Cold Response Transcriptome Analysis of the Alternative Splicing Events Induced by the Cold Stress in D. catenatum
Abstract
:1. Introduction
2. Results
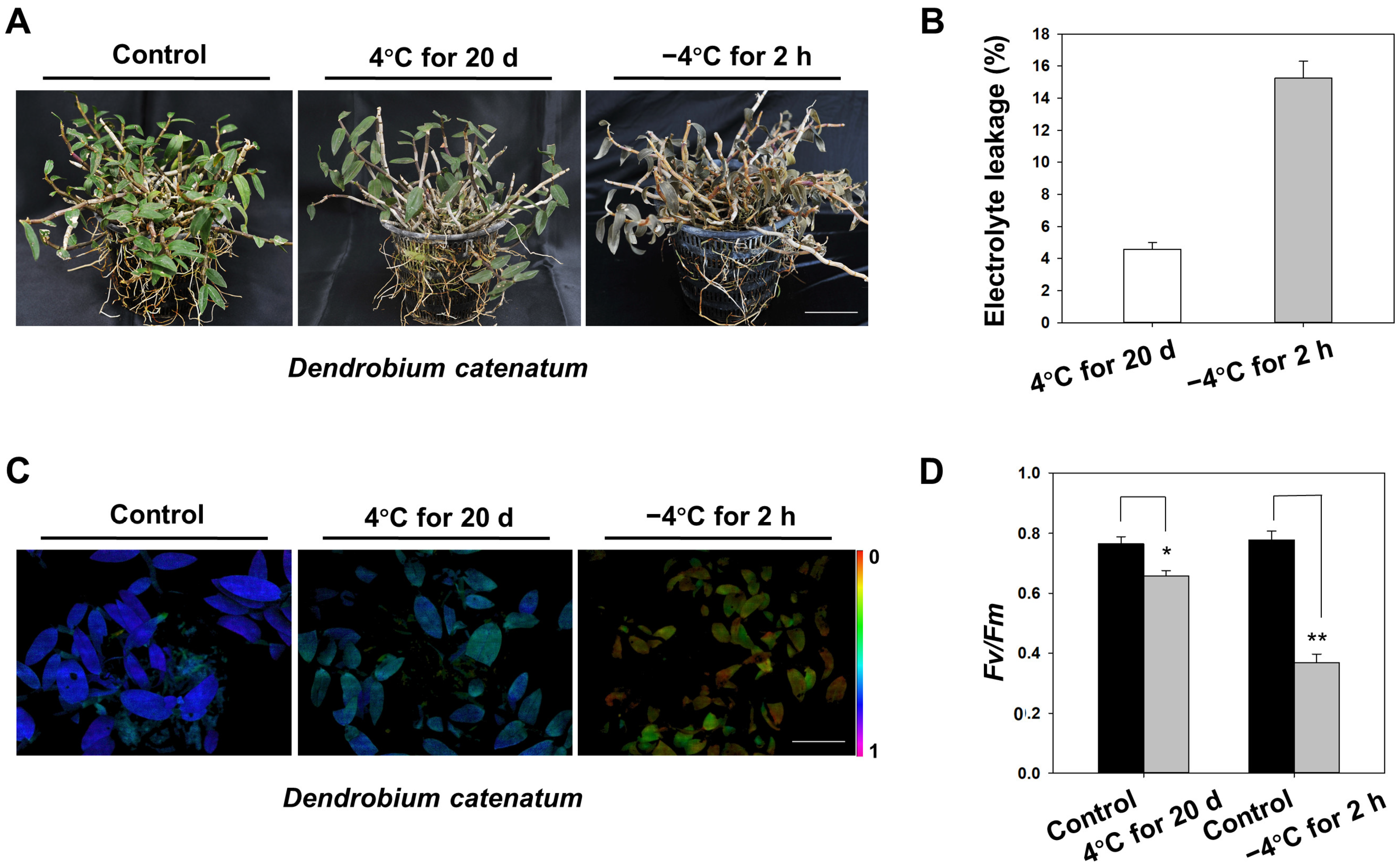
2.1. Damage to D. catenatum under Cold Stress
2.2. Transcriptional Changes Related to Chilling Stress in D. catenatum
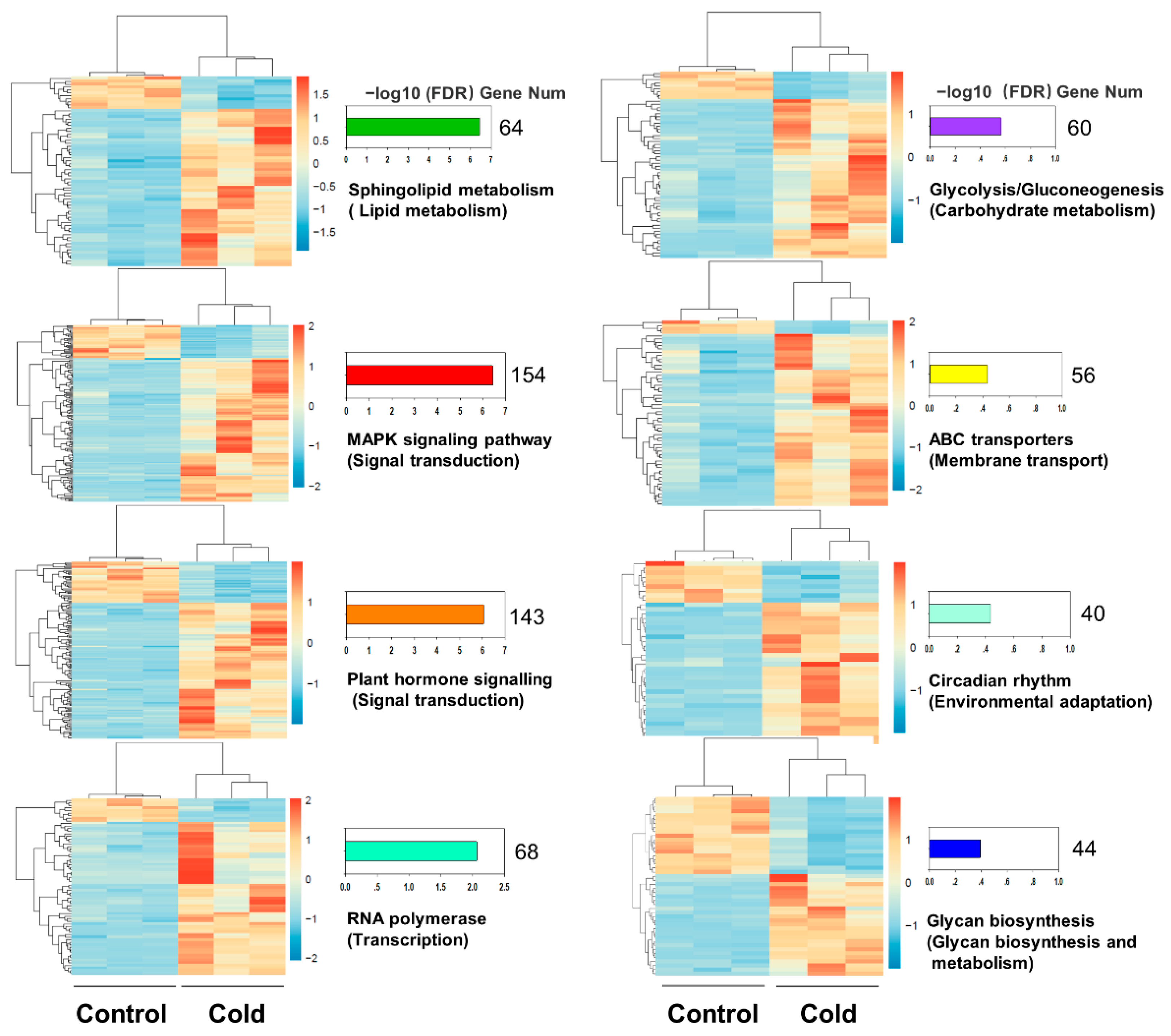
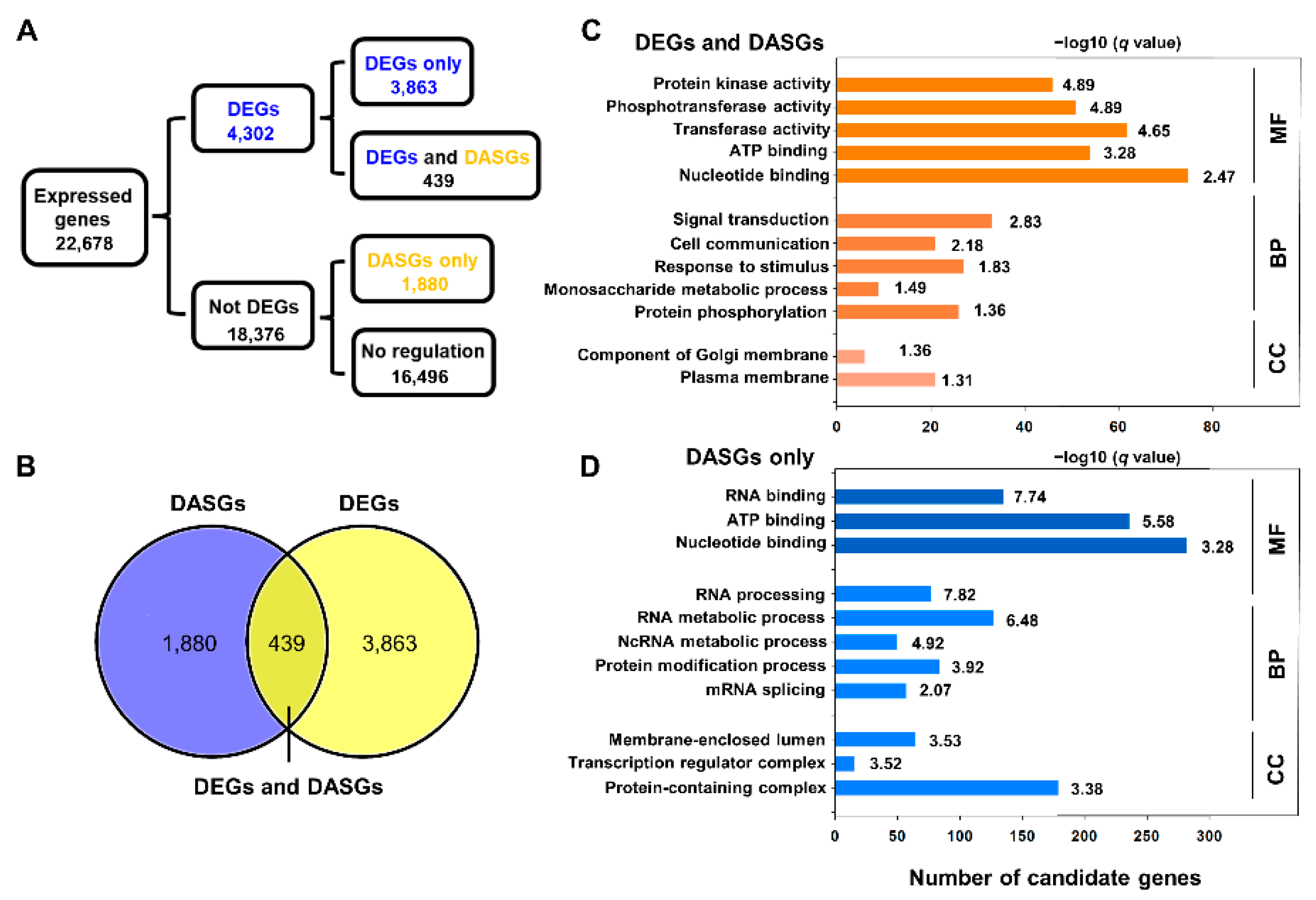
2.3. Analysis of Differential Expressed Genes (DEGs) in Cold Stress
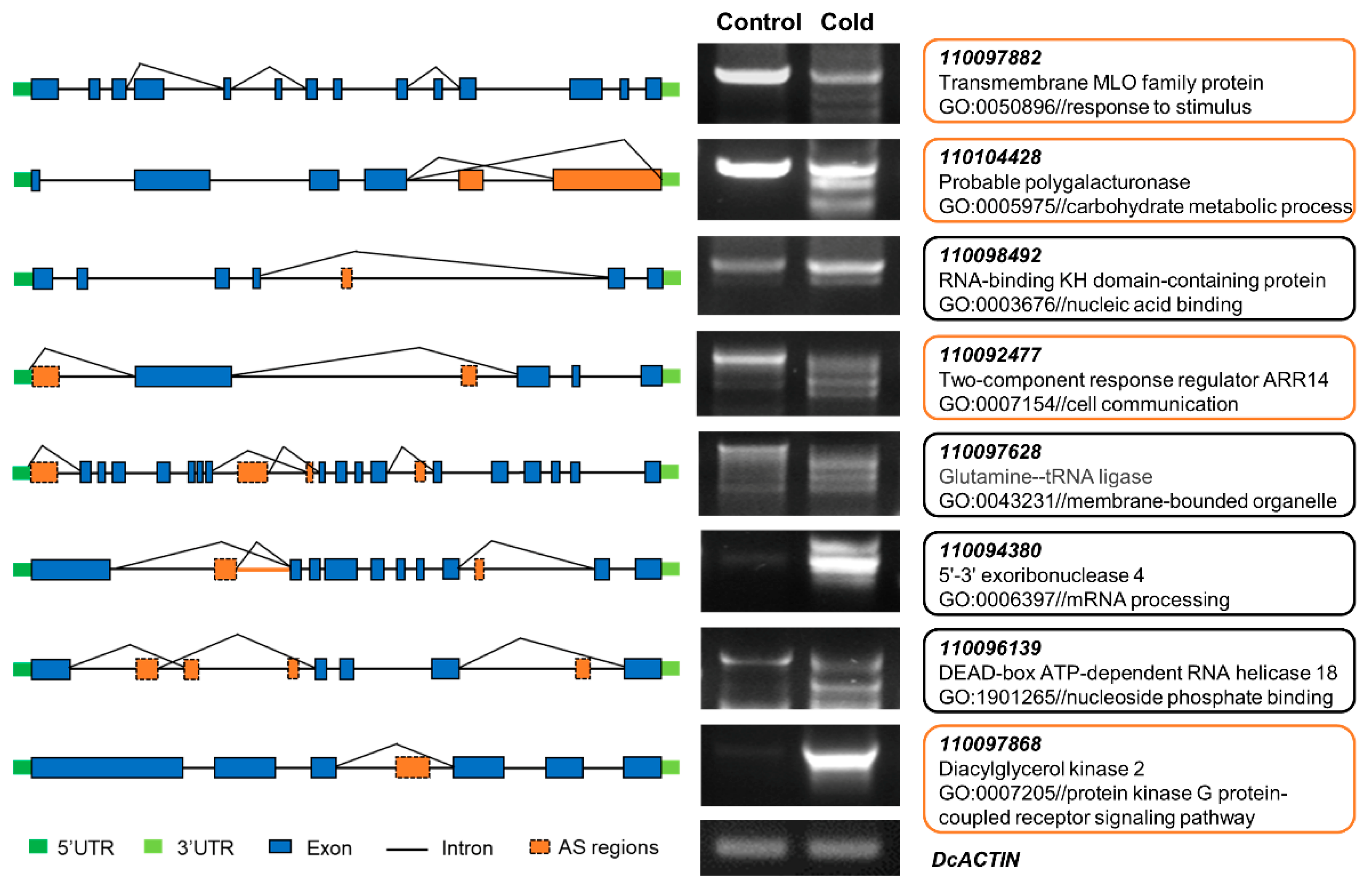
2.4. Analysis of Differential AS (DAS) in Response to Cold Stress
2.5. Comparative Analysis of DEGs and DASGs in Response to Cold Stress
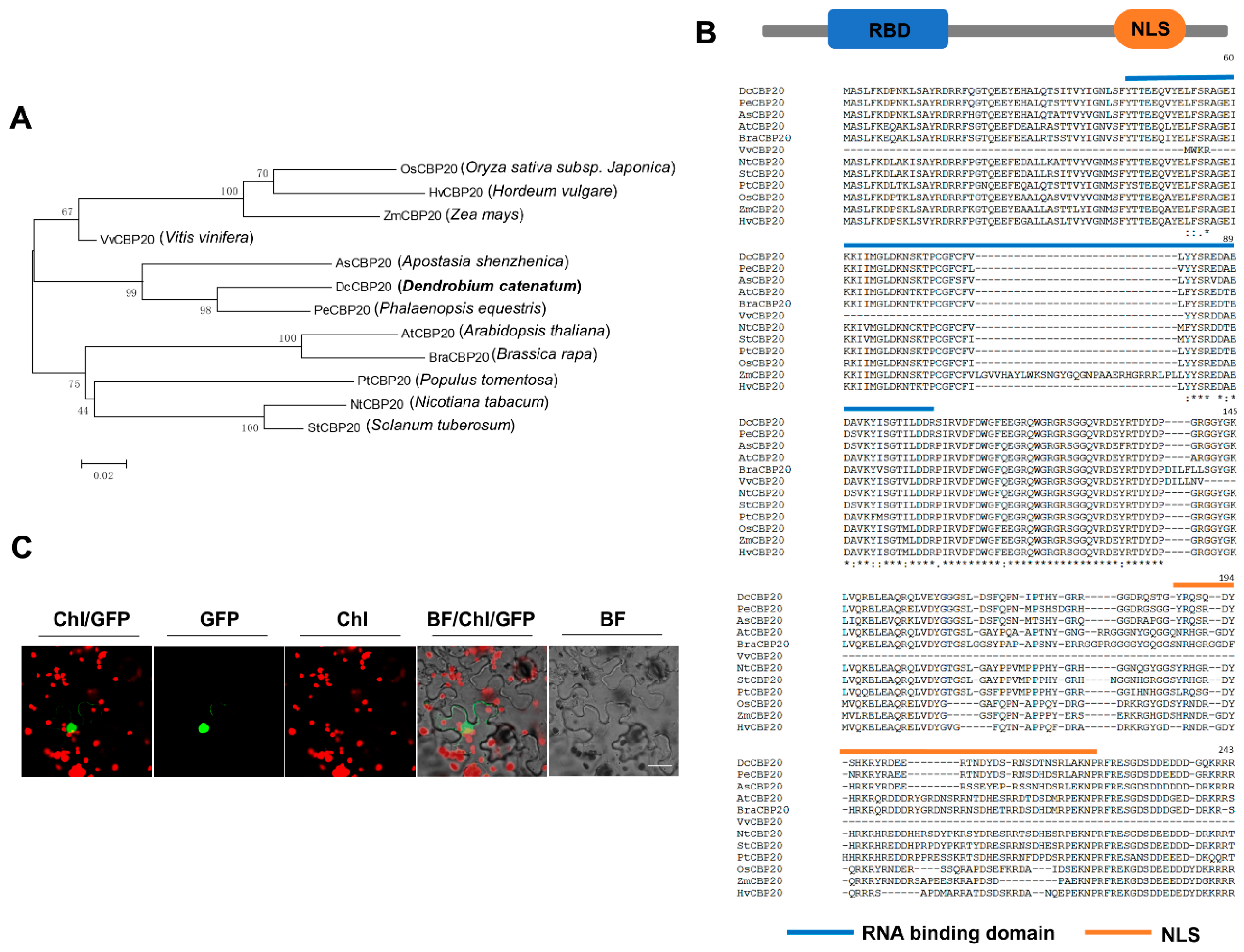
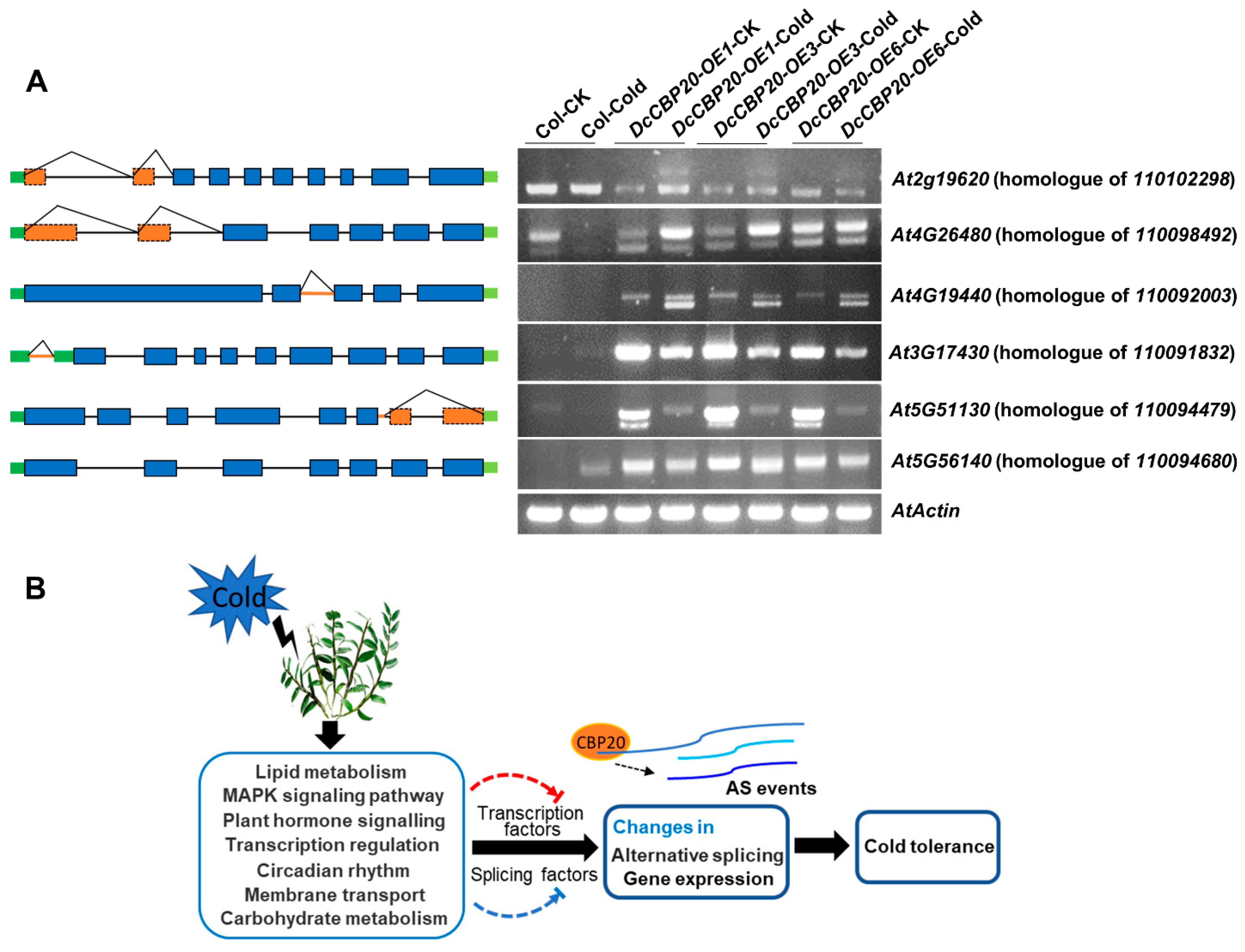
2.6. DcCBP20 of D. Catenatum Modulates the Alternative Splicing of Genes Responding to Cold Stress
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Cold Treatments
4.2. Measurements of Relative Electrolyte Leakage and Chl Fluorescence
4.3. RNA Isolation, Sequencing and Transcriptome
4.4. Analysis of Functional Enrichment of DEGs
4.5. Analysis of Functional Enrichment of DAS
4.6. RT–qPCR and RT–PCR
4.7. Generation of Transgenic Lines
4.8. Protein Immunoblotting
4.9. Transient Expression Assays
4.10. Phylogenetic Analysis and Conserved Motifs Prediction of CBP20
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhu, J.K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [Green Version]
- Gong, Z.Z.; Xiong, L.M.; Shi, H.Z.; Yang, S.H.; Herrera-Estrella, L.R.; Xu, G.H.; Chao, D.Y.; Li, J.R.; Wang, P.Y.; Qin, F.; et al. Plant abiotic stress response and nutrient use efficiency. Sci. China Life Sci. 2020, 63, 635–674. [Google Scholar] [CrossRef]
- Ding, Y.L.; Shi, Y.T.; Yang, S.H. Advances and challenges in uncovering cold tolerance regulatory mechanisms in plants. New Phytol. 2019, 222, 1690–1704. [Google Scholar] [CrossRef] [Green Version]
