBM), Plutella xylostella L. (Lepidoptera: Plutellidae), is estimated to cause approximately US$0.77 billion management tool and crop losses annually in China alone []. Chemical insecticides remain the main management route to control the DBM. However, the abuse of various insecticides, coupled with a short generation time and the large overlapping of generations in DBM [], have promoted DBM field resistance to all major kinds of insecticides, including organophosphates, carbamates, pyrethroids, and Bacillus thuringiensis (Bt) Cry toxins []. Chlorantraniliprole was the first commercialized anthranilic diamide insecticide widely used for pest control, especially for Lepidoptera pests []. However, within three years of its introduction in Guangdong in 2008, DBM had evolved a high level of resistance to chlorantraniliprole in the field []. In general, insecticide-resistance mechanisms have been described as three major types, (i) metabolic resistance that involves overexpression and elevated catalytic activity of detoxification enzymes; (ii) target resistance that involves mutation of the insecticide target site; and (iii) penetration resistance that involves modifications of the cuticle [,]. Previous research on the chlorantraniliprole-resistance mechanism in DBM mainly focused on the mutations in the target site, the ryanodine receptors (RyRs) [,,,]. More recently, the role of metabolism resistance in DBM that accompanies chlorantraniliprole resistance has been highlighted [,,,]. However, there has been no genome-wide scan of selection performed in P. xylostella to identify candidate chlorantraniliprole resistance-related genes.
The development of high-throughput sequencing technology, combined with the availability of genome-scale genetic data and statistical methods, is increasingly providing a general framework for genome-wide scans of selection (GWSS) to identify positively selected loci associated with several phenotypic traits. Methods based on population genetics’ statistics such as including FST, π, iHS, Tajima’s D, XP-CLR, and XP-EHH have been widely used to seek out the genetic targets of artificial selection. Examples include genes associated with the domestic yak’s behavior and tameness [], fat tail genes of Chinese indigenous sheep [], meat and milk quality traits in cattle [,], and other economic traits of domestic animals [,,]. In addition to domesticated traits, GWSS also has been used to identify adaptation to natural selective pressures, such as the adaptation of Dehong humped cattle to heat stress [], Yanbian cattle to a cold climate [], goats and sheep to a hot, arid environment [], and Tibetan pigs to a high altitude []. There are also successful applications of GWSS to insecticide resistance, such as detecting genomic regions affected by DDT selection in Drosophila melanogaster [] and genes associated with pyrthroid and DDT resistance in Amyelois transitella [].
In this study, whole-genome resequencing was performed on two DBM strains (chlorantraniliprole-susceptible and -resistant strains), and FST for GWSS was used to detect genes with a signature of positive selection. The susceptible strain was maintained in laboratory conditions without exposure to any insecticide for 5 years, while the resistant strain originates from a chlorantraniliprole-susceptible population through six generations of chlorantraniliprole selection and backcrossing (Figure 1a). Secondly, transcriptional data were obtained, and analysis was performed to study the potential response to insecticide via altered gene expression of candidate genes. It is hypothesized that through these diverse approaches focusing on changes at the DNA and RNA level, many resistance-related genes may be detected. Among these candidates, CYP6B6-like was confirmed to be involved in chlorantraniliprole resistance by RNAi. Such knowledge will prove essential to the future management and use of insecticides if the genetic and biochemical mechanisms of insecticide resistance are better understood.
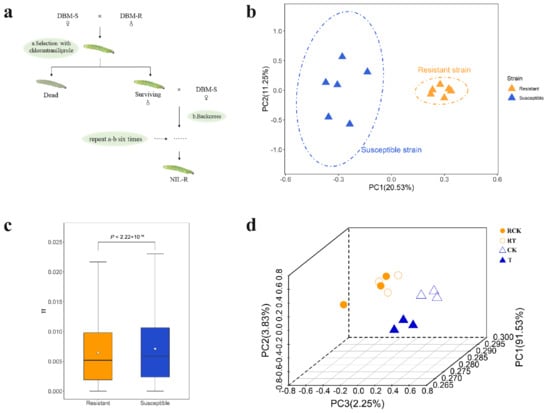
Figure 1.
Genetic differentiation between susceptible and resistant strains of Plutella xylostella. (a) Construction of near-isogenic Plutella xylostella strains resistant to chlorantraniliprole. (b) Principal component analysis of SNPs. (c) Significant difference in population nucleotide diversity (θπ), as evaluated using the t-test (P < 2.22 × 10−16). (d) Principal component analysis of gene counts. Two colors represent two strains of Plutella xylostella, orange for the resistant strain and blue for the susceptible strain. Different graphs represent samples, RCK, Resistant stain without treatment by chlorantraniliprole (LC50); RT, Resistant stain after treated with chlorantraniliprole (LC50); CK, Susceptible strain without exposure to any insecticide; and T, Susceptible strain exposed to chlorantraniliprole (LC50).
2. Results
2.1. Genome Resequencing and Genetic Variation
Genome resequencing of 6 resistance and 6 susceptible DBM individuals yielded 131.03 G of clean data. A total of 1.07 million high-quality SNPs were detected among all samples, and these SNPs were annotated using SnpEff (v4.3). Principal component analysis (PCA) divided the 12 DBM genome samples into two populations, matching the resistant and susceptible strains (Figure 1b). The first PC and the second PC explained 20.53% and 11.25% of the total variation separating the two populations, respectively. Reduced genetic diversity was observed among the resistant strain samples, by estimation of average θπ (P < 2.22 × 10−16) in both strains (Figure 1c). RNA-Seq expression data came to the same result (Figure 1d, see below for details). These results implicated higher diversity in the susceptible strain than the resistant strain, whose tolerance to chlorantraniliprole differs by 401.4-fold (Table 1). This pattern is in accordance with the expectation of selection on the resistant strain by selecting only the most pesticide-tolerant insects in subsequent generations for the continuation of the strain.

Table 1.
Toxicity of chlorantraniliprole to two strains of Plutella xylostella.
2.2. Selective-Sweep Analysis Identifies Candidate Genes Associated with Insecticide Resistance
To detect signatures of positive selection associated with insecticide resistance, we searched the DBM genome for regions with high FST between susceptible strains and resistant strains (FST > 0.50), yielding 1501 genes. All 1501 genes were used for KEGG analysis and gained a total of 132 enrichment pathways (Table S1), with 7 pathways significantly enriched associated with signal transduction (MAPK signaling pathway and Wnt signaling pathway), lipid metabolism (fatty acid degradation), and development and regeneration (cytoskeleton proteins) (Figure 2). Because insecticide resistance is known to be related to metabolic resistance, target resistance, and penetration resistance according to previous studies, we focused our analysis on detoxifying metabolic enzymes, insecticide receptors, cuticle proteins, ion channels, digestive enzymes, and the ubiquitinase system (Table S2). In this reduced dataset containing 63 genes, the metabolic detoxifying enzymes were overrepresented, including 13 P450s, 8 ABC transporters, and 1 UDP-glucuronosyltransferase, followed by ion channels. In addition, 41 genes coding insecticide receptors, cuticle proteins, ion channels, digestive enzymes, and the ubiquitinase system were also detected as positively selected genes (PSGs) (Table S2).
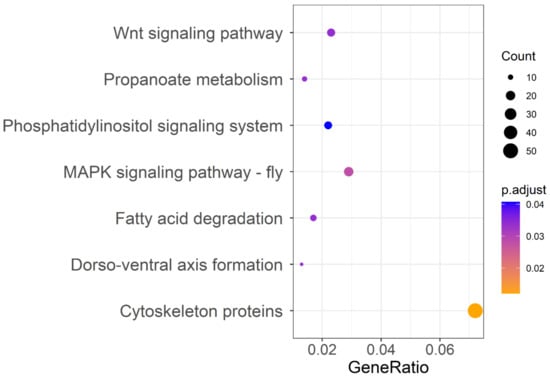
Figure 2.
KEGG pathway annotation classification of genes located in FST > 0.50 region of the Plutella xylostella genome of strains differing in susceptibility to chlorantraniliprole. The ordinate left is the KEGG classification, and the abscissa is the ratio of gene enrichment in the pathway. The size of the round spot represents the number of genes enriched in the pathway, and the color represents the size of the Padjust value.
In order to reduce false positives, we then calculated the ratio of nucleotide diversity (θπ (susceptible/resistant)). Using only those regions in the top 5% of both FST (>0.42) and top 5% of log2 (θπ (susceptible/resistant)) (>3.10) as the threshold, we identified 208 genes (Figure 3). Among 63 PSGs (FST > 0.50, Table S2), seven (LOC105381077, LOC105395328, LOC105386242, LOC105381169, LOC105394494, LOC105381143, and LOC105385997) were in the top 5% of the FST and log2 (θπ ratio. In particular, CYP6B6-like (LOC105395328) showed both high FST (0.65) and θπ ratio values (6.09) compared to neighboring regions, suggesting functional importance (Figure 4a). In resistant individuals of DBM (n = 6), we identified eight SNPs in the intragenic region of CYP6B6-like, including four intron variants (SNP48704, SNP48706, SNP48716, and SNP48729), one missense variant (SNP48968) and three synonymous variants (SNP48570, SNP48597, and SNP48999), which were absent in susceptible individuals (n = 6) (Figure 4b). These results indicate that these mutations of CYP6B6-like may be associated with insecticide resistance. Furthermore, the genomic sequence of CYP6B6-like was used as input in the DBM-DB blastn query (http://iae.fafu.edu.cn/DBM, accessed on 10 July 2020), and the result showed that CYP6B6-like was the gene CYP6BG5 (Gene ID: Px014216) described in Yu et al. [], which is over-expressed in chlorpyrifos- and fipronil-resistant strains of DBM.
