Validating Reference Gene Expression Stability in Human Ovarian Follicles, Oocytes, Cumulus Cells, Ovarian Medulla, and Ovarian Cortex Tissue
Abstract
:1. Introduction
2. Results
2.1. General Expression Levels of Reference Genes
2.2. Stability of Reference Genes in Human Oocytes, Preantral Follicles, Cumulus Cells, and Medulla Tissue
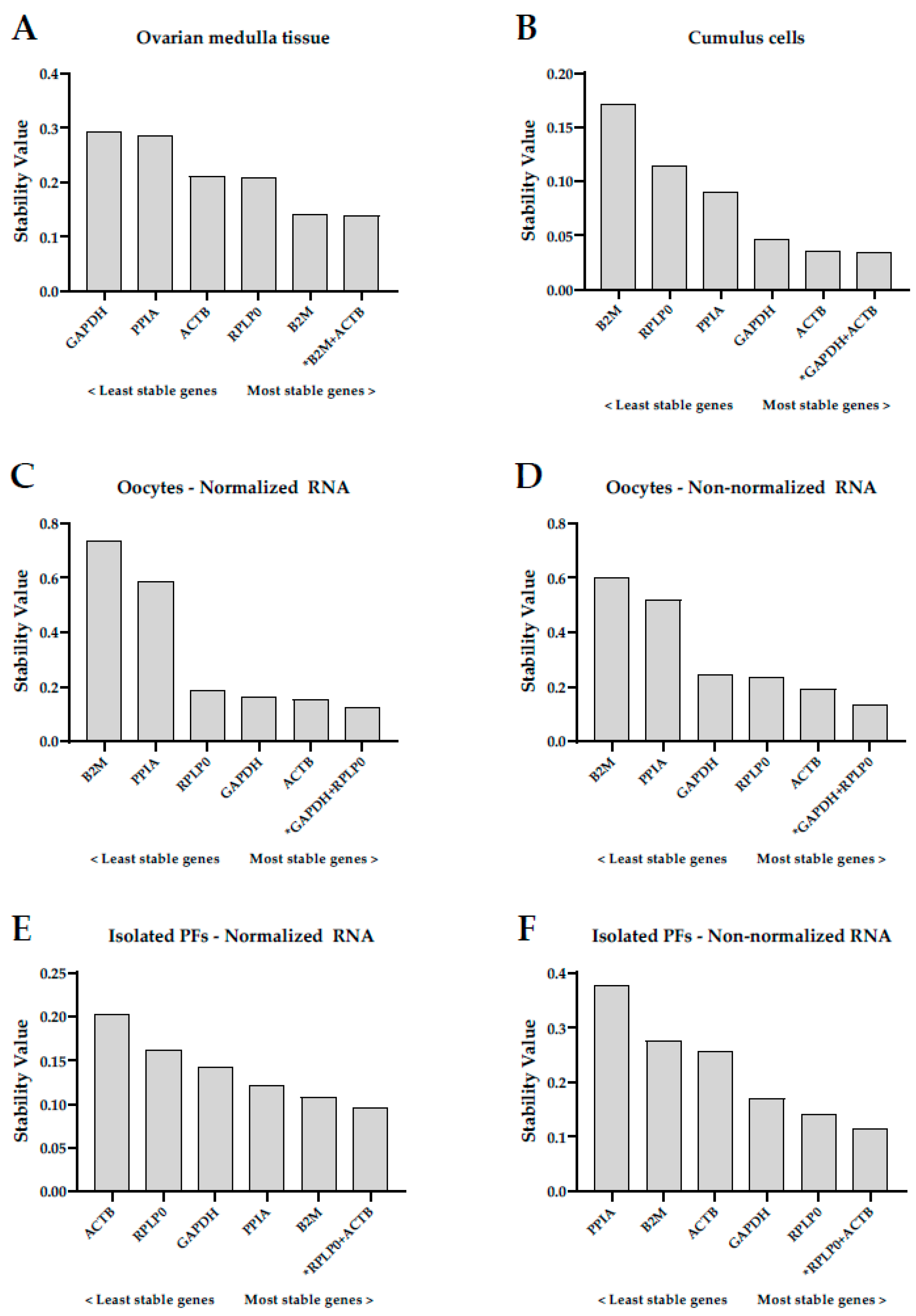
2.2.1. Ovarian Medulla Tissue
2.2.2. Cumulus Cells
2.2.3. Oocytes
2.2.4. Isolated Preantral Follicles
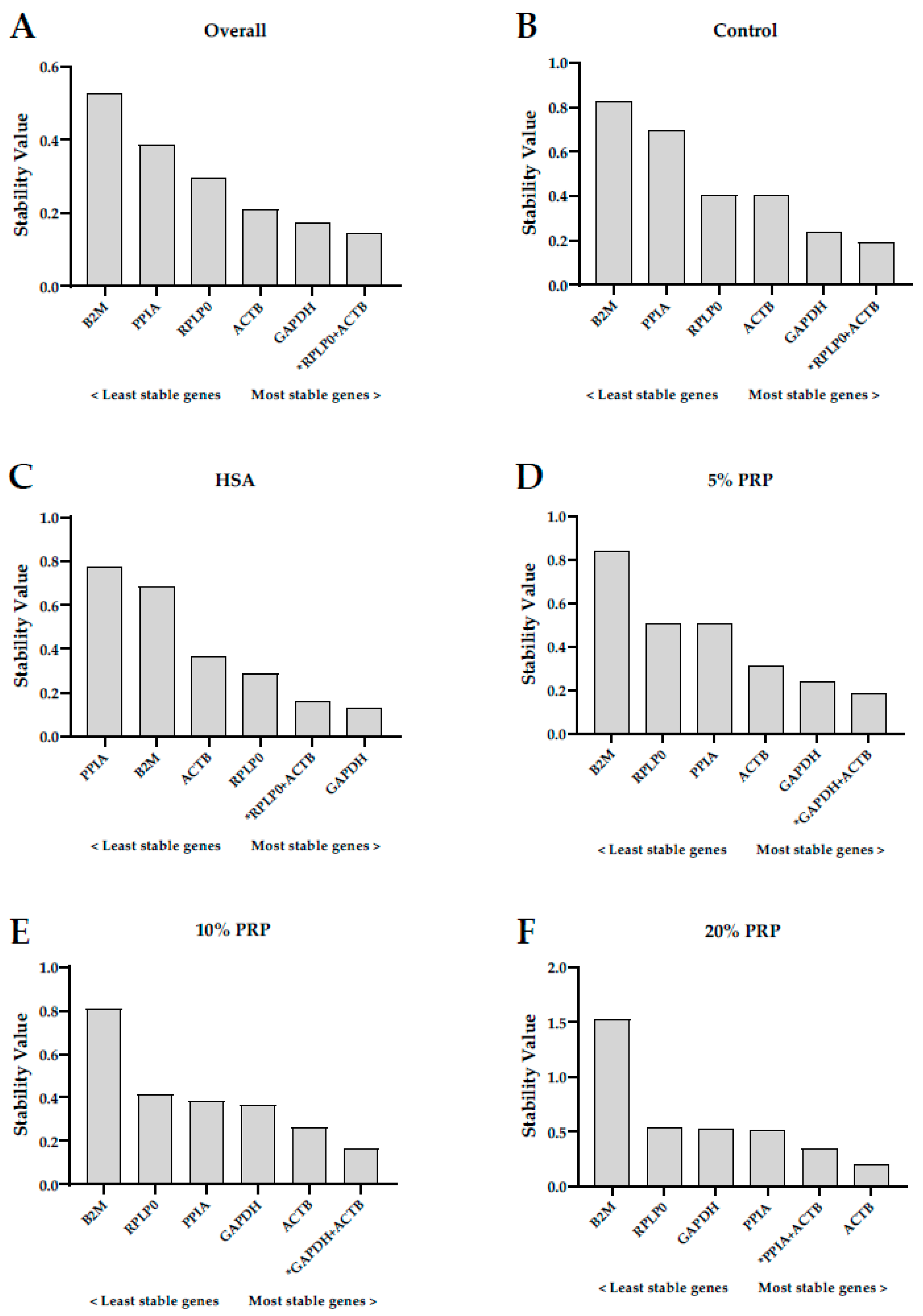
2.3. Stability of Reference Genes in Human Ovarian Cortex Cultured with Different Serum-Derived Supplements
3. Discussion
4. Materials and Methods
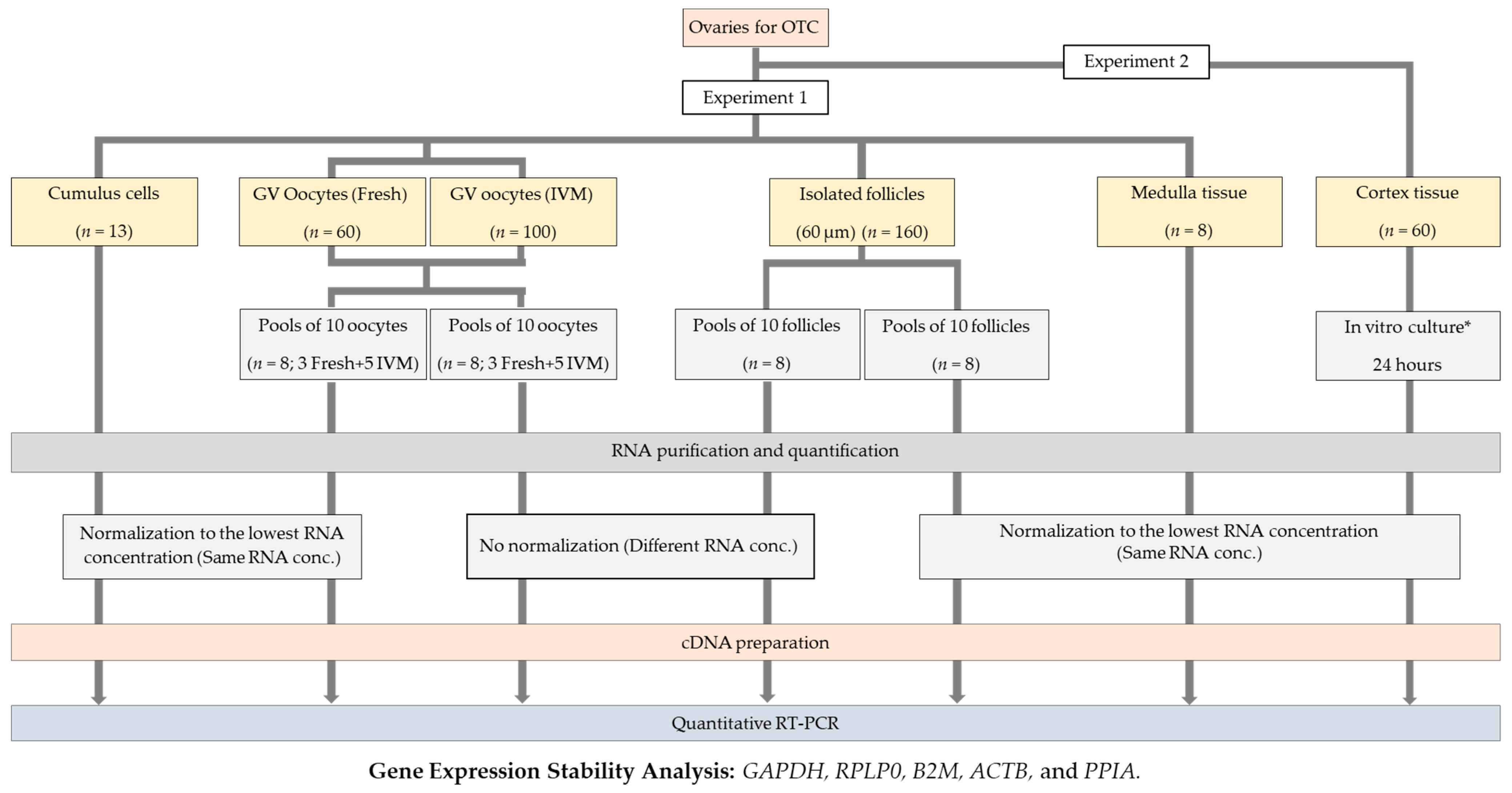
4.1. Experimental Design
4.2. Patients and Collected Material
4.3. Ovary Transport and Oocyte Collection
4.4. Medulla Tissue Collection and Follicle Isolation
4.5. In Vitro Culture of Human Ovarian Cortex (Experiment 2)
4.6. Quantitative Real-Time PCR (RT-qPCR)
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gremeau, A.S.; Andreadis, N.; Fatum, M.; Craig, J.; Turner, K.; McVeigh, E.; Child, T. In vitro maturation or in vitro fertilization for women with polycystic ovaries? A case-control study of 194 treatment cycles. Fertil. Steril. 2012, 98, 355–360. [Google Scholar] [CrossRef]
- Imesch, P.; Scheiner, D.; Xie, M.; Fink, D.; Macas, E.; Dubey, R.; Imthurn, B. Developmental potential of human oocytes matured in vitro followed by vitrification and activation. J. Ovarian Res. 2013, 6, 30. [Google Scholar] [CrossRef] [Green Version]
- Walls, M.L.; Hunter, T.; Ryan, J.P.; Keelan, J.A.; Nathan, E.; Hart, R.J. In vitro maturation as an alternative to standard in vitro fertilization for patients diagnosed with polycystic ovaries: A comparative analysis of fresh, frozen and cumulative cycle outcomes. Hum. Reprod. 2015, 30, 88–96. [Google Scholar] [CrossRef] [Green Version]