- Guy, C.L. Cold acclimation and freezing stress tolerance: Role of protein metabolism. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1990, 41, 187–223. [Google Scholar] [CrossRef]
- Thomashow, M.F. Plant cold acclimation: Freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 571–599. [Google Scholar] [CrossRef] [Green Version]
- Sangwan, V.; Foulds, I.; Singh, J.; Dhindsa, R.S. Cold-activation of Brassica napus Bn115 promoter is mediated by structural changes in membranes and cytoskeleton, and requires Ca2+ influx. Plant J. 2010, 27, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knight, H.; Trewavas, A.J.; Knight, M.R. Cold calcium signaling in Arabidopsis involves two cellular pools and a change in calcium signature after acclimation. Plant Cell 1996, 8, 489–503. [Google Scholar] [PubMed] [Green Version]
- Jaglo-Ottosen, K.R.; Gilmour, S.J.; Zarka, D.G.; Schabneberger, O.; Thomashow, M.F. Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Ence 1998, 280, 104–106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Q.; Kasuga, M.; Sakuma, Y.; Abe, H.; Miura, R.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 1998, 10, 1391–1406. [Google Scholar] [CrossRef] [Green Version]
- Stockinger, E.J.; Gilmour, S.J.; Thomashow, M.F. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-Repeat/Dre, a Cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. USA 1997, 94, 1035–1040. [Google Scholar] [CrossRef] [Green Version]
- Shi, Y.T.; Ding, Y.L.; Yang, S.H. Molecular regulation of CBF signaling in cold acclimation. Trends Plant Sci. 2018, 23, 623–637. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Ding, Y.L.; Shi, Y.T.; Zhang, X.Y.; Zhang, S.Q.; Gong, Z.Z.; Yang, S.H. MPK3- and MPK6-mediated ICE1 phosphorylation negatively regulates ICE1 stability and freezing tolerance in Arabidopsis. Dev. Cell 2017, 43, 630–642. [Google Scholar] [CrossRef]