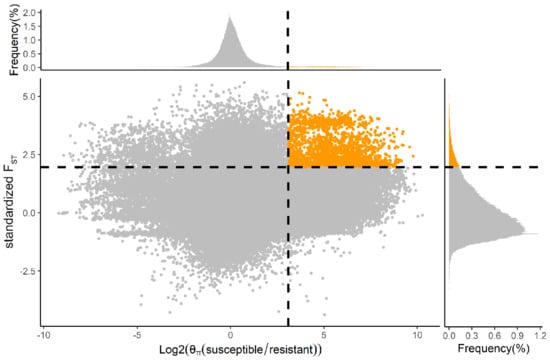
Figure 3.
Distribution of FST and θπ ratio (θπ (susceptible/resistant)) across, respectively, the X and Y axes with associated frequency plots. The top-right corner where orange points are located represents the genome region under strong selection pressure for insecticide. The horizontal and vertical gray dashed lines represent the top 5% value of standardized FST (1.96, FST = 0.42) and log2 (θπ (susceptible/resistant)) (3.10, θπ (susceptible/resistant) = 8.56). The FST value in the figure is standardized using scale () function in R.
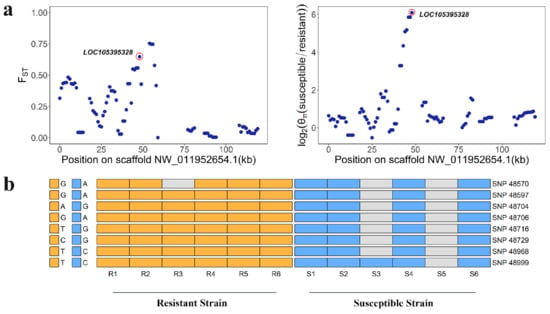
Figure 4.
CYP6B6-like (LOC105395328) shows different genetic signatures between resistant-strain and susceptible-strain Plutella xylostella. (a) FST and log2 (θπ (susceptible/resistant)) plot around the CYP6B6-like locus. The FST and log2 (θπ (susceptible/resistant)) value of CYP6B6-like is almost highest for scaffold NW_011952654.1, circled in red. (b) The eight SNPs were identified in resistant strains of Plutella xylostella and absent in susceptible strains of Plutella xylostella. SNPs were named according to their position on the scaffold. The gray cell represents data missing.
To further analyze the effect of mutation on the CYP6B6-like protein, we focused on the missense variant (SNP48968). SNP48968 caused an amino acid mutation (A486V) at the protein level (Figure S1). Sequence alignments show that CYP6B6-like has five conserved motifs common to insect CYP6s: The WxxxR motif, the GxE/DTT/S motif, the ExLR motif, the PxxFxPE/DRF motif, and the PFxxGxRxCxG/A motif (Figure S2). The three-dimensional structure of CYP6B6-like (Figure S3) was then predicted using I-TASSER (C-score = 0.56, TM-score = 0.79 ± 0.09). The quality assessment of VERIFY, ProQ, and ModFOLD indicated that the resulting model is of good quality. The missense mutation A486V of CYP6B6-like is not in the conserved region, but in the highly variable region adjacent to the SRS6 (SRS: Substrate recognition site) (Figures S2 and S3).
2.3. Differentially Expressed Genes (DEGs) and KEGG Enrichment Analysis
To investigate the effects of acute insecticide exposure (‘treatment’) and directional selection via insecticide tolerance (‘strain’) on the gene expression level of DBM, four pairwise comparisons were carried out between libraries to determine the DEGs (RT vs. T, RCK vs. CK, RT vs. RCK, and T vs. CK, with samples described in Table 2). The normalized RNA-Seq expression data were used to perform principal component analysis, and PC1, PC2, and PC3 explained 91.53%, 3.83%, and 2.25% of the total variation, respectively (Figure 1d). Primarily, the 12 individuals were divided into two main groups, which matched the resistant and susceptible populations. Secondarily, for the susceptible strain of DBM, individuals also clustered by treatment, while individuals representing the resistant strain of DBM did not show significant clustering by treatment. For further interpretation, genes with |log2FoldChange| > 1.5 and Padjust < 0.05 were recognized as differentially expressed. Our results showed that there were 518 (365 up-regulated and 153 down-regulated, CK vs. RCK), 615 (453 up-regulated and 162 down-regulated, T vs. RT), 52 (47 up-regulated and 5 down-regulated, RT vs. RCK), and 221 (94 up-regulated and 127 down-regulated, T vs. CK) DEGs by comparison of different strains or treatments (Figure S4). We identified 117 genes related to insecticide resistance from four pairwise comparisons based on previous research (Table S3), and the top 10 significantly expressed genes are listed in Table S4. Except for DEGs in the comparison of insecticide-treated susceptible DBM with control susceptible DBM, the majority involved up-regulation, including the cytochrome P450s, acetylcholinesterase, ion channels, and cuticle proteins. The overlap of DEGs between treatments and strains was analyzed and is displayed in Venn diagrams (Figure S5). Only three genes were differentially expressed (DE) after acute insecticide exposure both in the susceptible and the resistant strain, while 201 common genes were DE between strains with or without exposure to the insecticides. Coupled with the expression pattern of DE P450s, which showed an obvious expression difference between strains instead of treatments (Figure S6), these data indicate a stronger response to long-time directional selection for the susceptible strain when comparing exposure to non-exposure, which was consistent with the instability of insecticide-resistance that the resistant level of insects would gradually reduce once without pesticide pressure. Further analysis of the DEGs between strains by association with known KEGG pathways categorized DEGs into 33 groups (Figure 5), including pathways related to metabolic regulation, exogenous detoxification (cytochrome P450, ABC transporter), and immune response (Toll and Imd signaling).

Table 2.
Sample information of Plutella xylostella RNA-Seq.
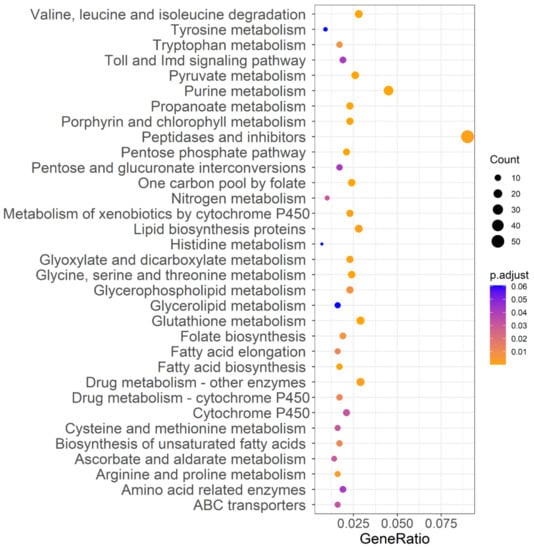
Figure 5.
KEGG pathway annotation classification of DEGs between strains. The ordinate left is the KEGG pathway, and the abscissa is the ratio of genes enriched in the pathway. The size of the round spot represents the number of genes enriched in the pathway, and the color represents the size of Padjust value.
To study whether the expression level of genes under positive selection (FST > 0.50) would be different between strains or between treatments, we investigated which genes with high FST were also listed as DEGs (Table S5). In total, 7.3% of PSGs were also DE between treatment or between strains, with more PSGs DE between strains. Among these candidate genes, we identified eight pesticide-resistance related genes, including cytochrome P450s, ubiquitin-protein ligase, and chymotrypsin (LOC105396609, LOC105395328, LOC105388375, LOC105382116, LOC105387005, LOC105397691, LOC105397997, and LOC105386494).
2.4. Identification of Hub Genes in Significant Modules Related to Resistance
Transcript data from all 12 samples from two strains of DBM (susceptible and resistant) were used to construct the module that consists of functionally related genes of a similar expression profile, according to the weighted gene co-expression network analysis (WGCNA) algorithm (Figure 6a). In the present study, the power of β = 6 was selected as the soft threshold to ensure a scale-free network (Figure 6b), and 16 modules were identified (Figure S7). Of these modules, the module eigengenes (ME) of the “magenta” and “darkred” module exhibited the high correlation with pesticide resistance (magenta: r = 0.94, P = 6 × 10−6; darkred: r = −0.79, P = 0.002) (Figure 6c). Thus, the two modules were considered the most meaningful module of co-expressed genes related to resistance. The set of genes was then further explored for functional enrichment analysis and the identification of key genes. A total of 5383 genes in the two modules were enriched for particular exogenous detoxification pathways including cytochrome P450 and ABC transporter, again highlighting the importance of metabolic resistance in chlorantraniliprole resistance (Figure 6d). Module membership vs. gene significance (MM vs. GS) in the two modules is highly correlated (Figures S8 and S9), illustrating that genes highly significantly associated with resistance are often also the most important elements of the significant module. We detect hub genes with GS > 0.80 and MM > 0.80 and show the expression pattern of the top 10 hub genes in “magenta” (Figure 7a) and “darkred” modules (Figure 7b). All of these hub genes show significant expression differences between strains, while showing no change in expression to chlorantraniliprole-treatment (Figure 7). In particular, there are two P450s, CYP6k1-like (LOC105386477) and CYP6B6-like (LOC105395328), in the two groups of hub genes showing a high expression level in the resistant insects, suggesting their important function in insecticide resistance.
