Molecular Events of Rice AP2/ERF Transcription Factors
Abstract
1. Introduction
2. Classification, Features and Binding Elements
3. Molecular Roles of AP2/ERF TFs in Rice
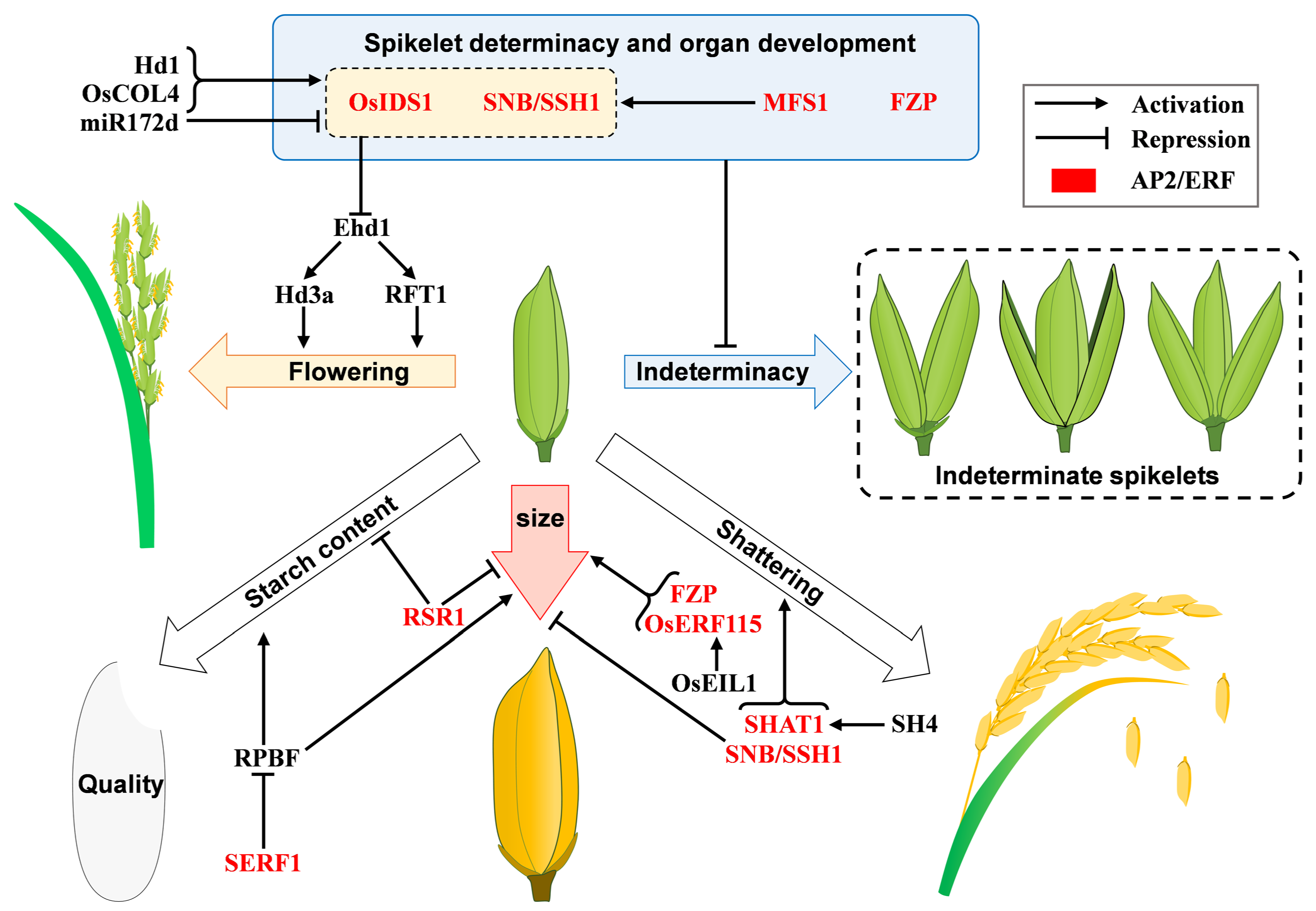
3.1. Spikelet Determinacy and Organ Development
3.2. Flowering Regulation
3.3. Role in Grain Size, Quality and Shattering
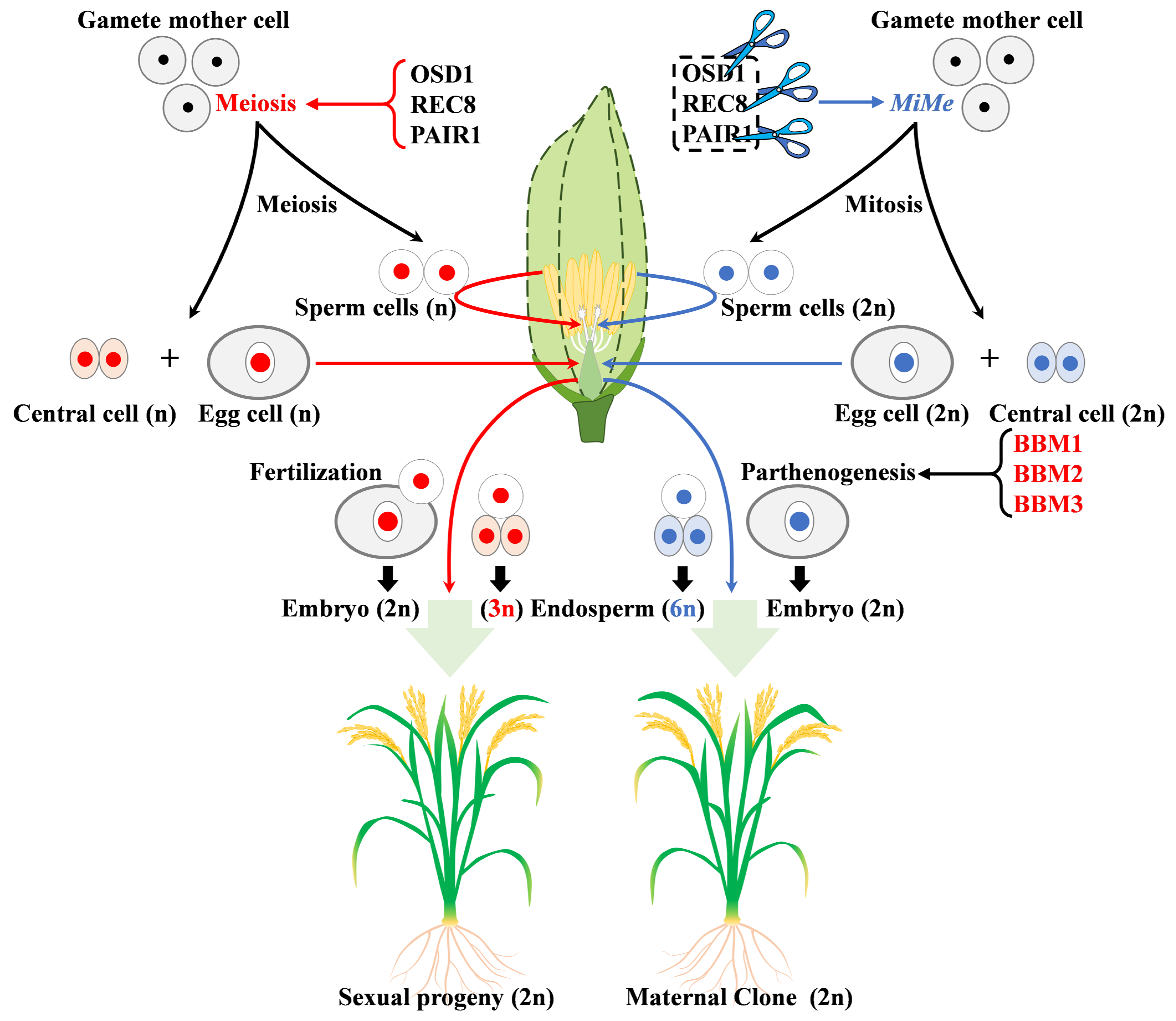
3.4. Role in Embryogenesis
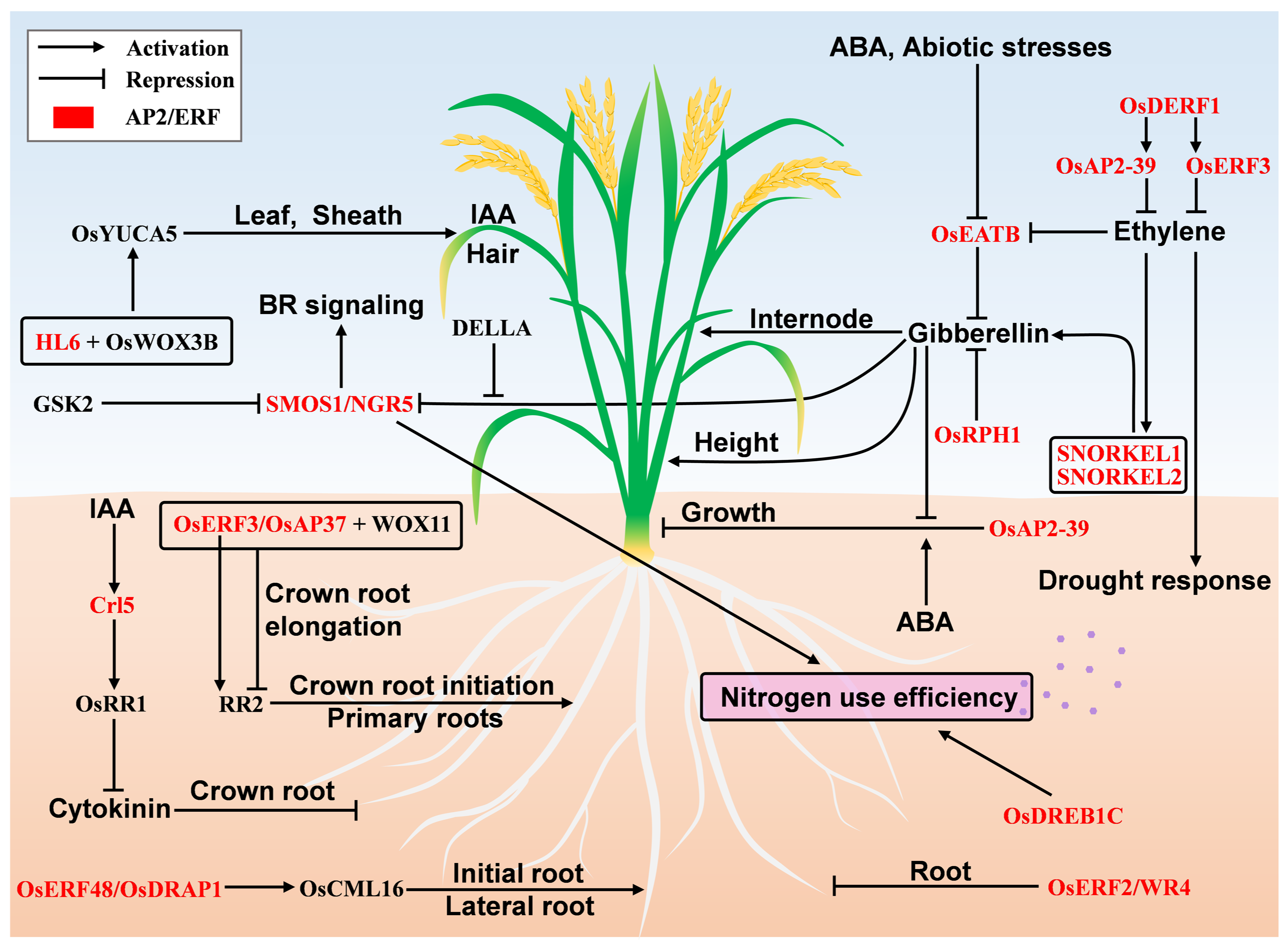
3.5. Root Initiation and Formation
3.6. AP2/ERF Regulatory Roles Mediated by Hormones
3.7. Regulation of Nutrient Use Efficiency
3.8. Role of AP2/ERF TFs in Stress Response
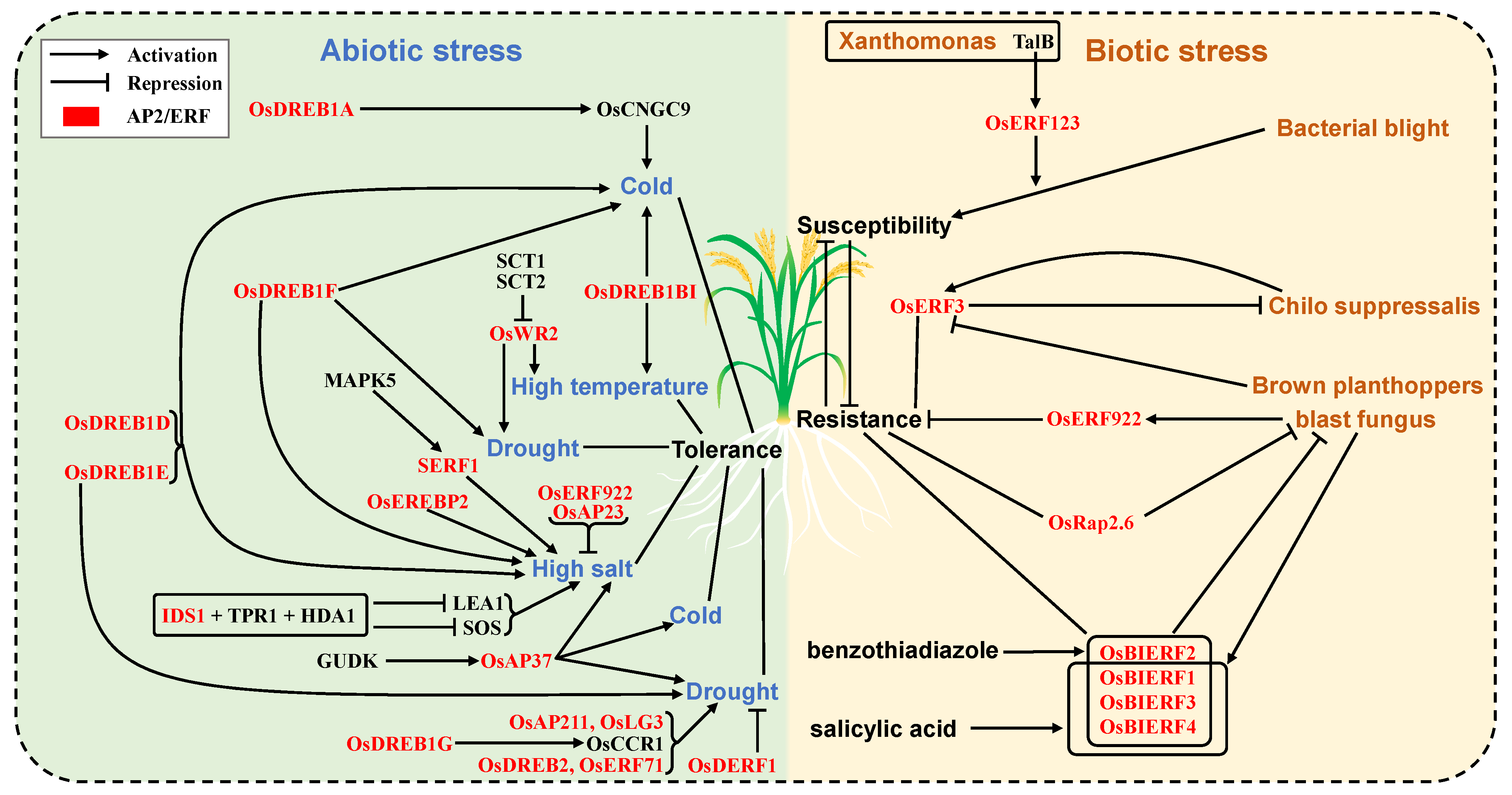
3.8.1. Abiotic Stress Response and Tolerance
3.8.2. Biotic Stress Response and Tolerance
4. Discussion and Prospects
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Feng, K.; Hou, X.L.; Xing, G.M.; Liu, J.X.; Duan, A.Q.; Xu, Z.S.; Li, M.Y.; Zhuang, J.; Xiong, A.S. Advances in AP2/ERF super-family transcription factors in plant. Crit. Rev. Biotechnol. 2020, 40, 750–776. [Google Scholar] [CrossRef] [PubMed]
- Jofuku, K.D.; den Boer, B.G.; Van Montagu, M.; Okamuro, J.K. Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 1994, 6, 1211–1225. [Google Scholar] [PubMed]
- Okamuro, J.K.; Caster, B.; Villarroel, R.; Van Montagu, M.; Jofuku, K.D. The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proc. Natl. Acad. Sci. USA 1997, 94, 7076–7081. [Google Scholar] [CrossRef] [PubMed]