- Yang, T.B.; Ali, G.S.; Yang, L.H.; Du, L.Q.; Reddy, A.S.N.; Poovaiah, B.W. Calcium/calmodulin-regulated receptor-like kinase CRLK1 interacts with MEKK1 in plants. Plant Signal. Behav. 2010, 5, 991–994. [Google Scholar] [CrossRef] [PubMed]
- Eremina, M.; Unterholzner, S.J.; Rathnayake, A.I.; Castellanos, M.; Khan, M.; Kugler, K.G.; May, S.T.; Mayer, K.F.X.; Rozhon, W.; Poppenberger, B. Brassinosteroids participate in the control of basal and acquired freezing tolerance of plants. Proc. Natl. Acad. Sci. USA 2016, 114, 1038–1039. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.R.; Jiang, L.Q.; Wang, F.; Yu, D.Q. Jasmonate regulates the inducer of CBF Expression-C-Repeat Binding Factor/Dre Binding Factor1 cascade and freezing tolerance in Arabidopsis. Plant cell 2013, 25, 2907–2924. [Google Scholar] [CrossRef] [Green Version]
- Shi, Y.T.; Tian, S.W.; Hou, L.Y.; Huang, X.Z.; Zhang, X.Y.; Guo, H.W.; Yang, S.H. Ethylene signaling negatively regulates freezing tolerance by repressing expression of CBF and Type-A ARR genes in Arabidopsis. Plant Cell 2012, 24, 2578–2595. [Google Scholar] [CrossRef] [Green Version]
- Kidokoro, S.; Hayashi, K.; Haraguchi, H.; Ishikawa, T.; Soma, F.; Konoura, I.; Toda, S.; Mizoi, J.; Suzuki, T.; Shinozaki, K.; et al. Posttranslational regulation of multiple clock-related transcription factors triggers cold-inducible gene expression in Arabidopsis. Proc. Natl. Acad. Sci. USA 2021, 118, 10. [Google Scholar] [CrossRef]
- Reddy, A.S.N.; Marquez, Y.; Kalyna, M.; Barta, A. Complexity of the alternative splicing landscape in plants. Plant Cell 2013, 25, 3657–3683. [Google Scholar] [CrossRef] [Green Version]