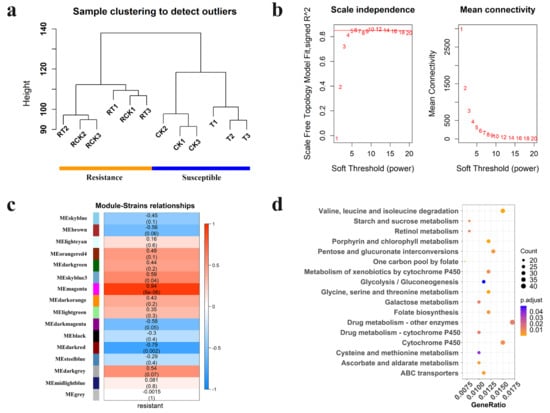
Figure 6.
Weighted gene co-expression network analysis of 12 samples from two strains of Plutella xylostella. (a) Clustering dendrogram of 12 samples and the trait (resistance). RCK, Resistant stain without treatment by chlorantraniliprole (LC50); RT, Resistant stain after treated with chlorantraniliprole (LC50); CK, Susceptible strain without exposure to any insecticide; and T, Susceptible strain exposed to chlorantraniliprole (LC50). (b) Analysis of the scale-free fit index for various soft-thresholding powers (β). (c) Heatmap of the correlation between module eigengenes and chlorantraniliprole-resistance (the color from blue to red (−1→1) represents the relationship between module and trait). Each row corresponds to a module eigengene, and each column to a trait. Each cell contains the corresponding correlation and p-value. (d) KEGG pathway annotation classification of genes in “magenta” and “darkred” module. The ordinate left is the KEGG pathway, and the abscissa is the ratio of genes enriched in the pathway. The size of the round spot represents the number of genes enriched in the pathway, and the color represents the size of Padjust value.
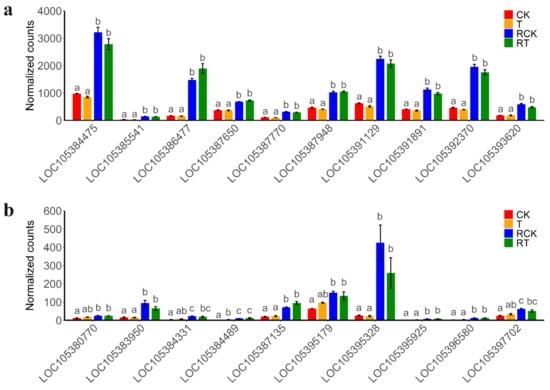
Figure 7.
The expression pattern of hub genes in “magenta” (a) and “darkred” (b) modules. Bars with different colors indicated different strains and insecticide-treatment (shown in Table 2). Values sharing the different letters are significantly different at P < 0.05 (one-way ANOVA followed by Duncan’s multiple comparison tests, P < 0.05). LOC105392370: Pyrroline-5-carboxylate reductase; LOC105391891: Adenylosuccinate lyase; LOC105387948: Probable methylcrotonoyl-CoA carboxylase beta chain, mitochondrial; LOC105385541: Uncharacterized; LOC105384475: Trifunctional purine biosynthetic protein adenosine-3; LOC105391129: Trifunctional purine biosynthetic protein adenosine-3-like; LOC105387770: Growth-blocking peptide, long form-like; LOC105386477: Cytochrome P450 6k1-like; LOC105393620: Senecionine N-oxygenase-like; LOC105387650: Cytochrome b5; LOC105380770: Tudor domain-containing protein 1; LOC105383950: Scidic amino acid decarboxylase GADL1-like; LOC105384331: Uncharacterized; LOC105384489: Putative inorganic phosphate cotransporter; LOC105387135: Lactase-like protein; LOC105395179: Adrenodoxin-like; LOC105395328: Cytochrome P450 6B6-like; LOC105395925: Juvenile hormone epoxide hydrolase-like; LOC105396580: Uncharacterized; LOC105397702: Phosphoglucomutase-2-like.
2.5. Knock-Down of CYP6B6-like Enhances the Sensibility to Chlorantraniliprole
Considering the strong selective signature, constitutive overexpression, and identification as a hub gene of CYP6B6-like, we focus the analysis on it. In addition to the coding region mutation as mentioned above, several SNPs were found in the 2000 nt regions upstream of CYP6B6-like (Figure 8a), and potential transcription factor binding sites at these regulation regions of CYP6B6-like were predicted using Jaspar (http://jaspar.genereg.net/, accessed on 20 September 2020). Interestingly, these SNPs landed at two predicted transcription factor binding sites, which were in the upstream fragment of the CYP6B6-like (NW_011952654.1:45630-47636 and NW_011952654.1:45641-45646). Furthermore, our RNA-seq data show a significantly higher expression level in the resistant DBM compared to the susceptible DBM (Figure 8b). These findings suggest that genetic divergence resulting from artificial selection for chlorantraniliprole resistance in DBM may influence the expression of CYP6B6-like to some extent. Therefore, to verify its expression level is indeed linked to chlorantraniliprole resistance, we next knocked down CYP6B6-like by feeding dsRNA to third-instar larvae. The expression level of CYP6B6-like decreased by 89% after 24 h feeding of dsCYP6B6 compared with the control (Figure 8c). We then used chlorantraniliprole (0.042 mg/L) to treat the RNAi individuals. The mortality rates of dsCYP6B6-feeding larvae were higher than those of the control group fed with the elution solution (Figure 8d and Figure S10) (Student t test, P < 0.01 in 72 h). Taking all evidence together, these results demonstrate that CYP6B6-like plays a critical role in chlorantraniliprole resistance.
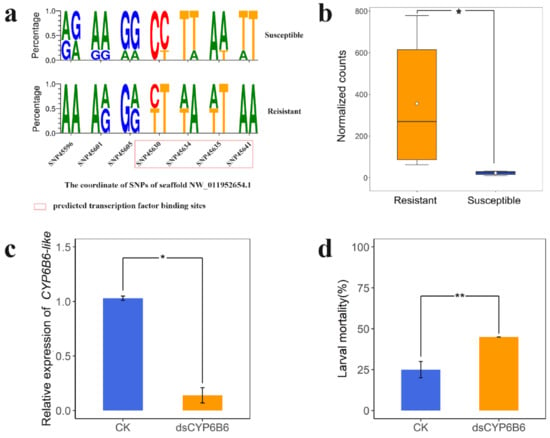
Figure 8.
Functional analysis of CYP6B6-like (LOC105395328). (a) The SeqLogo plots show the diploid genotypes’ frequency of potential transcriptional regulatory region of CYP6B6-like (2000 nt regions upstream of gene), which were significantly different between resistant strains and the susceptible strain. (b) The expression levels of CYP6B6-like in susceptible and resistant strains. Normalized counts: Counts normalized with DEseq2. * indicates significant influences (Student’s t test, P = 0.026, n = 6). (c) The expression levels of the CYP6B6-like were significantly reduced after 24 h dsRNA feeding. * indicates significant influences (Student’s t test, P = 0.037, n = 3). CK indicates individuals fed with elution solution. Bars indicate ± SD. (d) The mortality rate of Plutella xylostella third instar larvae treated with LC50 concentrations of chlorantraniliprole (0.042 mg/L) after 72 h dsRNA feeding. ** indicates significant influences (Student’s t test, P = 0.0023, n = 3). Twenty individuals were used for each group. Bars indicate ± SD.
3. Discussion
Insect resistance to pesticides is an urgent problem in integrated pest management. In the present study, we compared whole genome data of DBM from a chlorantraniliprole-resistant strain with a chlorantraniliprole-susceptible strain and identified (a) lower nucleotide diversity in the resistant strain, consistent with directional selection, and (b) several PSGs related to insecticide resistance using a population differentiation index (FST). From this comparison, several detoxification enzyme genes, pesticide target genes, digestive enzyme genes, ubiquitin-protein ligase, and cuticle protein genes were found to be subject to positive selection. After further correction for false positives using the nucleotide diversity ratio (θπ ratio, θπ (susceptible/resistant)), CYP6B6-like (LOC105395328) was identified as showing among the highest differentiation between resistant strains and susceptible strains, indicating it was under strong selective pressure. Transcriptome results also indicated that expression of many resistance genes was upregulated in the resistant strain and DEGs were enriched in pathways related to the detoxification metabolism.