- Zhao, P.; Qiao, J.; Huang, S.; Zhang, Y.; Liu, S.; Yan, L.Y.; Hsueh, A.J.; Duan, E.K. Gonadotrophin-induced paracrine regulation of human oocyte maturation by BDNF and GDNF secreted by granulosa cells. Hum. Reprod. 2011, 26, 695–702. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Liu, H.; Yu, Q.; Liu, H.; Huang, T.; Zhao, S.; Ma, J.; Zhao, H. Growth hormone promotes in vitro maturation of human oocytes. Front. Endocrinol. 2019, 10, 485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cadenas, J.; Nikiforov, D.; Pors, S.E.; Zuniga, L.A.; Wakimoto, Y.; Ghezelayagh, Z.; Mamsen, L.S.; Kristensen, S.G.; Andersen, C.Y. A threshold concentration of FSH is needed during IVM of ex vivo collected human oocytes. J. Assist. Reprod. Genet. 2021, 38, 1341–1348. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Nolan, T.; Pfaffl, M.W. Quantitative real-time RT-PCR—A perspective. J. Mol. Endocrinol. 2005, 34, 597–601. [Google Scholar] [CrossRef] [Green Version]
- Bustin, S.A. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): Trends and problems. J. Mol. Endocrinol. 2002, 29, 23–39. [Google Scholar] [CrossRef] [PubMed]
- Gebeh, A.K.; Marczylo, E.L.; Amoako, A.A.; Willets, J.M.; Konje, J.C. Variation in stability of endogenous reference genes in fallopian tubes and endometrium from healthy and ectopic pregnant women. Int. J. Mol. Sci. 2012, 13, 2810–2826. [Google Scholar] [CrossRef]
- Asiabi, P.; Ambroise, J.; Giachini, C.; Coccia, M.E.; Bearzatto, B.; Chiti, M.C.; Dolmans, M.M.; Amorim, C.A. Assessing and validating housekeeping genes in normal, cancerous, and polycystic human ovaries. J. Assist. Reprod. Genet. 2020, 37, 2545–2553. [Google Scholar] [CrossRef]
- Dheda, K.; Huggett, J.F.; Chang, J.S.; Kima, L.U.; Bustin, S.A.; Johnson, M.A.; Rook, G.A.W.; Zumla, A. The implications of using an inappropriate reference gene for real-time reverse transcription PCR data normalization. Anal. Biochem. 2005, 344, 141–143. [Google Scholar] [CrossRef] [PubMed]