- Marquez, Y.; Brown, J.W.S.; Simpson, C.; Barta, A.; Kalyna, M. Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res. 2012, 22, 1184–1195. [Google Scholar] [CrossRef] [Green Version]
- Shen, Y.T.; Zhou, Z.K.; Wang, Z.; Li, W.Y.; Fang, C.; Wu, M.; Ma, Y.M.; Liu, T.F.; Kong, L.A.; Peng, D.L.; et al. Global dissection of alternative splicing in Paleopolyploid soybean. Plant Cell 2014, 26, 996–1008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Z.S.; Qin, J.X.; Tian, X.J.; Xu, S.B.; Wang, Y.; Li, H.X.; Wang, X.M.; Peng, H.R.; Yao, Y.Y.; Hu, Z.R.; et al. Global profiling of alternative splicing landscape responsive to drought, heat and their combination in wheat (Triticum aestivum L.). Plant Biotechnol. J. 2017, 16, 714–726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Filichkin, S.; Priest, H.D.; Megraw, M.; Mockler, T.C. Alternative splicing in plants: Directing traffic at the crossroads of adaptation and environmental stress. Curr. Opin. Plant Biol. 2015, 24, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Calixto, C.P.G.; Guo, W.B.; James, A.B.; Tzioutziou, N.A.; Entizne, J.C.; Panter, P.E.; Knight, H.; Nimmo, H.G.; Zhang, R.X.; Brown, J.W.S. Rapid and dynamic alternative splicing impacts the Arabidopsis cold response transcriptome. Plant Cell 2018, 30, 1424–1444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Erkelenz, S.; Mueller, W.F.; Evans, M.S.; Busch, A.; Schoneweis, K.; Hertel, K.J.; Schaal, H. Position-dependent splicing activation and repression by SR and hnRNP proteins rely on common mechanisms. RNA—A Publ. RNA Soc. 2013, 19, 96–102. [Google Scholar] [CrossRef] [Green Version]