The KEGG pathway analysis of PSGs filtered with FST > 0.50 demonstrated that many candidate genes were enriched in the mitogen-activated protein kinase (MAPK) signaling pathway. The MAPKs regulate various cellular programs in response to extracellular signals, and have many endogenous and exogenous substrates []. The MAPK-signaling pathway is involved in the drug resistance of cancer cells [,,], a crucial factor of which is the reactivation of the MAPK-signaling pathway [,]. Although studies on the role of the MAPK signaling pathway in insecticide resistance are few, some exist. For example, the MAPK p38 pathway has been implicated in insect defense against Bt Cry toxins []. Furthermore, another study indicates that the MAPK signaling pathway confers Bacillus thuringiensis Cry1Ac resistance by altering the expression of midgut ALP and ABCC genes []. Additionally, recent research has revealed the role of the MAPK-signaling pathway in the regulation of P450-mediated insecticide resistance []. Our findings further confirm the potential role of the MAPK signaling pathway in chlorantraniliprole insecticide resistance, and also specify P450-mediated insecticide resistance. Similarly, studies have indicated the role of the Wnt signaling pathway in the chemoresistance of numerous cancer types by regulating the expression of resistance genes [,,]. More specific to insecticide resistance, studies have shown differential expression of genes associated with the Wnt signaling pathway in chlorantraniliprole- and flubendiamide-resistant DBM [,]. The present GWSS analysis provides valuable information at the genome level regarding the role of signal transduction in the development of chlorantraniliprole resistance, although biological functions of the pathways linked to insecticide resistance need to be confirmed experimentally and regulation mechanisms in resistant strains also need further elucidation. Our results from the transcriptomic analysis show that DEGs are enriched in multiple pathways associated with resistance, including the metabolism of xenobiotics by cytochrome P450, drug metabolism of other enzymes, drug metabolism of cytochrome P450, ABC transporters, and glutathione metabolism. Detoxification metabolism to exogenous toxic substances is the most commonly described mechanism of insecticide resistance [,]. Strong correlations between the detoxifying enzyme activity and chlorantraniliprole resistance have been confirmed [], and several studies have indeed also observed their overexpression in resistant DBM and verified the role of some detoxifying enzymes in chlorantraniliprole resistance of DBM by RNAi [,,]. These findings are consistent with our results of selective-sweep analysis and transcriptome analysis (Figure 5, Tables S1–S3) by pointing to the same pathways involved in pesticide resistance.
Among these detoxifying enzymes above, the genetic variation and expression difference of P450s are particularly prominent in the present study. P450s play an important role in endogenous and exogenous substance metabolism, the latter including drugs, pesticides, and plant toxins []. P450s in DBM are distributed in four main groups including CYP2, CYP3, CYP4, and a mitochondrial cluster. Members of the CYP3, mainly CYP6s and CYP9s, are shown to participate in xenobiotics metabolism and insecticide resistance, and members of the CYP4 clan are known to be induced by xenobiotics []. Our study comes to similar conclusions; the positively selected P450s and differentially expressed P450s are mainly CYP6s, CYP9s, and CYP4s (Tables S2 and S3). The transcriptional up-regulation of P450 genes and metabolic capacities of P450 proteins were regarded as important mechanisms of enhanced metabolic detoxification to insecticide []. At the transcript level, both the P450 constitutive overexpression caused by mutations in promoter sequence and the inducible expression caused by mutations in trans-acting factors or by their signaling cascades have been specifically associated with insecticide resistance []. At the protein level, mutations in catalytic site residues (SRS1, SRS4, SRS5, and SRS6), substrate access channel residues (SRS2 and SRS3), proximal surface residues, and interacting partners may account for the enhanced metabolic capacities of P450s []. In DBM, the overexpression of CYP6BG1 in resistant strains has been reported to be involved in chlorantraniliprole resistance []. In the present study, we not only also observed overexpression of CYP6BG1 (named CYP6k1-like, LOC105386477) in resistant DBM but also identified it as the hub gene in a significant module of co-expression associated with resistance (Figure 7a). In other lepidoptera, such as Chilo suppressalis [] and Spodoptera exigua [], the constitutive overexpression of multiple P450s in chlorantraniliprole-resistance strains has been confirmed. In our study, the expression pattern of P450s also shows constitutive overexpression of P450s in resistant strains (Figure S6), and we suggest the potential role of these overexpressed P450s in resistance to chlorantraniliprole, because their overexpression has been reported in chlorpyrifos- and fiprinol-, and chlorantraniliprole-resistance in DBM. However, while acute chlorantraniliprole-treatment does not induce increased expression of P450s, it could be connected to the insect growth stage, insecticide exposure time, insecticide type, and insecticide concentration []. These enzymes may not be sensitive to the inducer or do not have enough time to control their biosynthesis and respond to insecticide.
Combined with the result of FST and θπ, we found that a P450 gene, CYP6B6-like (LOC105395328), was under strong positive selection and had a high genetic divergence between resistant and susceptible DBM strains. As a member of CYP6s, it may participate in the metabolic detoxication of insecticide. A previous study showed that CYP6B6-like, also named CYP6BG5, was transcriptionally up-regulated in the chlorpyrifos- and fiprinol-resistance strains compared to susceptible strains []. Although the target site of chlorantraniliprole is different from chlorpyrifos and fiprinol, cross-resistance in insects is a common phenomenon as metabolic mechanisms of resistance could confer broad-spectrum resistance []. Moreover, CYP6B6-like shares a high identity with CYP6BG1, which has confirmed involvement in chlorantraniliprole resistance in DBM. Interestingly, it was also predicted as one of the target genes of 11 lncRNAs linked to chlorantraniliprole resistance in DBM []. Taken together, these data may be regarded as evidence that CYP6B6-like is involved in the detoxification metabolism of chlorantraniliprole. Furthermore, we uncovered a missense mutation in the resistant strain, causing an amino acid mutation A486V, which is located in a highly variable region near the SRS6. SRSs are reported to be located in regions of extensive sequence variation, and even a single amino acid mutation in the SRS regions could affect metabolic capacity [,]. For example, a variant in SRS6 (A484V) of CYP6B3 has been reported to force substrates to bind closer to the reactive oxygen on heme []. However, catalytic properties are not only associated with SRSs. Three point mutations in CYP6A2, located between SRS4 and SRS5 (R335S and L336V) and before SRS6 (V476L), are reported to have a prominent role in DDT-insecticide resistance []. In our study, the missense mutation A486V is located before SRS6, but its potential role in chlorantraniliprole resistance needs further study.
Alongside the possible change of CYP6B6-like protein function in the resistant strain due to directional selection, we also observed its overexpression in resistant strains, which may be associated with mutations in the transcriptional regulatory region (2000 nt upstream region of gene). As mentioned above, the promoter variations of P450s are related to constitutive overexpression []. For example, some transposon insertions of P450s are reported to be associated with insecticide resistance in Drosophila melanogaster [,]. Here, several substitutions (SNP45630, SNP45634, SNP45635, and SNP45641) were observed to be located at two predicted transcription factor binding sites (NW_011952654.1:45630-45636 and NW_011952654.1:45641-45646) (Figure 8a). However, the specific role of these predicted transcription factors in the transcriptional regulation of P450s needs further confirmation (Table S6).
Alongside several P450s, we also identified several other genes with strong signs of selective sweeps (Table S2). This finding is not entirely surprising, since metabolism resistance is a complex physiological process involving a number of enzymes and transporters. P450s, esterases, and UDP–glycosyltransferases (UGTs) are three critical enzyme categories participating in the metabolism of xenobiotics and ABC transporters transfer and excrete their products [,]. In DBM, overexpression of UGT2B17 has been reported to be involved in chlorantraniliprole resistance []. The contribution of ABC transporters to chloramphenicol resistance is rarely reported, but a recent study revealed that ABC transporters were involved in the transport of chlorantraniliprole []. Interestingly, the overexpression of P450s and ABC transporters is also known to change the thickness and composition of i”sect’cuticles [,,], which may also be caused by the overexpression of cuticle proteins [,]. The insect cuticle is a critical determinant of insecticide resistance by reducing insecticide penetration, which has been reported in various insects [,,], though studies on this aspect are fewer than those that support metabolism resistance. It also appears that proteasomes are associated with the response to insecticides, because they increased energy and amino acid production [,]. In addition, trypsin has been confirmed to catalyze the degradation of deltamethrin [], which may lead to a deeper understanding of the role of proteasomes in insecticide resistance. These studies mentioned above are in agreement with the possible role of candidate genes found in our study in the development of chlorantraniliprole resistance.
In summary, we provide insights into the genomic basis of chlorantraniliprole resistance of DBM. The abundance of information from genome and transcriptome analysis could facilitate further investigations of the detailed resistance mechanisms of DBM against chlorantraniliprole.