- Riechmann, J.L.; Meyerowitz, E.M. The AP2/EREBP family of plant transcription factors. Biol. Chem. 1998, 379, 633–646. [Google Scholar] [PubMed]
- Magnani, E.; Sjölander, K.; Hake, S. From endonucleases to transcription factors: Evolution of the AP2 DNA binding domain in plants. Plant Cell 2004, 16, 2265–2277. [Google Scholar] [CrossRef] [PubMed]
- Nakano, T.; Suzuki, K.; Fujimura, T.; Shinshi, H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 2006, 140, 411–432. [Google Scholar] [CrossRef] [PubMed]
- Sakuma, Y.; Liu, Q.; Dubouzet, J.G.; Abe, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 2002, 290, 998–1009. [Google Scholar] [CrossRef]
- Sharoni, A.M.; Nuruzzaman, M.; Satoh, K.; Shimizu, T.; Kondoh, H.; Sasaya, T.; Choi, I.R.; Omura, T.; Kikuchi, S. Gene structures, classification and expression models of the AP2/EREBP transcription factor family in rice. Plant Cell Physiol. 2011, 52, 344–360. [Google Scholar] [CrossRef]
- Rashid, M.; Guangyuan, H.; Guangxiao, Y.; Hussain, J.; Xu, Y. AP2/ERF Transcription Factor in Rice: Genome-Wide Canvas and Syntenic Relationships between Monocots and Eudicots. Evol. Bioinform. 2012, 8, 321–355. [Google Scholar] [CrossRef]