- Gonatopoulos-Pournatzis, T.; Cowling, V.H. Cap-Binding Complex (CBC). Biochem. J. 2014, 457, 231–242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kierzkowski, D.; Kmieciak, M.; Piontek, P.; Wojtaszek, P.; Szweykowska-Kulinska, Z.; Jarmolowski, A. The Arabidopsis CBP20 targets the cap-binding complex to the nucleus, and is stabilized by CBP80. Plant J. 2010, 59, 814–825. [Google Scholar] [CrossRef]
- Daszkowska-Golec, A. Emerging roles of the nuclear cap-binding complex in abiotic stress responses. Plant Physiol. 2018, 176, 242–253. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Guo, Q.H.; Liu, P.; Huang, J.G.; Zhang, S.Z.; Yang, G.D.; Wu, C.G.; Zheng, C.C.; Yan, K. Dual roles of the serine/arginine-rich splicing factor SR45a in promoting and interacting with nuclear cap-binding complex to modulate the salt-stress response in Arabidopsis. New Phytol. 2021, 230, 641–655. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.X.; Ma, L.; Yang, L.M.; Chen, Q.; Xiang, N.; Yang, Y.P.; Hu, X.Y. Quantitative proteomics analysis reveals that the nuclear cap-binding complex proteins Arabidopsis CBP20 and CBP80 modulate the salt stress response. J. Proteome Res. 2014, 13, 2495–2510. [Google Scholar] [CrossRef]
- Zhang, G.Q.; Xu, Q.; Bian, C.; Tsai, W.C.; Yeh, C.M.; Liu, K.W.; Yoshida, K.; Zhang, L.S.; Chang, S.B.; Chen, F.; et al. The Dendrobium Catenatum Lindl. genome sequence provides insights into polysaccharide synthase, floral development and adaptive evolution. Sci. Rep. 2016, 6, 19029. [Google Scholar] [CrossRef] [Green Version]
- Tang, X.G.; Yuan, Y.D.; Zhang, J.C. How climate change will alter the distribution of suitable Dendrobium habitats. Front. Ecol. Evol. 2020, 8, 8. [Google Scholar] [CrossRef]
- Zhang, S.H.; Li, J.; Shen, Y.C.; Nartey, L.N.; Pu, Q.; Lu, J.; Shakeela, B.; Kong, D.D.; Li, O.; Zeng, G.H.; et al. Physiological responses of Dendrobium Officinale under exposure to cold stress with two cultivars. Phyton 2020, 89, 599–617. [Google Scholar] [CrossRef]