4. Materials and Methods
4.1. Insect Sample and Treatment
The susceptible strain (DBM-S) was initially purchased from the Pilot-Scale Base of Bio-Pesticides, Institute of Zoology, Chinese Academy of Sciences in 2014, and was maintained in the laboratory for 5 years without exposure to any insecticide. The resistant strain (DBM-R) was collected in 2017 from a field at Fuluo, Guangzhou, China. Resistant insects used for sequencing were from a near-isogenic line (NIL) constructed with the backcross method (Figure 1a), which referred to Zhu et al. []. Specifically, the resistant males (DBM-R) were crossed with susceptible females (DBM-S), and their offspring were reared on fresh cabbage leaves with a discriminating dose of chlorantraniliprole, resulting in 40–70% mortality. The surviving larvae were reared on chlorantraniliprole cabbage leaves to pupae, and then surviving males were backcrossed with DBM-S females, and the same selection was performed for their offspring using a discriminating dose of chlorantraniliprole. The backcross and the selection process were repeated for six generations to obtain the NIL resistant to chlorantraniliprole (NIL-R).
The toxicity of chlorantraniliprole to DBM-S and NIL-R (LC50) was determined by the leaf dipping method. Cabbage leaves measuring 5 cm × 5 cm were immersed for 10 s in different concentrations of chlorantraniliprole diluted with distilled water. We tested 8 concentrations using two-fold serial dilutions. The range of tested concentrations for susceptible and resistant strains was 0.003125–0.4 mg and 0.78125–100 mg/L, respectively. The leaves were then air-dried for 0.5 h and then placed individually into a Petri dish with filter paper. A total of 20 third instars of different populations were then put into the Petri dishes (with three replications). We recorded the mortality of insects 96 h after treatment. Larvae that did not move when pushed gently with a brush were considered dead. LC50 of DBM-R and NIL-R populations were analyzed by probit analysis using POLOPLUS 2.0 (LeOra Software, Petaluma, CA, USA). The resistance ratio (RR) was determined as the ratio of the NIL-R LC50/DBM-S LC50. Finally, DBM-S and NIL-R were used for DNA extraction, where the whole body of larvae was a sample, with six samples for each strain.
Subsequently, DBM-S and NIL-R were divided into two groups. One group (treatment) was reared with cabbage leaves treated with an LC50 concentration of chlorantraniliprole, whereas the cabbage leaves in the other group (control) were treated with water. The whole body of surviving larvae was used for RNA extraction, with three replicates per treatment.
4.2. Sequencing and Library Preparation
Genomic DNA was extracted using TIANGEN Magnetic Universal Genomic DNA Kit. Sequencing libraries were generated using NEB Next® Ultra DNA Library Prep Kit for Illumina® (NEB, Ipswich, MA, USA) following the manufacturer’s recommendations and sequenced on an Illumina Hiseq 4000 platform. We generated 131.03 G of paired-end reads of 150 bp length (Table S7).
RNA was extracted using the TRIzol (Ambion, Austin, TX, USA) extraction method. The mRNA Sequencing libraries were generated using the NEB Next® Ultra™ RNA Library Prep Kit for Illumina® (NEB, Ipswich, MA, USA) following the manufacturer’s recommendations, and sequenced on an Illumina NovaSeq 6000 platform. We generated 112.93 G of paired-end reads of 150 bp length (Table S8).
4.3. Read Alignment and SNP Calling
Because the quality of the NGS data is very important for the downstream sequence analysis, we filtered low-quality reads using the NGS QC Tool kit (v2.3.3) with default parameters []. The clean data were aligned using BWA-MEM (v0.7.15) to the P. xylostella reference genome (RefSeq accession: GCF_000330985.1). Sequence Alignment/Map (SAM) format files were dealt with using Picard tools SortSam (v2.2.4) for sorting, outputting them as Binary sequence Alignment/Map (BAM) format files. Duplicate reads were removed from individual sample alignments using Picard tools MarkDuplicates (v2.2.4) [].
Before SNP calling, the Genome Analysis ToolKit (v3.6), RealignerTargetCreator, and IndelRealigner were used for global realignment. SNPs were called using GATK UnifiedGenotyper with the parameters min_base_quality_score of 20, stand_call_conf of 30, and stand_emit_conf of 30. Then, GATK VariantFiltration was used to filter the unconfident variant sites, with the filter (a) QUAL < 30.0; (b) QD < 5.0; (c) FS > 60.0; (d) MQ < 40.0; (e) MQRankSum < −12.5; (f) ReadPosRankSum < −8.0.
4.4. Selective-Sweep Analysis
To detect PSGs related to insecticide resistance to chlorantraniliprole, we scanned the genome for regions with a population differentiation index (FST) and nucleotide diversity (θπ) ratio. FST and nucleotide diversity (θπ) were calculated with VCFtools (v0.1.13) using a 5 kb window with a 1 kb step. The negative and missing FST values were discarded, because these values have no biological interpretation []. The θπ ratio was calculated as θπ (susceptible)/θπ (resistant).
4.5. Sequence Alignment and 3D Modeling of CYP6B6-like
The multiple alignments were constructed with MEGA-X [] and Jalview (v2.11.1.0) []. 3D models of CYP6B6-like were generated by I-TASSER [,,] and model quality was assessed with Verify 3D, ProQ, and ModFOLD [,].
4.6. RNA Interference
The cDNA fragments of CYP6B6-like (LOC105395328) were amplified by RT-PCR using the gene-specific primers conjugated with 20 bases of the T7 RNA polymerase promoter listed in Table S9. The PCR products were used as templates for double-stranded RNA (dsRNA) synthesis using a MEGAscript RNAi kit (Ambion Inc., Vilnius, Lithuania) following the manufacturer’s instructions. The synthesized dsRNA was dissolved in elution solution, quantified with a NanoDrop2000 (Thermo Scientific, Wilmington, DE, USA), and stored at −20 °C until use. Third-instars P. xylostella larvae were individually placed in a Petri dish to avoid cannibalism, starved for 20 h, and then fed with a 5 mm × 5 mm cabbage leaf disc. Then, 500 ng dsCYP6B6 (dissolved in elution solution, provided in the RNAi kit) or the same volume of elution solution only were distributed to the cabbage leaf discs. After 24 h, the larvae that had completely eaten the cabbage leaf discs were chosen for the subsequent experiments. A total of 10 larvae were randomly collected after another 24 h for total RNA isolation and qRT-PCR analysis of gene expression. The larvae fed with the elution solution were used as controls. The elution solution used in this study was used as a negative control because our preliminary experiments showed that the CYP6B6-like transcript level had no significant difference between being fed dsGFP and being fed the elution solution (Figure S11). The primers used for qRT-PCR are listed in Table S9.
To evaluate the effect of RNAi of CYP6B6-like on the susceptibility of P. xylostella to chlorantraniliprole, the larvae that had eaten dsRNA were raised on cabbage leaf discs treated with the LC50 of chlorantraniliprole, and mortality was recorded at 24 h, 48 h, 72 h, and 96 h post-treatment. Approximately 20 dsRNA-feeding larvae were tested in every three replicates.
4.7. RNA-Seq, Data Processing, and Differentially Expressed Gene (DEG) Analysis
To further identify whether the potential selective-sweep regions (FST > 0.50) could also affect gene expression, we compared the gene expression between the susceptible and resistant strains of DBM. Three susceptible and three resistant DBM samples were used for RNA-seq. Sample information is listed in Table 2.
We used fastp (v0.20.0) to filter out the bad reads and cut adapters with the default parameters []. After building a HISAT2 index using hisat2-build, we mapped the clean reads to the DBM reference genome using HISAT2 (v2.1.0) [], with the output files of the SAM format. Then, the SAM format files were imported to SAMtools for sorting and building index files, and the index of the fasta file of the reference genome was built with SAMtools (v1.9). We used the htseq-count tool to calculate the counts of the reads mapping to each gene [], These count data were used to determine gene expression variation by analyzing each resistant sample to a susceptible sample with DESeq2 (v1.24.0) [] in R (v3.6.1) []. Genes with log2FC > 1.5 and Padjust < 0.05 were considered to be upregulated after insecticide treatment, while those with log2FC > −1.5 and Padjust < 0.05 considered down-regulated genes. We plotted the volcano plots and a Venn diagram of differentially expressed genes related to insecticide resistance in DBM using ggplot2 (v3.2.1) and VennDiagram (v1.6.20) in R [,], respectively.
We used the clusterProfiler (v3.12.0) [], an R package, to determine significant Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways enriched within the DEG dataset using Padjust < 0.05 as a threshold.
4.8. Weighted Gene Co-Expression Network Analysis and Identification of Hub Genes in Significant Module
WGCNA is a systems biology method for identifying patterns that have relevance among genes in microarray samples. In the WGCNA network, interesting gene modules related to sample traits and key genes can be identified. In this study, the expression data of 12 samples were log-transformed using log2 (x + 1) and then used for WGCNA. The weighted co-expression network was constructed in accordance with the protocol of the WGCNA package in R []. We chose the power of 6, which is the lowest power for which the scale-free topology fit index reaches 0.90. We merged highly similar modules (correlation > 0.75) and obtained 16 modules. To identify modules that were significantly associated with the trait (resistance), we used module eigengenes (MEs) as principal components to correlate them with external traits and look for the most significant module. Gene Significance (GS) is defined as the correlation between the gene and the trait, and module membership (MM) is a measure of intra-modular connectivity. We identified hub genes as those genes with both a high Gene Significance (GS > 0.80) for resistance as well as a high Module Membership (MM > 0.80) in significant modules.