- Ruan, W.; Lai, M. Actin, a reliable marker of internal control? Clin. Chim. Acta 2007, 385, 1–5. [Google Scholar] [CrossRef]
- Caradec, J.; Sirab, N.; Keumeugni, C.; Moutereau, S.; Chimingqi, M.; Matar, C.; Revaud, D.; Bah, M.; Manivet, P.; Conti, M.; et al. Desperate house genes’: The dramatic example of hypoxia. Br. J. Cancer 2010, 102, 1037–1043. [Google Scholar] [CrossRef] [Green Version]
- Caradec, J.; Sirab, N.; Revaud, D.; Keumeugni, C.; Loric, S. Is GAPDH a relevant housekeeping gene for normalisation in colorectal cancer experiments? Br. J. Cancer 2010, 103, 1475–1476. [Google Scholar] [CrossRef]
- Krzystek-Korpacka, M.; Hotowy, K.; Czapinska, E.; Podkowik, M.; Bania, J.; Gamian, A.; Bednarz-Misa, I. Serum availability affects expression of common house-keeping genes in colon adenocarcinoma cell lines: Implications for quantitative real-time PCR studies. Cytotechnology 2016, 68, 2503–2517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nikishin, D.A.; Filatov, M.A.; Kiseleva, M.V.; Bagaeva, T.S.; Konduktorova, V.V.; Khramova, Y.V.; Malinova, I.V.; Komarova, E.V.; Semenova, M.L. Selection of stable expressed reference genes in native and vitrified/thawed human ovarian tissue for analysis by qRT-PCR and Western blot. J. Assist. Reprod. Genet. 2018, 35, 1851–1860. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.; de Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; de Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034.1. [Google Scholar] [CrossRef] [Green Version]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Orntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [Green Version]
- Jeong, Y.J.; Choi, H.W.; Shin, H.S.; Cui, X.S.; Kim, N.H.; Gerton, G.L.; Jun, J.H. Optimization of real time RT-PCR methods for the analysis of gene expression in mouse eggs and preimplantation embryos. Mol. Reprod. Dev. 2005, 71, 284–289. [Google Scholar] [CrossRef]