- Liang, S.; Ye, Q.S.; Li, R.H.; Leng, J.Y.; Li, M.R.; Wang, X.J.; Li, H.Q. Transcriptional regulations on the low-temperature-induced floral transition in an Orchidaceae species, Dendrobium nobile: An expressed sequence tags analysis. Comp. Funct. Genom. 2012, 2012, 757801. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.N.; Mount, S.M. Two alternatively spliced isoforms of the Arabidopsis SR45 protein have distinct roles during normal plant development. Plant Physiol. 2009, 150, 1450–1458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, S.X.; Yu, X.; Cheng, Z.H.; Zeng, C.Y.; Li, W.B.; Zhang, L.S.; Peng, M. Large-scale analysis of the cassava transcriptome reveals the impact of cold stress on alternative splicing. J. Exp. Bot. 2020, 71, 422–434. [Google Scholar] [CrossRef]
- Thatcher, S.R.; Danilevskaya, O.N.; Meng, X.; Beatty, M.; Zastrow-Hayes, G.; Harris, C.; Allen, B.V.; Habben, J.; Li, B. Genome-wide analysis of alternative splicing during development and drought stress in maize. Plant Physiol. 2016, 170, 586–599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brannan, K.; Kim, H.; Erickson, B.; Glover-Cutter, K.; Bentley, D.L. mRNA decapping factors and the exonuclease Xrn2 function in widespread premature termination of RNA polymerase ii transcription. Mol. Cell 2012, 46, 311–324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, Y.Q.; Yang, B.; Harris, N.S.; Deyholos, M.K. Comparative proteomic analysis of NACL stress-responsive proteins in Arabidopsis roots. J. Exp. Bot. 2007, 8, 3591–3607. [Google Scholar] [CrossRef] [Green Version]