- Licausi, F.; Ohme-Takagi, M.; Perata, P. APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factors: Mediators of stress responses and developmental programs. New Phytol. 2013, 199, 639–649. [Google Scholar] [CrossRef]
- Shigyo, M.; Ito, M. Analysis of gymnosperm two-AP2-domain-containing genes. Dev. Genes Evol. 2004, 214, 105–114. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 1994, 6, 251–264. [Google Scholar] [PubMed]
- Ohme-Takagi, M.; Shinshi, H. Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 1995, 7, 173–182. [Google Scholar] [PubMed]
- Agarwal, P.K.; Agarwal, P.; Reddy, M.K.; Sopory, S.K. Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Rep. 2006, 25, 1263–1274. [Google Scholar] [CrossRef]
- Sharma, M.K.; Kumar, R.; Solanke, A.U.; Sharma, R.; Tyagi, A.K.; Sharma, A.K. Identification, phylogeny, and transcript profiling of ERF family genes during development and abiotic stress treatments in tomato. Mol. Genet. Genomics 2010, 284, 455–475. [Google Scholar] [CrossRef]
- Gu, C.; Guo, Z.H.; Hao, P.P.; Wang, G.M.; Jin, Z.M.; Zhang, S.L. Multiple regulatory roles of AP2/ERF transcription factor in angiosperm. Bot. Stud. 2017, 58, 6. [Google Scholar] [CrossRef]
- Ren, D.Y.; Li, Y.F.; He, G.H.; Qian, Q. Multifloret spikelet improves rice yield. New Phytol. 2020, 225, 2301–2306. [Google Scholar] [CrossRef]
