Label-Free Enrichment of Circulating Tumor Plasma Cells: Future Potential Applications of Dielectrophoresis in Multiple Myeloma
Abstract
1. Introduction
2. The Emerging Prognostic Role of CTCs in Multiple Myeloma
2.1. Clinical Relevance of CTC Detection in Multiple Myeloma
2.2. Platforms Available to Detect CTPCs in Multiple Myeloma
2.2.1. Immune Phenotype of CTPCs
2.2.2. Liquid Biopsy to Analyze ctDNA and cfDNA by NGS
2.2.3. Limitations for Clinical Application of CTPC Detection in Multiple Myeloma
3. Dielectrophoresis (DEP)
3.1. DEP: Phisical Principles
3.2. DEP to Separate Plasma Cells in Peripheral Blood
3.3. DEP Applications in Cancer
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kyle, R.A.; Remstein, E.D.; Therneau, T.M.; Dispenzieri, A.; Kurtin, P.J.; Hodnefield, J.M.; Larson, D.R.; Plevak, M.F.; Jelinek, D.F.; Fonseca, R.; et al. Clinical Course and Prognosis of Smoldering (Asymptomatic) Multiple Myeloma. N. Engl. J. Med. 2007, 356, 2582–2590. [Google Scholar] [CrossRef] [PubMed]
- Manz, R.A.; Thiel, A.; Radbruch, A. Lifetime of plasma cells in the bone marrow. Nature 1997, 388, 133–134. [Google Scholar] [CrossRef] [PubMed]
- Binder, M.; Nandakumar, B.; Rajkumar, S.V.; Kapoor, P.; Buadi, F.K.; Dingli, D.; Lacy, M.Q.; Gertz, M.A.; Hayman, S.R.; Leung, N.; et al. Mortality trends in multiple myeloma after the introduction of novel therapies in the United States. Leukemia 2022, 36, 801–808. [Google Scholar] [CrossRef]
- Rasche, L.; Chavan, S.S.; Stephens, O.W.; Patel, P.H.; Tytarenko, R.; Ashby, C.; Bauer, M.; Stein, C.; Deshpande, S.; Wardell, C.; et al. Spatial genomic heterogeneity in multiple myeloma revealed by multi-region sequencing. Nat. Commun. 2017, 8, 268. [Google Scholar] [CrossRef]
- Tschautscher, M.A.; Jevremovic, A.; Rajkumar, V.; Dispenzieri, A.; Lacy, M.Q.; Gertz, M.A.; Buadi, F.K.; Dingli, D.; Hwa, Y.L.; Fonder, A.L.; et al. Prognostic value of minimal residual disease and polyclonal plasma cells in myeloma patients achieving a complete response to therapy. Am. J. Hematol. 2019, 94, 751–756. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Paiva, B.; Anderson, K.C.; Durie, B.; Landgren, O.; Moreau, P.; Munshi, N.; Lonial, S.; Bladé, J.; Mateos, M.-V.; et al. International Myeloma Working Group consensus criteria for response and minimal residual disease assessment in multiple myeloma. Lancet Oncol. 2016, 17, e328–e346. [Google Scholar] [CrossRef]
- Romano, A.; Palumbo, G.A.; Parrinello, N.L.; Conticello, C.; Martello, M.; Terragna, C. Minimal Residual Disease Assessment within the Bone Marrow of Multiple Myeloma: A Review of Caveats, Clinical Significance and Future Perspectives. Front. Oncol. 2019, 9, 699. [Google Scholar] [CrossRef] [PubMed]