- Gasic, K.; Hernandez, A.; Korban, S.S. RNA extraction from different apple tissues rich in polyphenols and polysaccharides for cDNA library construction. Plant Mol. Biol. Rep. 2004, 22, 437–438. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, Y.; Shi, C.; Huang, Z.; Zhang, Y.; Li, S.; Li, Y.; Ye, J.; Yu, C.; Li, Z.; et al. SOAPnuke: A MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. Gigascience 2018, 7, gix120. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast splices aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Method 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Deway, C.N.; Li, B. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mi, H.; Huang, X.; Muruganujan, A.; Tang, H.; Mils, C.; Kang, D.; Thomas, P.D. PANTHER version 14: More genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019, 47, 419–426. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Furumichi, M.; Morishima, K.; Tanabe, M. New approach for understanding genome variations in KEGG. Nucleic Acid Res. 2019, 47, D590–D595. [Google Scholar] [CrossRef] [Green Version]
- Shen, S.; Park, J.W.; Lu, Z.; Lin, L.; Henry, M.D.; Wu, Y.N.; Zhou, Q.; Xing, Y. rMATS: Robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl Acad. Sci. USA 2014, 111, 5593–5601. [Google Scholar] [CrossRef] [Green Version]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative realtime PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sparkes, I.A.; Runions, J.; Kearns, A.; Hawes, C. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 2006, 1, 2019–2025. [Google Scholar] [CrossRef]

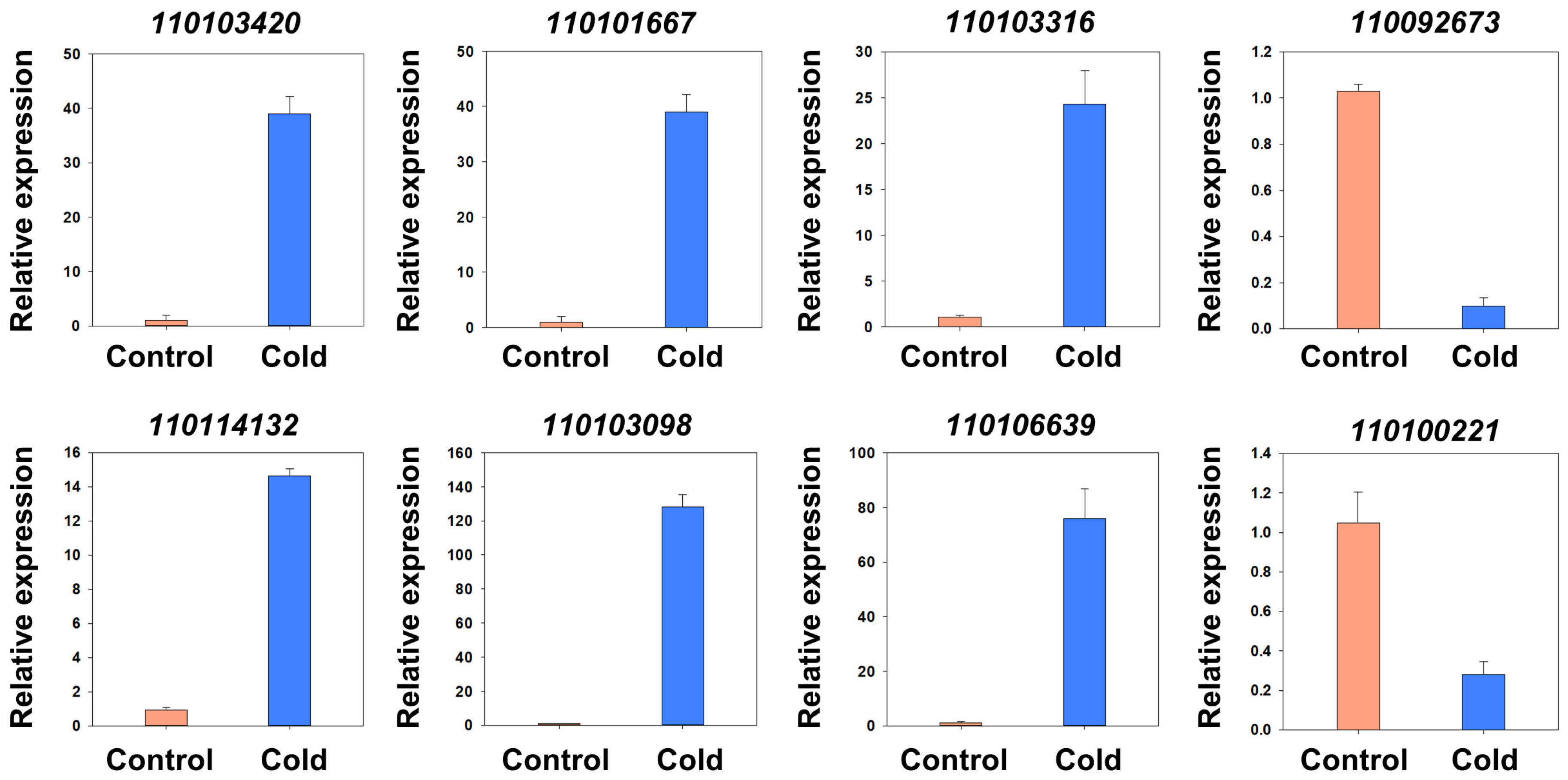

| Gene ID | Gene Annotation | Log2 (Cold/ck) | q Value (Cold-vs-ck) |
|---|---|---|---|
| 110103420 | Protein TIFY 5A | 5.882954842 | 1.15 × 10−16 |
| 110101667 | Two-component response regulator-like APRR9 | 5.75307647 | 1.14 × 10−148 |