Supplementary Materials
The supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms232012245/s1.
Author Contributions
L.B. conceived the project and acquired the funding. B.Z. carried out the construction of susceptible and resistant insects. D.S. participated in study design and coordination. W.D. carried out the DNA and RNA sequencing data analysis, functional verification of the candidate gene, and wrote the manuscript. T.Z. helped sequencing data analysis. L.B., M.v.T., P.A., P.L. and H.U. helped to draft the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Beijing Agriculture Innovation Consortium (BAIC02-2022) and the National Natural Science Foundation of China (31971759).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The 12 DBMs used in whole-genome resequencing analysis and the 12 DBMs used in RNA-seq analysis are accessible at the National Center for Biotechnology Information (NCBI) under Bio-Project accession numbers PRJNA664309. The whole-genome resequencing data and the transcriptome data have been deposited in SRA under SRR12674258-SRR12674281.
Acknowledgments
We are grateful to Lujiang Qu (Department of Animal Genetics and Breeding, China Agricultural University, Beijing, China) for providing the data analysis platform for free.
Conflicts of Interest
The authors declare that they have no competing interests.
References
- Li, Z.; Feng, X.; Liu, S.-S.; You, M.; Furlong, M.J. Biology, Ecology, and Management of the Diamondback Moth in China. Annu. Rev. Èntomol. 2016, 61, 277–296. [Google Scholar] [CrossRef] [PubMed]
- Talekar, N.S.; Shelton, A.M. Biology, ecology, and management of the diamondback moth. Annu. Rev. Entomol. 1993, 38, 275–301. [Google Scholar] [CrossRef]
- Furlong, M.J.; Wright, D.J.; Dosdall, L.M. Diamondback Moth Ecology and Management: Problems, Progress, and Prospects. Annu. Rev. Èntomol. 2013, 58, 517–541. [Google Scholar] [CrossRef] [PubMed]
- Selby, T.P.; Lahm, G.P.; Stevenson, T.M. A retrospective look at anthranilic diamide insecticides: Discovery and lead optimization to chlorantraniliprole and cyantraniliprole. Pest Manag. Sci. 2016, 73, 658–665. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.D.; Chen, H.Y.; Li, Z.Y.; Zhang, D.Y.; Yin, F.; Lin, Q.S.; Bao, H.L.; Zhou, X.M.; Feng, X. Found a field population of diamondback moth, Plutella xylostella (L.), with high-level resistance to chlorantraniliprole in South China. Guangdong Agric. Science 2012, 39, 79–81. [Google Scholar] [CrossRef]
- Balabanidou, V.; Grigoraki, L.; Vontas, J. Insect cuticle: A critical determinant of insecticide resistance. Curr. Opin. Insect Sci. 2018, 27, 68–74. [Google Scholar] [CrossRef]
- Khan, S.; Uddin, M.N.; Rizwan, M.; Khan, W.; Farooq, M.; Shah, A.S.; Subhan, F.; Aziz, F.; Rahman, K.U.; Khan, A.; et al. Mechanism of Insecticide Resistance in Insects/Pests. Pol. J. Environ. Stud. 2020, 29, 2023–2030. [Google Scholar] [CrossRef]
- Troczka, B.; Zimmer, C.T.; Elias, J.; Schorn, C.; Bass, C.; Davies, T.E.; Field, L.M.; Williamson, M.S.; Slater, R.; Nauen, R. Resistance to diamide insecticides in diamondback moth, Plutella xylostella (Lepidoptera: Plutellidae) is associated with a mutation in the membrane-spanning domain of the ryanodine receptor. Insect Biochem. Mol. Biol. 2012, 42, 873–880. [Google Scholar] [CrossRef]
- Steinbach, D.; Gutbrod, O.; Lümmen, P.; Matthiesen, S.; Schorn, C.; Nauen, R. Geographic spread, genetics and functional characteristics of ryanodine receptor based target-site resistance to diamide insecticides in diamondback moth, Plutella xylostella. Insect Biochem. Mol. Biol. 2015, 63, 14–22. [Google Scholar] [CrossRef]
- Guo, L.; Wang, Y.; Zhou, X.G.; Li, Z.Y.; Liu, S.Z.; Pei, L.; Gao, X. Function alanalysis of a point mutation in the ryanodine receptor of Plutella xylostella (L.) associated with resistance to chlorantraniliprole. Pest Manag. Sci. 2014, 7, 1083–1089. [Google Scholar] [CrossRef]
- Guo, L.; Liang, P.; Zhou, X.; Gao, X. Novel mutations and mutation combinations of ryanodine receptor in a chlorantraniliprole resistant population of Plutella xylostella (L.). Sci. Rep. 2014, 4, 6924. [Google Scholar] [CrossRef] [PubMed]
- Etebari, K.; Afrad, M.H.; Tang, B.; Silva, R.; Furlong, M.J.; Asgari, S. Involvement of microRNA miR-2b-3p in regulation of metabolic resistance to insecticides in Plutella xylostella. Insect Mol. Biol. 2018, 27, 478–491. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhu, B.; Gao, X.; Liang, P. Over-expression of UDP-glycosyltransferase gene UGT2B17 is involved in chlorantraniliprole resistance in Plutella xylostella (L.). Pest Manag. Sci. 2017, 73, 1402–1409. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, R.; Zhu, B.; Gao, X.; Liang, P. Overexpression of cytochrome P450 CYP6BG1 may contribute to chlorantraniliprole resistance in Plutella xylostella (L.). Pest Manag. Sci. 2017, 74, 1386–1393. [Google Scholar] [CrossRef]
- Lin, Q.; Jin, F.; Hu, Z.; Chen, H.; Yin, F.; Li, Z.; Dong, X.; Zhang, D.; Ren, S.; Feng, X. Transcriptome Analysis of Chlorantraniliprole Resistance Development in the Diamondback Moth Plutella xylostella. PLoS ONE 2013, 8, e72314. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Q.; Wang, L.; Wang, K.; Yang, Y.; Ma, T.; Wang, Z.; Zhang, X.; Ni, Z.; Hou, F.; Long, R.; et al. Yak whole-genome resequencing reveals domestication signatures and prehistoric population expansions. Nat. Commun. 2015, 6, 10283. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Z.; Liu, E.; Liu, Z.; Kijas, J.W.; Zhu, C.; Hu, S.; Ma, X.; Zhang, L.; Du, L.; Wang, H.; et al. Selection signature analysis reveals genes associated with tail type in Chinese indigenous sheep. Anim. Genet. 2016, 48, 55–66. [Google Scholar] [CrossRef]
- Mei, C.; Wang, H.; Liao, Q.; Khan, R.; Raza, S.H.A.; Zhao, C.; Wang, H.; Cheng, G.; Tian, W.; Li, Y.; et al. Genome-wide analysis reveals the effects of artificial selection on production and meat quality traits in Qinchuan cattle. Genomics 2018, 111, 1201–1208. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; McParland, S.; Kearney, F.; Du, L.; Berry, D.P. Detection of selection signatures in dairy and beef cattle using high-density genomic information. Genet. Sel. Evol. 2015, 47, 49. [Google Scholar] [CrossRef] [PubMed]
- Bertolini, F.; Servin, B.; Talenti, A.; Rochat, E.; Kim, E.; Oget, C.; Palhiere, I.; Crisa, A.; Catillo, G.; Steri, R. Signatures of selection and environmental adaptation across the goat genome post-domestication. Genet. Sel. Evol. 2018, 50, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Bickhart, D.M.; Cole, J.B.; Schroeder, S.G.; Song, J.; Van Tassell, C.P.; Sonstegard, T.S.; Liu, G.E. Genomic Signatures Reveal New Evidences for Selection of Important Traits in Domestic Cattle. Mol. Biol. Evol. 2015, 32, 711–725. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Jia, Y.; Almeida, P.; Mank, J.E.; van Tuinen, M.; Wang, Q.; Jiang, Z.; Chen, Y.; Zhan, K.; Hou, S.; et al. Whole-genome resequencing reveals signatures of selection and timing of duck domestication. GigaScience 2018, 7, giy027. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Li, C.; Chen, H.; Li, R.; Chong, Q.; Xiao, H.; Chen, S. Genome-wide scan of selection signatures in Dehong humped cattle for heat tolerance and disease resistance. Anim. Genet. 2020, 51, 292–299. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Hanif, Q.; Cao, Y.; Yu, Y.; Lei, C.; Zhang, G.; Zhao, Y. Whole Genome Scan and Selection Signatures for Climate Adaption in Yanbian Cattle. Front. Genet. 2020, 11, 94. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.-S.; Elbeltagy, A.; Aboul-Naga, A.M.; Rischkowsky, B.; Sayre, B.; Mwacharo, J.M.; Rothschild, M.F. Multiple genomic signatures of selection in goats and sheep indigenous to a hot arid environment. Heredity 2015, 116, 255–264. [Google Scholar] [CrossRef] [PubMed]