- Huhn, S.; Weinhold, N.; Nickel, J.; Pritsch, M.; Hielscher, T.; Hummel, M.; Bertsch, U.; Huegle-Doerr, B.; Vogel, M.; Angermund, R.; et al. Circulating tumor cells as a biomarker for response to therapy in multiple myeloma patients treated within the GMMG-MM5 trial. Bone Marrow Transplant. 2017, 52, 1194–1198. [Google Scholar] [CrossRef]
- Lohr, J.G.; Kim, S.; Gould, J.; Knoechel, B.; Drier, Y.; Cotton, M.J.; Gray, D.; Birrer, N.; Wong, B.; Ha, G.; et al. Genetic interrogation of circulating multiple myeloma cells at single-cell resolution. Sci. Transl. Med. 2016, 8, 363ra147. [Google Scholar] [CrossRef]
- Manier, S.; Park, J.; Capelletti, M.; Bustoros, M.; Freeman, S.; Ha, G.; Rhoades, J.; Liu, C.J.; Huynh, D.; Reed, S.; et al. Whole-exome sequencing of cell-free DNA and circulating tumor cells in multiple myeloma. Nat. Commun. 2018, 9, 1691. [Google Scholar] [CrossRef]
- Benet, Z.; Jing, Z.; Fooksman, D.R. Plasma cell dynamics in the bone marrow niche. Cell Rep. 2021, 34, 108733. [Google Scholar] [CrossRef] [PubMed]
- Aaron, T.S.; Fooksman, D.R. Dynamic organization of the bone marrow plasma cell niche. FEBS J. 2022, 289, 4228–4239. [Google Scholar] [CrossRef] [PubMed]
- Sui, C.; Zilberberg, J.; Lee, W. Microfluidic device engineered to study the trafficking of multiple myeloma cancer cells through the sinusoidal niche of bone marrow. Sci. Rep. 2022, 12, 1439. [Google Scholar] [CrossRef] [PubMed]
- Ponti, G.; Manfredini, M.; Tomasi, A. Non-blood sources of cell-free DNA for cancer molecular profiling in clinical pathology and oncology. Crit. Rev. Oncol. 2019, 141, 36–42. [Google Scholar] [CrossRef]
- Paiva, B.; Paino, T.; Sayagues, J.-M.; Garayoa, M.; San-Segundo, L.; Martín, M.; Mota, I.; Sanchez, M.-L.; Bárcena, P.; Aires-Mejia, I.; et al. Detailed characterization of multiple myeloma circulating tumor cells shows unique phenotypic, cytogenetic, functional, and circadian distribution profile. Blood 2013, 122, 3591–3598. [Google Scholar] [CrossRef]
- Allegra, A.; Cancemi, G.; Mirabile, G.; Tonacci, A.; Musolino, C.; Gangemi, S. Circulating Tumour Cells, Cell Free DNA and Tumour-Educated Platelets as Reliable Prognostic and Management Biomarkers for the Liquid Biopsy in Multiple Myeloma. Cancers 2022, 14, 4136. [Google Scholar] [CrossRef]
- Sanoja-Flores, L.; Flores-Montero, J.; Garcés, J.J.; Paiva, B.; Puig, N.; García-Mateo, A.; García-Sánchez, O.; Corral-Mateos, A.; Burgos, L.; Blanco, E.; et al. Next generation flow for minimally-invasive blood characterization of MGUS and multiple myeloma at diagnosis based on circulating tumor plasma cells (CTPC). Blood Cancer J. 2018, 8, 117. [Google Scholar] [CrossRef]