- Ren, D.Y.; Li, Y.F.; Zhao, F.M.; Sang, X.C.; Shi, J.Q.; Wang, N.; Guo, S.; Ling, Y.H.; Zhang, C.; Yang, Z.; et al. MULTI-FLORET SPIKELET1, which encodes an AP2/ERF protein, determines spikelet meristem fate and sterile lemma identity in rice. Plant Physiol. 2013, 162, 872–884. [Google Scholar] [CrossRef]
- Ren, D.Y.; Hu, J.; Xu, Q.K.; Cui, Y.J.; Zhang, Y.; Zhou, T.T.; Rao, Y.C.; Xue, D.W.; Zeng, D.L.; Zhang, G.H.; et al. FZP determines grain size and sterile lemma fate in rice. J. Exp. Bot. 2018, 69, 4853–4866. [Google Scholar] [CrossRef]
- Komatsu, M.; Chujo, A.; Nagato, Y.; Shimamoto, K.; Kyozuka, J. FRIZZY PANICLE is required to prevent the formation of axillary meristems and to establish floral meristem identity in rice spikelets. Development 2003, 130, 3841–3850. [Google Scholar] [CrossRef]
- Lee, D.Y.; Lee, J.; Moon, S.; Park, S.Y.; An, G. The rice heterochronic gene SUPERNUMERARY BRACT regulates the transition from spikelet meristem to floral meristem. Plant J. 2007, 49, 64–78. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.Y.; An, G. Two AP2 family genes, SUPERNUMERARY BRACT (SNB) and OsINDETERMINATE SPIKELET 1 (OsIDS1), synergistically control inflorescence architecture and floral meristem establishment in rice. Plant J. 2012, 69, 445–461. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Lee, D.Y.; Cho, L.H.; An, G. Rice miR172 induces flowering by suppressing OsIDS1 and SNB, two AP2 genes that negatively regulate expression of Ehd1 and florigens. Rice 2014, 7, 31. [Google Scholar] [CrossRef]