- Dong, K.; Yao, N.; Pu, Y.; He, X.; Zhao, Q.; Luan, Y.; Guan, W.; Rao, S.; Ma, Y. Genomic Scan Reveals Loci under Altitude Adaptation in Tibetan and Dahe Pigs. PLoS ONE 2014, 9, e110520. [Google Scholar] [CrossRef] [PubMed]
- Steele, L.D.; Coates, B.; Valero, M.C.; Sun, W.; Seong, K.M.; Muir, W.M.; Clark, J.M.; Pittendrigh, B.R. Selective Sweep Analysis in the Genomes of the 91-R and 91-C Drosophila melanogaster Strains Reveals Few of the ‘Usual Suspects’ in Dichlorodiphenyltrichloroethane (DDT) Resistance. PLoS ONE 2015, 10, e0123066. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Calla, B.; Demkovich, M.; Siegel, J.P.; Viana, J.P.G.; Walden, K.K.O.; Robertson, H.M.; Berenbaum, M.R. Selective Sweeps in a Nutshell: The Genomic Footprint of Rapid Insecticide Resistance Evolution in the Almond Agroecosystem. Genome Biol. Evol. 2020, 13, evaa234. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Tang, W.; He, W.; Ma, X.; Vasseur, L.; Baxter, S.W.; Yang, G.; Huang, S.; Song, F.; You, M. Characterization and expression of the cytochrome P450 gene family in diamondback moth, Plutella xylostella (L.). Sci. Rep. 2015, 5, srep08952. [Google Scholar] [CrossRef] [PubMed]
- Cargnello, M.; Roux, P.P. Activation and Function of the MAPKs and Their Substrates, the MAPK-Activated Protein Kinases. Microbiol. Mol. Biol. Rev. 2011, 75, 50–83. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Lu, Y.; Chen, Y.; Cheng, J.; Zhang, W. Transcripts 202 and 205 of IL-6 confer resistance to Vemurafenib by reactivating the MAPK pathway in BRAF(V600E) mutant melanoma cells. Exp. Cell Res. 2020, 390, 111942. [Google Scholar] [CrossRef]
- Liao, X.; Gao, Y.; Sun, L.; Liu, J.; Chen, H.; Yu, L.; Chen, Z.; Chen, W.; Lin, L. Rosmarinic acid reverses non-small cell lung cancer cisplatin resistance by activating the MAPK signaling pathway. Phytother. Res. 2020, 34, 1142–1153. [Google Scholar] [CrossRef] [PubMed]
- Kuroshima, K.; Yoshino, H.; Okamura, S.; Tsuruda, M.; Osako, Y.; Sakaguchi, T.; Sugita, S.; Tatarano, S.; Nakagawa, M.; Enokida, H. Potential new therapy of Rapalink-1, a new generation mammalian target of rapamycin inhibitor, against sunitinib-resistant renal cell carcinoma. Cancer Sci. 2020, 111, 1607–1618. [Google Scholar] [CrossRef]
- Corcoran, R.B.; Ebi, H.; Turke, A.B.; Coffee, E.M.; Nishino, M.; Cogdill, A.P.; Brown, R.D.; Della Pelle, P.; Dias-Santagata, D.; Hung, K.E.; et al. EGFR-Mediated Reactivation of MAPK Signaling Contributes to Insensitivity of BRAF-Mutant Colorectal Cancers to RAF Inhibition with Vemurafenib. Cancer Discov. 2012, 2, 227–235. [Google Scholar] [CrossRef] [PubMed]
- Ahronian, L.G.; Sennott, E.M.; Van Allen, E.M.; Wagle, N.; Kwak, E.L.; Faris, J.E.; Godfrey, J.T.; Nishimura, K.; Lynch, K.D.; Mermel, C.H.; et al. Clinical Acquired Resistance to RAF Inhibitor Combinations in BRAF-Mutant Colorectal Cancer through MAPK Pathway Alterations. Cancer Discov. 2015, 5, 358–367. [Google Scholar] [CrossRef] [PubMed]
- Cancino-Rodezno, A.; Alexander, C.; Villaseñor, R.; Pacheco, S.; Porta, H.; Pauchet, Y.; Soberón, M.; Gill, S.S.; Bravo, A. The mitogen-activated protein kinase p38 is involved in insect defense against Cry toxins from Bacillus thuringiensis. Insect Biochem. Mol. Biol. 2010, 40, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Kang, S.; Chen, D.; Wu, Q.; Wang, S.; Xie, W.; Zhu, X.; Baxter, S.W.; Zhou, X.; Jurat-Fuentes, J.L.; et al. MAPK Signaling Pathway Alters Expression of Midgut ALP and ABCC Genes and Causes Resistance to Bacillus thuringiensis Cry1Ac Toxin in Diamondback Moth. PLoS Genet. 2015, 11, e1005124. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Deng, S.; Wei, X.; Yang, J.; Zhao, Q.; Yin, C.; Du, T.; Guo, Z.; Xia, J.; Yang, Z.; et al. MAPK-directed activation of the whitefly transcription factor CREB leads to P450-mediated imidacloprid resistance. Proc. Natl. Acad. Sci. USA 2020, 117, 10246–10253. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Zhu, H.; Zhou, S.; Wu, T.; Wu, H.; Yang, H.; Mao, H.; SekharKathera, C.; Janardhan, A.; Edick, A.; et al. Wnt pathway is involved in 5-FU drug resistance of colorectal cancer cells. Exp. Mol. Med. 2018, 50, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Wang, J.; Bu, X.; Yang, B.; Wang, B.; Hu, S.; Yan, Z.; Gao, Y.; Han, S.; Qu, M. Resveratrol restores sensitivity of glioma cells to temozolamide through inhibiting the activation of Wnt signaling pathway. J. Cell. Physiol. 2018, 234, 6783–6800. [Google Scholar] [CrossRef]
- Yamamoto, T.M.; McMellen, A.; Watson, Z.L.; Aguilera, J.; Ferguson, R.; Nurmemmedov, E.; Thakar, T.; Moldovan, G.L.; Kim, H.; Cittelly, D.M.; et al. Activation of Wnt signaling promotes olaparib resistant ovarian cancer. Mol. Carcinog. 2019, 58, 1770–1782. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.H.; Sun, S.Q.; Xu, J.L.; Zhao, X.L.; Xue, C.B. Differential expressed genes and their pathways of the resistance to flubendiamide in Plutella xylostella. Sci. Agric. Sin. 2018, 51, 2106–2115. [Google Scholar] [CrossRef]
- Hemingway, J.; Hawkes, N.J.; McCarroll, L.; Ranson, H. The molecular basis of insecticide resistance in mosquitoes. Insect Biochem. Mol. Biol. 2004, 34, 653–665. [Google Scholar] [CrossRef]
- Liu, N.N.; Zhu, F.; Xu, Q.; Pridgeon, J.W.; Gao, X.W. Behavioral change, physiological modification, and metabolic detoxification: Mechanisms of insecticide resistance. Acta Entomol. Sin. 2006, 49, 671–679. [Google Scholar]
- Hu, Z.-D.; Feng, X.; Lin, Q.-S.; Chen, H.-Y.; Li, Z.-Y.; Yin, F.; Liang, P.; Gao, X.-W. Biochemical Mechanism of Chlorantraniliprole Resistance in the Diamondback Moth, Plutella xylostella Linnaeus. J. Integr. Agric. 2014, 13, 2452–2459. [Google Scholar] [CrossRef]
- Hu, Z.; Lin, Q.; Chen, H.; Li, Z.; Yin, F.; Feng, X. Identification of a novel cytochrome P450 gene, CYP321E1from the diamondback moth, Plutella xylostella (L.) and RNA interference to evaluate its role in chlorantraniliprole resistance. Bull. Èntomol. Res. 2014, 104, 716–723. [Google Scholar] [CrossRef] [PubMed]
- Scott, J.G. Cytochromes P450 and insecticide resistance. Insect Biochem. Mol. Biol. 1999, 29, 757–777. [Google Scholar] [CrossRef]
- Schuler, M.A.; Berenbaum, M.R. Structure and Function of Cytochrome P450S in Insect Adaptation to Natural and Synthetic Toxins: Insights Gained from Molecular Modeling. J. Chem. Ecol. 2013, 39, 1232–1245. [Google Scholar] [CrossRef]
- Liu, N.; Li, M.; Gong, Y.; Liu, F.; Li, T. Cytochrome P450s—Their expression, regulation, and role in insecticide resistance. Pestic. Biochem. Physiol. 2015, 120, 77–81. [Google Scholar] [CrossRef]
- Xu, L.; Zhao, J.; Sun, Y.; Xu, D.; Xu, G.; Xu, X.; Zhang, Y.; Huang, S.; Han, Z.; Gu, Z. Constitutive overexpression of cytochrome P450 monooxygenase genes contributes to chlorantraniliprole resistance in Chilo suppressalis (Walker). Pest Manag. Sci. 2018, 75, 718–725. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Chen, Y.; Gong, C.; Yao, X.; Jiang, C.; Yang, Q. Molecular identification of four novel cytochrome P450 genes related to the development of resistance of Spodoptera exigua (Lepidoptera: Noctuidae) to chlorantraniliprole. Pest Manag. Sci. 2018, 74, 1938–1952. [Google Scholar] [CrossRef] [PubMed]