- Garcés, J.-J.; Cedena, M.-T.; Puig, N.; Burgos, L.; Perez, J.J.; Cordon, L.; Flores-Montero, J.; Sanoja-Flores, L.; Calasanz, M.-J.; Ortiol, A.; et al. Circulating Tumor Cells for the Staging of Patients with Newly Diagnosed Transplant-Eligible Multiple Myeloma. J. Clin. Oncol. 2022, 40, 3151–3161. [Google Scholar] [CrossRef]
- Bertamini, L.; Oliva, S.; Rota-Scalabrini, D.; Paris, L.; Morè, S.; Corradini, P.; Ledda, A.; Gentile, M.; De Sabbata, G.; Pietrantuono, G.; et al. High Levels of Circulating Tumor Plasma Cells as a Key Hallmark of Aggressive Disease in Transplant-Eligible Patients with Newly Diagnosed Multiple Myeloma. J. Clin. Oncol. 2022, 40, 3120–3131. [Google Scholar] [CrossRef]
- De Rubis, G.; Krishnan, S.R.; Bebawy, M. Liquid Biopsies in Cancer Diagnosis, Monitoring, and Prognosis. Trends Pharmacol. Sci. 2019, 40, 172–186. [Google Scholar] [CrossRef]
- Stella, S.; Vitale, S.R.; Stagno, F.; Massimino, M.; Puma, A.; Tomarchio, C.; Pennisi, M.S.; Tirrò, E.; Romano, C.; Di Raimondo, F.; et al. Impact of different cell counting methods in molecular monitoring in chronic myeloid leukemia patients. Diagnostics 2022, 12, 1051. [Google Scholar] [CrossRef] [PubMed]
- Flores-Montero, J.; Sanoja-Flores, L.; Paiva, B.; Puig, N.; García-Sánchez, O.; Böttcher, S.; Van Der Velden, V.H.J.; Pérez-Morán, J.-J.; Vidriales, M.-B.; García-Sanz, R.; et al. Next Generation Flow for highly sensitive and standardized detection of minimal residual disease in multiple myeloma. Leukemia 2017, 31, 2094–2103. [Google Scholar] [CrossRef] [PubMed]
- Anderson, K.C.; Auclair, D.; Adam, S.J.; Agarwal, A.; Anderson, M.; Avet-Loiseau, H.; Bustoros, M.; Chapman, J.; Connors, D.E.; Dash, A.; et al. Minimal Residual Disease in Myeloma: Application for Clinical Care and New Drug Registration. Clin. Cancer Res. 2021, 27, 5195–5212. [Google Scholar] [CrossRef] [PubMed]
- Sanoja-Flores, L.; Flores-Montero, J.; Puig, N.; Contreras-Sanfeliciano, T.; Pontes, R.; Corral-Mateos, A.; García-Sánchez, O.; Díez-Campelo, M.; Pessoa de Magalhães, R.J.; García-Martín, L.; et al. Blood monitoring of circulating tumor plasma cells by next generation flow in multiple myeloma after therapy. Blood 2019, 134, 2218–2222. [Google Scholar] [CrossRef]
- Botta, C.; Maia, C.D.S.; Garcés, J.-J.; Termini, R.; Perez, C.; Manrique, I.; Burgos, L.; Zabaleta, A.; Alignani, D.; Sarvide, S.; et al. FlowCT for the analysis of large immunophenotypic data sets and biomarker discovery in cancer immunology. Blood Adv. 2022, 6, 690–703. [Google Scholar] [CrossRef]