- Yuanyuan, S.; Qin, S.; Rongrong, X.; Yujing, G.; Chengbin, P.; Jianjun, M.; Yanzhou, Y.; Xiuying, P. Reference gene selection for real-time quantitative PCR analysis on ovarian cryopreservation by vitrification in mice. J. Assist. Reprod. Genet. 2015, 32, 1277–1284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caetano, L.C.; Miranda-Furtado, C.L.; Batista, L.A.; Pitangui-Molina, C.P.; Higa, T.T.; Padovan, C.C.; Rosa-E-Silva, A. Validation of reference genes for gene expression studies in bovine oocytes and cumulus cells derived from in vitro maturation. Anim Reprod. 2019, 16, 290–296. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Li, T.; Huang, B.; Cheng, H.; Ding, H.; Dong, W.; Xiao, M.; Liu, L.; Wang, Z. The use of laser microdissection in the identification of suitable reference genes for normalization of quantitative real-time PCR in human FFPE epithelial ovarian tissue samples. PLoS ONE 2014, 9, e95974. [Google Scholar] [CrossRef] [PubMed]
- Ofinran, O.; Bose, U.; Hay, D.; Abdul, S.; Tufatelli, C.; Khan, R. Selection of suitable reference genes for gene expression studies in normal human ovarian tissues, borderline ovarian tumours and ovarian cancer. Mol. Med. Rep. 2016, 14, 5725–5731. [Google Scholar] [CrossRef]
- Li, Y.L.; Ye, F.; Hu, Y.; Lu, W.G.; Xie, X. Identification of suitable reference genes for gene expression studies of human serous ovarian cancer by real-time polymerase chain reaction. Anal. Biochem. 2009, 394, 110–116. [Google Scholar] [CrossRef]
- Fu, J.; Bian, L.; Zhao, L.; Dong, Z.; Gao, X.; Luan, H.; Sun, Y.; Song, H. Identification of genes for normalization of quantitative real-time PCR data in ovarian tissues. Acta Biochim. Biophys. Sin. 2010, 42, 568–574. [Google Scholar] [CrossRef] [Green Version]