- Terriere, L. Induction of detoxication enzymes in insects. Annu. Rev. Entomol. 1984, 29, 71–88. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Xu, M.; Shi, H.; Gao, X.; Liang, P. Genome-wide identification of lncRNAs associated with chlorantraniliprole resistance in diamondback moth Plutella xylostella (L.). BMC Genom. 2017, 18, 380. [Google Scholar] [CrossRef] [PubMed]
- Gotoh, O. Substrate recognition sites in cytochrome P450 family 2 (CYP2) proteins inferred from comparative analyses of amino acid and coding nucleotide sequences. J. Biol. Chem. 1992, 267, 83–90. [Google Scholar] [CrossRef]
- Wen, Z.; Rupasinghe, S.; Niu, G.; Berenbaum, M.R.; Schuler, M.A. CYP6B1 and CYP6B3 of the Black Swallowtail (Papilio polyxenes): Adaptive Evolution through Subfunctionalization. Mol. Biol. Evol. 2006, 23, 2434–2443. [Google Scholar] [CrossRef]
- Amichot, M.; Tarès, S.; Brun-Barale, A.; Arthaud, L.; Bride, J.; Bergé, J. Point mutations associated with insecticide resistance in the Drosophila cytochrome P450 Cyp6a2 enable DDT metabolism. Eur. J. Biochem. 2004, 271, 1250–1257. [Google Scholar] [CrossRef]
- Schmidt, J.M.; Good, R.T.; Appleton, B.; Sherrard, J.; Raymant, G.C.; Bogwitz, M.R.; Martin, J.; Daborn, P.J.; Goddard, M.E.; Batterham, P.; et al. Copy Number Variation and Transposable Elements Feature in Recent, Ongoing Adaptation at the Cyp6g1 Locus. PLoS Genet. 2010, 6, e1000998. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Li, X. Transposable elements are enriched within or in close proximity to xenobiotic-metabolizing cytochrome P450 genes. BMC Evol. Biol. 2007, 7, 46. [Google Scholar] [CrossRef] [PubMed]
- Merzendorfer, H. ABC Transporters and their role in protecting insects from pesticides and their metabolites. In Advances in Insect Physiology; Cohen, E., Ed.; Academic Press: London, UK, 2014; pp. 1–73. [Google Scholar] [CrossRef]
- Li, X.; Schuler, M.A.; Berenbaum, M.R. Molecular Mechanisms of Metabolic Resistance to Synthetic and Natural Xenobiotics. Annu. Rev. Èntomol. 2007, 52, 231–253. [Google Scholar] [CrossRef]
- Meng, X.; Yang, X.; Wu, Z.; Shen, Q.; Miao, L.; Zheng, Y.; Qian, K.; Wang, J. Identification and transcriptional response of ATP -binding cassette transporters to chlorantraniliprole in the rice striped stem borer, Chilo suppressalis. Pest Manag. Sci. 2020, 76, 3626–3635. [Google Scholar] [CrossRef]
- Balabanidou, V.; Kampouraki, A.; MacLean, M.; Blomquist, G.J.; Tittiger, C.; Juárez, M.P.; Mijailovsky, S.J.; Chalepakis, G.; Anthousi, A.; Lynd, A.; et al. Cytochrome P450 associated with insecticide resistance catalyzes cuticular hydrocarbon production in Anopheles gambiae. Proc. Natl. Acad. Sci. USA 2016, 113, 9268–9273. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Pei, X.; Li, S.; Fan, Y.; Liu, T. Involvement of integument-rich CYP4G19 in hydrocarbon biosynthesis and cuticular penetration resistance in Blattella germanica (L.). Pest Manag. Sci. 2019, 76, 215–226. [Google Scholar] [CrossRef]
- Pignatelli, P.; Ingham, V.A.; Balabanidou, V.; Vontas, J.; Lycett, G.; Ranson, H. The Anopheles gambiae ATP-binding cassette transporter family: Phylogenetic analysis and tissue localization provide clues on function and role in insecticide resistance. Insect Mol. Biol. 2017, 27, 110–122. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Guo, J.; Ye, W.; Guo, Q.; Huang, Y.; Ma, L.; Zhou, D.; Shen, B.; Sun, Y.; Zhu, C. Cuticle genes CpCPR63 and CpCPR47 may confer resistance to deltamethrin in Culex pipiens pallens. Parasitol. Res. 2017, 116, 2175–2179. [Google Scholar] [CrossRef]
- Huang, Y.; Guo, Q.; Sun, X.; Zhang, C.; Xu, N.; Xu, Y.; Zhou, D.; Sun, Y.; Ma, L.; Zhu, C.; et al. Culex pipiens pallens cuticular protein CPLCG5 participates in pyrethroid resistance by forming a rigid matrix. Parasites Vectors 2018, 11, 6. [Google Scholar] [CrossRef] [PubMed]
- Fang, F.; Wang, W.; Zhang, D.; Lv, Y.; Zhou, D.; Ma, L.; Shen, B.; Sun, Y.; Zhu, C. The cuticle proteins: A putative role for deltamethrin resistance in Culex pipiens pallens. Parasitol. Res. 2015, 114, 4421–4429. [Google Scholar] [CrossRef]
- Simma, E.A.; Dermauw, W.; Balabanidou, V.; Snoeck, S.; Bryon, A.; Clark, R.M.; Yewhalaw, D.; Vontas, J.; Duchateau, L.; Van Leeuwen, T. Genome-wide gene expression profiling reveals that cuticle alterations and P450 detoxification are associated with deltamethrin and DDT resistance in Anopheles arabiensis populations from Ethiopia. Pest Manag. Sci. 2019, 75, 1808–1818. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Jin, T.; Zeng, L.; Lu, Y. Cuticular penetration of β-cypermethrin in insecticide-susceptible and resistant strains of Bactrocera dorsalis. Pestic. Biochem. Physiol. 2012, 103, 189–193. [Google Scholar] [CrossRef]
- Wilkins, R.M. Insecticide resistance and intracellular proteases. Pest Manag. Sci. 2017, 73, 2403–2412. [Google Scholar] [CrossRef]
- Hou, M.-Z.; Shen, G.-M.; Wei, D.; Li, Y.-L.; Dou, W.; Wang, J.-J. Characterization of Bactrocera dorsalis Serine Proteases and Evidence for Their Indirect Role in Insecticide Tolerance. Int. J. Mol. Sci. 2014, 15, 3272–3286. [Google Scholar] [CrossRef] [PubMed]
- Xiong, C.; Fang, F.; Chen, L.; Yang, Q.; He, J.; Zhou, D.; Shen, B.; Ma, L.; Sun, Y.; Zhang, D.; et al. Trypsin-Catalyzed Deltamethrin Degradation. PLoS ONE 2014, 9, e89517. [Google Scholar] [CrossRef]
- Zhu, X.; Lei, Y.; Yang, Y.; Baxter, S.W.; Li, J.; Wu, Q.; Wang, S.; Xie, W.; Guo, Z.; Fu, W.; et al. Construction and characterisation of near-isogenic Plutella xylostella (Lepidoptera: Plutellidae) strains resistant to Cry1Ac toxin. Pest Manag. Sci. 2014, 71, 225–233. [Google Scholar] [CrossRef]
- Patel, R.K.; Jain, M. NGS QC Toolkit: A Toolkit for Quality Control of Next Generation Sequencing Data. PLoS ONE 2012, 7, e30619. [Google Scholar] [CrossRef] [PubMed]
- Picard Toolkit. Broad Institute, GitHub Repository. 2019. Available online: http://broadinstitute.github.io/picard/ (accessed on 10 January 2020).
- Akey, J.M.; Zhang, G.; Zhang, K.; Jin, L.; Shriver, M.D. Interrogating a High-Density SNP Map for Signatures of Natural Selection. Genome Res. 2002, 12, 1805–1814. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef]
- Roy, A.; Yang, J.; Zhang, Y. COFACTOR: An accurate comparative algorithm for structure-based protein function annotation. Nucleic Acids Res. 2012, 40, W471–W477. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, Y. I-TASSER server: New development for protein structure and function predictions. Nucleic Acids Res. 2015, 43, W174–W181. [Google Scholar] [CrossRef]
- Zhang, Y. I-TASSER: Fully automated protein structure prediction in CASP8. Proteins Struct. Funct. Bioinform. 2009, 77 (Suppl. S9), 100–113. [Google Scholar] [CrossRef] [PubMed]
- Maghrabi, A.H.A.; McGuffin, L. ModFOLD6: An accurate web server for the global and local quality estimation of 3D protein models. Nucleic Acids Res. 2017, 45, W416–W421. [Google Scholar] [CrossRef]
- Wallner, B.; Elofsson, A. Can correct protein models be identified? Protein Sci. 2003, 12, 1073–1086. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 5, 550. [Google Scholar] [CrossRef]
- Gentleman, R.C.; Carey, V.J.; Bates, D.M.; Bolstad, B.; Dettling, M.; Dudoit, S.; Ellis, B.; Gautier, L.; Ge, Y.; Gentry, J.; et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004, 5, R80. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Chen, H.; Boutros, P.C. VennDiagram: A package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinform. 2011, 12, 35. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).