- Biancon, G.; Gimondi, S.; Vendramin, A.; Carniti, C.; Corradini, P. Noninvasive Molecular Monitoring in Multiple Myeloma Patients Using Cell-Free Tumor DNA: A Pilot Study. J. Mol. Diagn. 2018, 20, 859–870. [Google Scholar] [CrossRef]
- Oberle, A.; Brandt, A.; Voigtlaender, M.; Thiele, B.; Radloff, J.; Schulenkorf, A.; Alawi, M.; Akyüz, N.; März, M.; Ford, C.T.; et al. Monitoring multiple myeloma by next-generation sequencing of V(D)J rearrangements from circulating myeloma cells and cell-free myeloma DNA. Haematologica 2017, 102, 1105–1111. [Google Scholar] [CrossRef]
- Garcés, J.-J.; Bretones, G.; Burgos, L.; Valdes-Mas, R.; Puig, N.; Cedena, M.-T.; Alignani, D.; Rodriguez, I.; Puente, D.Á.; Álvarez, M.G.; et al. Circulating tumor cells for comprehensive and multiregional non-invasive genetic characterization of multiple myeloma. Leukemia 2020, 34, 3007–3018. [Google Scholar] [CrossRef]
- Russo, G.I.; Musso, N.; Romano, A.; Caruso, G.; Petralia, S.; Lanzanò, L.; Broggi, G.; Camarda, M. The Role of Dielectrophoresis for Cancer Diagnosis and Prognosis. Cancers 2022, 14, 198. [Google Scholar] [CrossRef]
- Fischer, J.C.; Niederacher, D.; Topp, S.A.; Honisch, E.; Schumacher, S.; Schmitz, N.; Föhrding, L.Z.; Vay, C.; Hoffmann, I.; Kasprowicz, N.S.; et al. Diagnostic leukapheresis enables reliable detection of circulating tumor cells of nonmetastatic cancer patients. Proc. Natl. Acad. Sci. USA 2013, 110, 16580–16585. [Google Scholar] [CrossRef]
- Rzhevskiy, A.S.; Bazaz, S.R.; Ding, L.; Kapitannikova, A.; Sayyadi, N.; Campbell, D.; Walsh, B.; Gillatt, D.; Warkiani, M.E.; Zvyagin, A.V. Rapid and Label-Free Isolation of Tumour Cells from the Urine of Patients with Localised Prostate Cancer Using Inertial Microfluidics. Cancers 2020, 12, 81. [Google Scholar] [CrossRef] [PubMed]
- Hoque, R.; Mostafid, H.; Hughes, M.P. Rapid, Low-Cost Dielectrophoretic Diagnosis of Bladder Cancer in a Clinical Setting. IEEE J. Transl. Eng. Health Med. 2020, 8, 1–5. [Google Scholar] [CrossRef]
- Maltoni, R.; Fici, P.; Amadori, D.; Gallerani, G.; Cocchi, C.; Zoli, M.; Rocca, A.; Cecconetto, L.; Folli, S.; Scarpi, E.; et al. Circulating tumor cells in early breast cancer: A connection with vascular invasion. Cancer Lett. 2015, 367, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Chiriac, E.; Aveam, M.; Bälan, C. Proceedings of the International Semoconductor Conference (CAS) 2020, Sinaia, Romania, 7–9 October 2020; IEEE: Piscataway, NJ, USA, 2020; pp. 2011–2014. [Google Scholar]