| 110103316 | WRKY transcription factor 50 | 5.270290384 | 7.89 × 10−17 |
| 110092673 | Cyclin-P4-1-like | −3.506757892 | 2.00 × 10−3 |
| 110114132 | WRKY transcription factor 48 | 5.583790123 | 3.73 × 10−47 |
| 110103098 | Gibberellin 2-β-dioxygenase 1 | 6.673964409 | 2.76 × 10−60 |
| 110106639 | polygalacturonase inhibitor 2-like | 8.403207013 | 3.38 × 10−117 |
| 110100221 | Alpha-humulene synthase-like | −3.539482498 | 2.38 × 10−6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, Y.; Luo, L.; Chen, Q.; Yang, D.; Gong, Y.; Yang, Y.; Qin, X.; Wang, Y.; Kong, X.; Yang, Y. Cold Response Transcriptome Analysis of the Alternative Splicing Events Induced by the Cold Stress in D. catenatum. Int. J. Mol. Sci. 2022, 23, 981. https://doi.org/10.3390/ijms23020981
Zheng Y, Luo L, Chen Q, Yang D, Gong Y, Yang Y, Qin X, Wang Y, Kong X, Yang Y. Cold Response Transcriptome Analysis of the Alternative Splicing Events Induced by the Cold Stress in D. catenatum. International Journal of Molecular Sciences. 2022; 23(2):981. https://doi.org/10.3390/ijms23020981
Chicago/Turabian StyleZheng, Yan, Landi Luo, Qian Chen, Danni Yang, Yuqiang Gong, Ya Yang, Xiangshi Qin, Yuhua Wang, Xiangxiang Kong, and Yongping Yang. 2022. "Cold Response Transcriptome Analysis of the Alternative Splicing Events Induced by the Cold Stress in D. catenatum" International Journal of Molecular Sciences 23, no. 2: 981. https://doi.org/10.3390/ijms23020981
APA StyleZheng, Y., Luo, L., Chen, Q., Yang, D., Gong, Y., Yang, Y., Qin, X., Wang, Y., Kong, X., & Yang, Y. (2022). Cold Response Transcriptome Analysis of the Alternative Splicing Events Induced by the Cold Stress in D. catenatum. International Journal of Molecular Sciences, 23(2), 981. https://doi.org/10.3390/ijms23020981