- Ma, X.S.; Feng, F.J.; Zhang, Y.; Elesawi, I.E.; Xu, K.; Li, T.F.; Mei, H.W.; Liu, H.Y.; Gao, N.N.; Chen, C.L.; et al. A novel rice grain size gene OsSNB was identified by genome-wide association study in natural population. PLoS Genet. 2019, 15, e1008191. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.Y.; Ma, X.; Zhao, S.S.; Tang, Y.Y.; Liu, F.X.; Gu, P.; Fu, Y.C.; Zhu, Z.F.; Cai, H.W.; Sun, C.Q.; et al. The APETALA2-Like Transcription Factor SUPERNUMERARY BRACT Controls Rice Seed Shattering and Seed Size. Plant Cell 2019, 31, 17–36. [Google Scholar] [CrossRef]
- Zhou, Y.; Lu, D.F.; Li, C.Y.; Luo, J.H.; Zhu, B.F.; Zhu, J.J.; Shangguan, Y.Y.; Wang, Z.X.; Sang, T.; Zhou, B.; et al. Genetic control of seed shattering in rice by the APETALA2 transcription factor shattering abortion1. Plant Cell 2012, 24, 1034–1048. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Ma, T.; Yuan, D.Y.; Zhou, Y.; Long, Y.; Li, Z.W.; Dong, Z.Y.; Duan, M.J.; Yu, D.; Jing, Y.Z.; et al. The OsEIL1-OsERF115-target gene regulatory module controls grain size and weight in rice. Plant Biotechnol. J. 2022, 20, 1470–1486. [Google Scholar] [CrossRef]
- Bai, X.F.; Huang, Y.; Hu, Y.; Liu, H.Y.; Zhang, B.; Smaczniak, C.; Hu, G.; Han, Z.M.; Xing, Y. Duplication of an upstream silencer of FZP increases grain yield in rice. Nat. Plants 2017, 3, 885–893. [Google Scholar] [CrossRef]