- Gascoyne, P.R.C.; Shim, S. Isolation of Circulating Tumor Cells by Dielectrophoresis. Cancers 2014, 6, 545–579. [Google Scholar] [CrossRef]
- Çağlayan, Z.; Yalçın, Y.D.; Külah, H. A Prominent Cell Manipulation Technique in BioMEMS: Dielectrophoresis. Micromachines 2020, 11, 990. [Google Scholar] [CrossRef] [PubMed]
- Camarda, M.; Fisicaro, G.; Anzalone, R.; Scalese, S.; Alberti, A.; La Via, F.; La Magna, A.; Ballo, A.; Giustolisi, G.; Minafra, L.; et al. Theoretical and experimental study of the role of cell-cell dipole interaction in dielectrophoretic devices: Application to polynomial electrodes. Biomed. Eng. Online 2014, 13, 71. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gascoyne, P.R.C.; Noshari, J.; Anderson, T.J.; Becker, F.F. Isolation of rare cells from cell mixtures by dielectrophoresis. Electrophoresis 2009, 30, 1388–1398. [Google Scholar] [CrossRef]
- Schwarz, G. R. Pethig: Dielectric and Electronic Properties of Biological Materials. John Wiley and Sons, New York, Chichester, Brisbane, Toronto 1979. 376 Seiten, Preis: £15.—. Ber. Bunsenges. Phys. Chem. 1980, 84, 110. [Google Scholar] [CrossRef]
- Shen, Y.; Elele, E.; Khusid, B. A novel concept of dielectrophoretic engine oil filter. Electrophoresis 2011, 32, 2559–2568. [Google Scholar] [CrossRef]
- Yao, J.; Zhu, G.; Zhao, T.; Takei, M. Microfluidic device embedding electrodes for dielectrophoretic manipulation of cells—A review. Electrophoresis 2019, 40, 1166–1177. [Google Scholar] [CrossRef]
- Çetin, B.; Li, D. Dielectrophoresis in microfluidics technology. Electrophoresis 2011, 32, 2410–2427. [Google Scholar] [CrossRef] [PubMed]
- Pethig, R. (Keynote) Dielectrophoresis: Exploring the ‘ 2nd Frontier’ of Its Application in the Biomedical Sciences. In ECS Meeting Abstracts MA2015-01; IOP Publishing: Bristol, UK, 2015. [Google Scholar] [CrossRef]
- Pethig, R.; Kell, D. The passive electrical properties of biological systems: Their significance in physiology, biophysics and biotechnology. Phys. Med. Biol. 1987, 32, 933–970. [Google Scholar] [CrossRef] [PubMed]
- Ratanachoo, K.; Gascoyne, P.R.; Ruchirawat, M. Detection of cellular responses to toxicants by dielectrophoresis. Biochim. Biophys. Acta (BBA) -Biomembr. 2002, 1564, 449–458. [Google Scholar] [CrossRef]
- Ho, S.N. Intracellular water homeostasis and the mammalian cellular osmotic stress response. J. Cell. Physiol. 2006, 206, 9–15. [Google Scholar] [CrossRef]
- Chung, C.; Waterfall, M.; Pells, S.; Menachery, A.; Smith, S.; Pethig, R. Dielectrophoretic Characterisation of Mammalian Cells above 100 MHz. J. Electr. Bioimpedance 2011, 2, 64–71. [Google Scholar] [CrossRef]