- Baddela, V.S.; Baufeld, A.; Yenuganti, V.R.; Vanselow, J.; Singh, D. Suitable housekeeping genes for normalization of transcript abundance analysis by real-time RT-PCR in cultured bovine granulosa cells during hypoxia and differential cell plating density. Reprod. Biol. Endocrinol. 2014, 46, 2941–2946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ståhlberg, A.; Håkansson, J.; Xian, X.; Semb, H.; Kubista, M. Properties of the reverse transcription reaction in mRNA quantification. Clin. Chem. 2004, 50, 509–515. [Google Scholar] [CrossRef] [Green Version]
- Andersen, C.Y.; Mamsen, L.S.; Kristensen, S.G. Freezing of ovarian tissue and clinical opportunities. Reproduction 2019, 158, F27–F34. [Google Scholar] [CrossRef]
- Pors, S.E.; Kristensen, S.G.; Nikiforov, D.; Mamsen, L.M.; Cadenas, J.; Greve, V.H.; Dueholm, M.; Andersen, C.Y. Histidine buffered media maintains pH stabile during cooled transportation of human ovarian tissue. J. Ovarian Res. 2021, 14, 116. [Google Scholar] [CrossRef]
- Yin, H.; Kristensen, S.G.; Jiang, H.; Rasmussen, A.; Andersen, C.Y. Survival and growth of isolated pre-antral follicles from human ovarian medulla tissue during long-term 3D culture. Hum. Reprod. 2016, 31, 1531–1539. [Google Scholar] [CrossRef] [PubMed]
| Preantral Follicles | Oocytes | Cumulus Cells | Medulla Tissue | Cortex Tissue | |||
|---|---|---|---|---|---|---|---|
| RGs | Normalized | Non-Normalized | Normalized | Non-Normalized | Normalized | Normalized | Normalized |
| GAPDH | 30.1 ± 0.5 a (1.6%) | 29.7 ± 1.1 a (3.7%) | 30.0 ± 2.2 a (7.3%) | 30.4 ± 2.5 a (8.2%) | 25.6 ± 1.2 a (4.7%) | 23.6 ± 1.5 a (6.4%) | 21.6 ± 1.0 a (4.6%) |
| RPLP0 | 27.7 ± 0.4 b (1.4%) | 27.6 ± 1.0 b (3.6%) | 28.2 ± 2.4 b (8.5%) | 28.7 ± 2.3 b (8.0%) | 24.1 ± 1.1 b (4.6%) | 20.9 ± 1.6 bc (7.4%) | 21.3 ± 0.7 a (3.3%) |
| B2M | 29.2 ± 0.6 c (2.0%) | 29.3 ± 1.6 a (5.5%) | 31.3 ± 3.3 c (10.5%) | 33.1 ± 3.1 c (9.4%) | 26.3 ± 1.3 a (4.9%) | 21.6 ± 1.6 c (7.4%) | 25.6 ± 2.5 b (9.8%) |
| ACTB | 26.7 ± 0.8 d (3.0%) | 26.7 ± 1.5 c (5.6%) | 26.9 ± 2.2 d (8.2%) | 28.1 ± 2.4 b (8.5%) | 22.5 ± 1.1 c (4.9%) | 20.6 ± 1.3 b (6.3%) | 23.4 ± 0.7 a (3.0%) |
| PPIA | 34.1 ± 0.4 e (1.2%) | 33.5 ± 0.8 d (2.4%) | 32.7 ± 1.6 e (4.9%) | 33.3 ± 1.7 c (5.1%) | 29.4 ± 1.5 d (5.1%) | 27.1 ± 1.7 d (6.3%) | 21.2 ± 1.4 c (6.6%) |