- Schmidt, R.; Schippers, J.H.; Mieulet, D.; Watanabe, M.; Hoefgen, R.; Guiderdoni, E.; Mueller-Roeber, B. SALT-RESPONSIVE ERF1 is a negative regulator of grain filling and gibberellin-mediated seedling establishment in rice. Mol. Plant 2014, 7, 404–421. [Google Scholar] [CrossRef]
- Fu, F.F.; Xue, H.W. Coexpression analysis identifies Rice Starch Regulator1, a rice AP2/EREBP family transcription factor, as a novel rice starch biosynthesis regulator. Plant Physiol. 2010, 154, 927–938. [Google Scholar] [CrossRef]
- Khanday, I.; Skinner, D.; Yang, B.; Mercier, R.; Sundaresan, V. A male-expressed rice embryogenic trigger redirected for asexual propagation through seeds. Nature 2019, 565, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Xing, Y.D.; Wang, N.; Zhang, T.Q.; Zhang, Q.L.; Du, D.; Chen, X.L.; Lu, X.; Zhang, Y.Y.; Zhu, M.D.; Liu, M.M.; et al. SHORT-ROOT 1 is critical to cell division and tracheary element development in rice roots. Plant J. 2021, 105, 1179–1191. [Google Scholar] [CrossRef] [PubMed]
- Kitomi, Y.; Kitano, H.; Inukai, Y. Molecular mechanism of crown root initiation and the different mechanisms between crown root and radicle in rice. Plant Signal. Behav. 2011, 6, 1270–1278. [Google Scholar] [CrossRef] [PubMed]