- Puttaswamy, S.V.; Sivashankar, S.; Chen, R.-J.; Chin, C.-K.; Chang, H.-Y.; Liu, C.H. Enhanced cell viability and cell adhesion using low conductivity medium for negative dielectrophoretic cell patterning. Biotechnol. J. 2010, 5, 1005–1015. [Google Scholar] [CrossRef] [PubMed]
- Lambert, E.; Manczak, R.; Barthout, E.; Saada, S.; Porcù, E.; Maule, F.; Bessette, B.; Viola, G.; Persano, L.; Dalmay, C.; et al. Microfluidic Lab-on-a-Chip Based on UHF-Dielectrophoresis for Stemness Phenotype Characterization and Discrimination among Glioblastoma Cells. Biosensors 2021, 11, 388. [Google Scholar] [CrossRef]
- Qian, C.; Huang, H.; Chen, L.; Li, X.; Ge, Z.; Chen, T.; Yang, Z.; Sun, L. Dielectrophoresis for Bioparticle Manipulation. Int. J. Mol. Sci. 2014, 15, 18281–18309. [Google Scholar] [CrossRef]
- Salmanzadeh, A.; Sano, M.B.; Shafiee, H.; Davalos, R.V. Isolation of rare cáncer cells from blood cells using dielectrophoresis. In Proceedings of the 2012 Annual International Conference of the IEEE Engineering in Medicine and Biology Society, San Diego, CA, USA, 28 August–1 September 2012. [Google Scholar]
- Shim, S.; Stemke-Hale, K.; Noshari, J.; Becker, F.F.; Gascoyne, P.R.C. Dielectrophoresis has broad applicability to marker-free isolation of tumor cells from blood by microfluidic systems. Biomicrofluidics 2013, 7, 011808. [Google Scholar] [CrossRef]
- Guido, I.; Jaeger, M.S.; Duschl, C. Dielectrophoretic stretching of cells allows for characterization of their mechanical properties. Eur. Biophys. J. 2011, 40, 281–288. [Google Scholar] [CrossRef]
- Cross, S.E.; Jin, Y.S.; Rao, J.; Gimzewski, J.K. Nanomechanical analysis of cells from cancer patients. Nat. Nanotechnol. 2007, 2, 780–783. [Google Scholar] [CrossRef] [PubMed]
- Pohl, H.A.; Crane, J.S. Dielectrophoresis of cells. Biophys. J. 1971, 11, 711–727. [Google Scholar] [CrossRef]
- Chung, C.; Pethig, R.; Smith, S.; Waterfall, M. Intracellular potassium under osmotic stress determines the dielectrophoresis cross-over frequency of murine myeloma cells in the MHz range. Electrophoresis 2018, 39, 989–997. [Google Scholar] [CrossRef] [PubMed]
- Liang, W.; Zhao, Y.; Liu, L.; Wang, Y.; Dong, Z.; Li, W.J.; Lee, G.-B.; Xiao, X.; Zhang, W. Rapid and Label-Free Separation of Burkitt’s Lymphoma Cells from Red Blood Cells by Optically-Induced Electrokinetics. PLoS ONE 2014, 9, e90827. [Google Scholar] [CrossRef] [PubMed]
- Alshareef, M.; Metrakos, N.; Perez, E.J.; Azer, F.; Yang, F.; Yang, X.; Wang, G. Separation of tumor cells with dielectrophoresis-based microfluidic chip. Biomicrofluidics 2013, 7, 011803. [Google Scholar] [CrossRef]