| Gene Symbol | Gene Name | TaqMan Probe |
|---|---|---|
| GAPDH | Glyceraldehyde 3-phosphate dehydrogenase | Hs02786624_g1 |
| ACTB | Beta-actin | Hs01060665_g1 |
| RPLP0 | Large ribosomal protein P0 | Hs00420895_gH |
| B2M | Beta-2-microglobulin | Hs00187842_m1 |
| PPIA | Peptidylprolyl isomerase A | Hs04194521_s1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cadenas, J.; Pors, S.E.; Nikiforov, D.; Zheng, M.; Subiran, C.; Bøtkjær, J.A.; Mamsen, L.S.; Kristensen, S.G.; Andersen, C.Y. Validating Reference Gene Expression Stability in Human Ovarian Follicles, Oocytes, Cumulus Cells, Ovarian Medulla, and Ovarian Cortex Tissue. Int. J. Mol. Sci. 2022, 23, 886. https://doi.org/10.3390/ijms23020886
Cadenas J, Pors SE, Nikiforov D, Zheng M, Subiran C, Bøtkjær JA, Mamsen LS, Kristensen SG, Andersen CY. Validating Reference Gene Expression Stability in Human Ovarian Follicles, Oocytes, Cumulus Cells, Ovarian Medulla, and Ovarian Cortex Tissue. International Journal of Molecular Sciences. 2022; 23(2):886. https://doi.org/10.3390/ijms23020886
Chicago/Turabian StyleCadenas, Jesús, Susanne Elisabeth Pors, Dmitry Nikiforov, Mengxue Zheng, Cristina Subiran, Jane Alrø Bøtkjær, Linn Salto Mamsen, Stine Gry Kristensen, and Claus Yding Andersen. 2022. "Validating Reference Gene Expression Stability in Human Ovarian Follicles, Oocytes, Cumulus Cells, Ovarian Medulla, and Ovarian Cortex Tissue" International Journal of Molecular Sciences 23, no. 2: 886. https://doi.org/10.3390/ijms23020886
APA StyleCadenas, J., Pors, S. E., Nikiforov, D., Zheng, M., Subiran, C., Bøtkjær, J. A., Mamsen, L. S., Kristensen, S. G., & Andersen, C. Y. (2022). Validating Reference Gene Expression Stability in Human Ovarian Follicles, Oocytes, Cumulus Cells, Ovarian Medulla, and Ovarian Cortex Tissue. International Journal of Molecular Sciences, 23(2), 886. https://doi.org/10.3390/ijms23020886