- An, J.; Lee, J.; Lee, S.H.; Park, J.; Kim, B. Separation of malignant human breast cancer epithelial cells from healthy epithelial cells using an advanced dielectrophoresis-activated cell sorter (DACS). Anal. Bioanal. Chem. 2009, 394, 801–809. [Google Scholar] [CrossRef]
- Cheng, J.; Liu, Y.; Zhao, Y.; Zhang, L.; Zhang, L.; Mao, H.; Huang, C. Nanotechnology-Assisted Isolation and Analysis of Circulating Tumor Cells on Microfluidic Devices. Micromachines 2020, 11, 774. [Google Scholar] [CrossRef]
- Ribeiro-Samy, S.; Oliveira, M.; Pereira-Veiga, T.; Muinelo-Romay, L.; Carvalho, S.; Gaspar, J.; Freitas, P.P.; López-López, R.; Costa, C.; Diéguez, L. Fast and efficient microfluidic cell filter for isolation of circulating tumor cells from unprocessed whole blood of colorectal cancer patients. Sci. Rep. 2019, 9, 8032. [Google Scholar] [CrossRef]
- Labeed, F.; Coley, H.M.; Thomas, H.; Hughes, M.P. Assessment of Multidrug Resistance Reversal Using Dielectrophoresis and Flow Cytometry. Biophys. J. 2003, 85, 2028–2034. [Google Scholar] [CrossRef]
- Cohen, K.; Emmanuel, R.; Kisin-Finfer, E.; Shabat, D.; Peer, D. Modulation of Drug Resistance in Ovarian Adenocarcinoma Using Chemotherapy Entrapped in Hyaluronan-Grafted Nanoparticle Clusters. ACS Nano 2014, 8, 2183–2195. [Google Scholar] [CrossRef]
- Yalçın, Y.D.; Töral, T.B.; Sukas, S.; Yıldırım, E.; Zorlu, Ö.; Gündüz, U.; Külah, H. A microfluidic device enabling drug resistance analysis of leukemia cells via coupled dielectrophoretic detection and impedimetric counting. Sci. Rep. 2021, 11, 13193. [Google Scholar] [CrossRef] [PubMed]
- Yalçın, Y.D.; Sukas, S.; Töral, T.B.; Gündüz, U.; Külah, H. Exploring the relationship between cytoplasmic ion content variation and multidrug resistance in cancer cells via ion-release based impedance spectroscopy. Sens. Actuators B Chem. 2019, 290, 180–187. [Google Scholar] [CrossRef]
- Duncan, L.; Shelmerdine, H.; Hughes, M.; Coley, H.M.; Hübner, Y.; Labeed, F. Dielectrophoretic analysis of changes in cytoplasmic ion levels due to ion channel blocker action reveals underlying differences between drug-sensitive and multidrug-resistant leukaemic cells. Phys. Med. Biol. 2008, 53, N1–N7. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhu, F.; Dan, W.; Fu, Y.; Liu, S. Construction of carbon nanotube based nanoarchitectures for selective impedimetric detection of cancer cells in whole blood. Analyst 2014, 139, 5086–5092. [Google Scholar] [CrossRef]
- Varmazyari, V.; Habibiyan, H.; Ghafoorifard, H.; Ebrahimi, M.; Ghafouri-Fard, S. A dielectrophoresis-based microfluidic system having double-sided optimized 3D electrodes for label-free cancer cell separation with preserving cell viability. Sci. Rep. 2022, 12, 12100. [Google Scholar] [CrossRef]
- Graham, K.A.; Mulhall, H.J.; Labeed, F.H.; Lewis, M.P.; Hoettges, K.F.; Kalavrezos, N.; McCaul, J.; Liew, C.; Porter, S.; Fedele, S.; et al. A dielectrophoretic method of discrimination between normal oral epithelium, and oral and oropharyngeal cancer in a clinical setting. Analyst 2015, 140, 5198–5204. [Google Scholar] [CrossRef]
- Velmanickam, L.; Fondakowski, M.; Nawarathna, D. Integrated dielectrophoresis and fluorescence-based platform for biomarker detection from serum samples. Biomed. Phys. Eng. Express 2018, 4, 025018. [Google Scholar] [CrossRef]

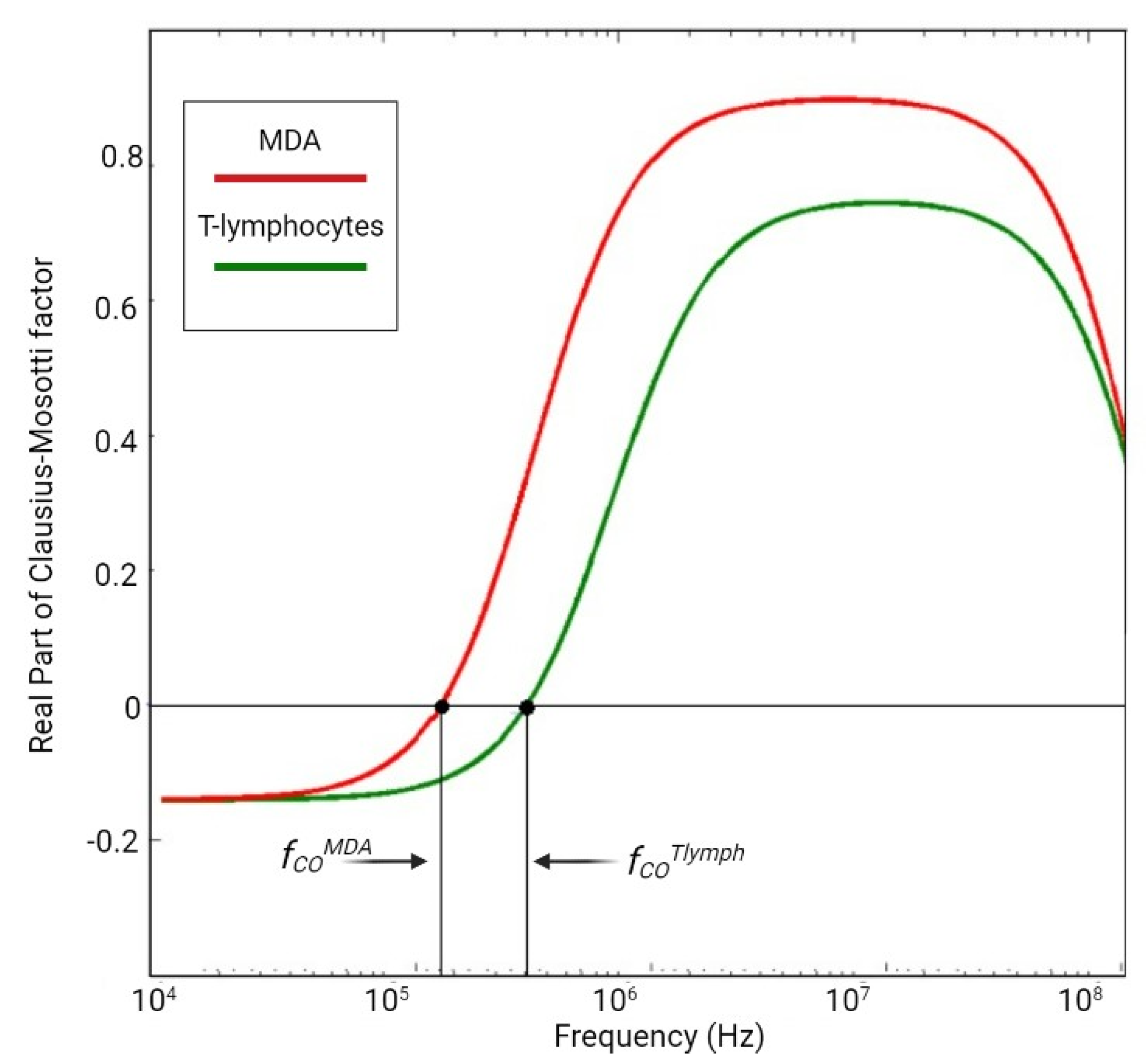
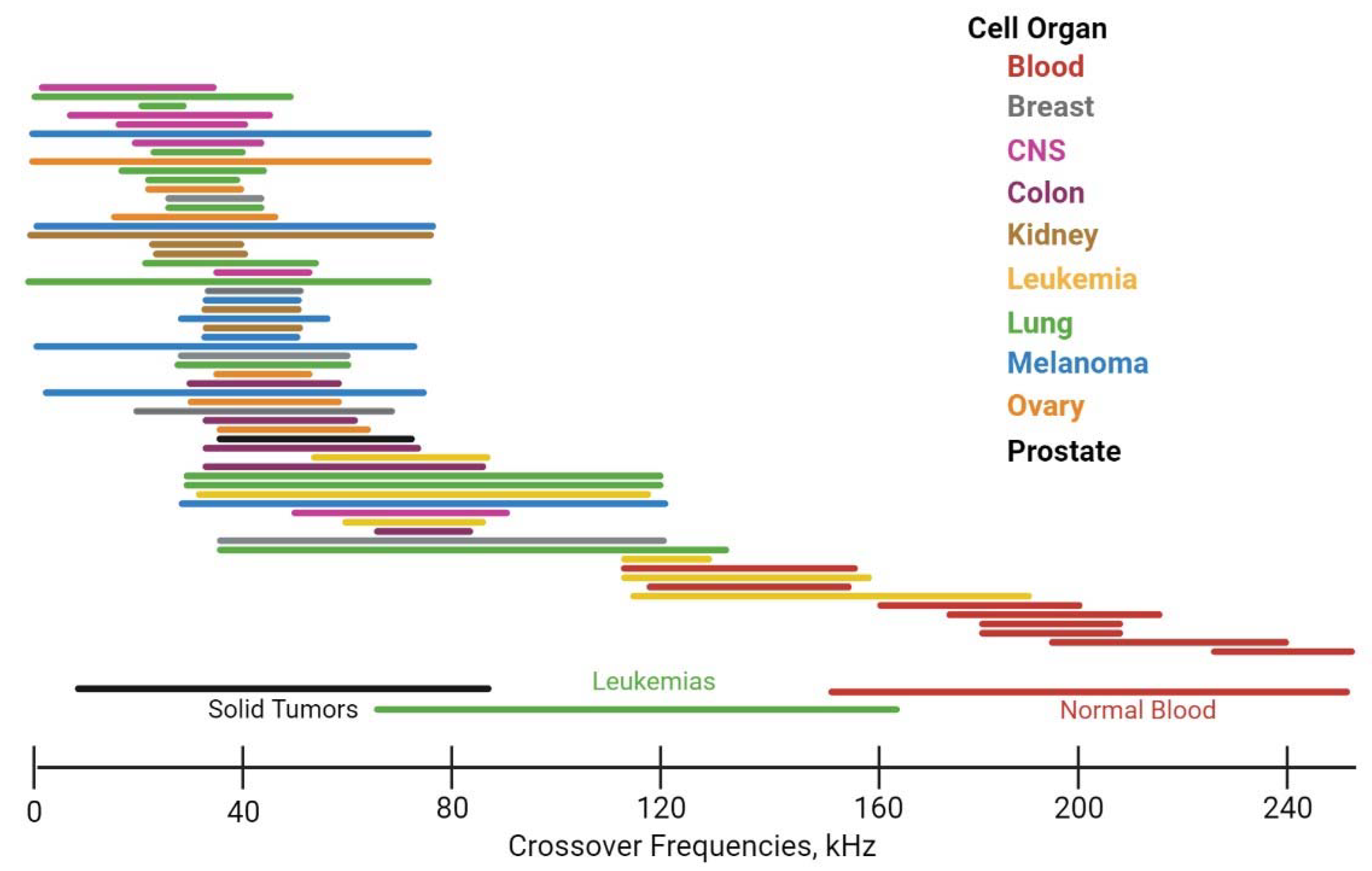
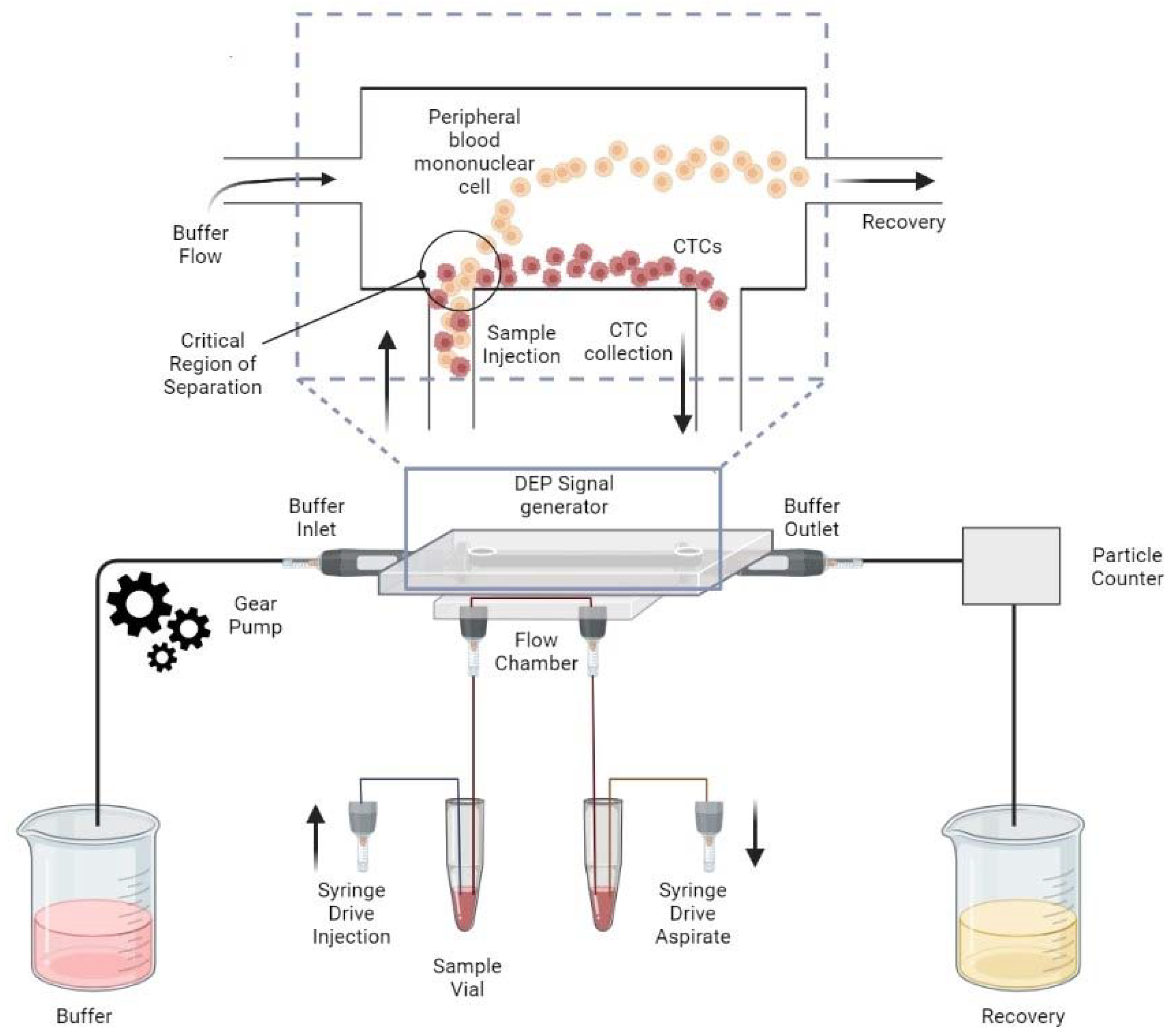
| Platform | Pros | Cons |
|---|---|---|
| Multicolour Flow Cytometry (MCF) |
|
|
| Next-Generation Flow Cytometry (NGF) |
|
|
| Next-generation sequencing (NGS) |
|
|
| GeneScanning |
|
|
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Musso, N.; Romano, A.; Bonacci, P.G.; Scandura, G.; Pandino, C.; Camarda, M.; Russo, G.I.; Di Raimondo, F.; Cacciola, E.; Cacciola, R. Label-Free Enrichment of Circulating Tumor Plasma Cells: Future Potential Applications of Dielectrophoresis in Multiple Myeloma. Int. J. Mol. Sci. 2022, 23, 12052. https://doi.org/10.3390/ijms231912052
Musso N, Romano A, Bonacci PG, Scandura G, Pandino C, Camarda M, Russo GI, Di Raimondo F, Cacciola E, Cacciola R. Label-Free Enrichment of Circulating Tumor Plasma Cells: Future Potential Applications of Dielectrophoresis in Multiple Myeloma. International Journal of Molecular Sciences. 2022; 23(19):12052. https://doi.org/10.3390/ijms231912052
Chicago/Turabian StyleMusso, Nicolò, Alessandra Romano, Paolo Giuseppe Bonacci, Grazia Scandura, Clarissa Pandino, Massimo Camarda, Giorgio Ivan Russo, Francesco Di Raimondo, Emma Cacciola, and Rossella Cacciola. 2022. "Label-Free Enrichment of Circulating Tumor Plasma Cells: Future Potential Applications of Dielectrophoresis in Multiple Myeloma" International Journal of Molecular Sciences 23, no. 19: 12052. https://doi.org/10.3390/ijms231912052
APA StyleMusso, N., Romano, A., Bonacci, P. G., Scandura, G., Pandino, C., Camarda, M., Russo, G. I., Di Raimondo, F., Cacciola, E., & Cacciola, R. (2022). Label-Free Enrichment of Circulating Tumor Plasma Cells: Future Potential Applications of Dielectrophoresis in Multiple Myeloma. International Journal of Molecular Sciences, 23(19), 12052. https://doi.org/10.3390/ijms231912052