- Kitomi, Y.; Ito, H.; Hobo, T.; Aya, K.; Kitano, H.; Inukai, Y. The auxin responsive AP2/ERF transcription factor CROWN ROOTLESS5 is involved in crown root initiation in rice through the induction of OsRR1, a type-A response regulator of cytokinin signaling. Plant J. 2011, 67, 472–484. [Google Scholar] [CrossRef]
- Jung, H.; Chung, P.J.; Park, S.H.; Redillas, M.; Kim, Y.S.; Suh, J.W.; Kim, J.K. Overexpression of OsERF48 causes regulation of OsCML16, a calmodulin-like protein gene that enhances root growth and drought tolerance. Plant Biotechnol. J. 2017, 15, 1295–1308. [Google Scholar] [CrossRef]
- Zhao, Y.; Cheng, S.F.; Song, Y.L.; Huang, Y.L.; Zhou, S.L.; Liu, X.Y.; Zhou, D.X. The Interaction between Rice ERF3 and WOX11 Promotes Crown Root Development by Regulating Gene Expression Involved in Cytokinin Signaling. Plant Cell 2015, 27, 2469–2483. [Google Scholar] [CrossRef]
- Wan, L.Y.; Zhang, J.F.; Zhang, H.W.; Zhang, Z.J.; Quan, R.D.; Zhou, S.R.; Huang, R.F. Transcriptional activation of OsDERF1 in OsERF3 and OsAP2-39 negatively modulates ethylene synthesis and drought tolerance in rice. PLoS ONE 2011, 6, e25216. [Google Scholar] [CrossRef]
- Hattori, Y.; Nagai, K.; Furukawa, S.; Song, X.J.; Kawano, R.; Sakakibara, H.; Wu, J.; Matsumoto, T.; Yoshimura, A.; Kitano, H.; et al. The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature 2009, 460, 1026–1030. [Google Scholar] [CrossRef]
- Xiao, G.Q.; Qin, H.; Zhou, J.H.; Quan, R.D.; Lu, X.Y.; Huang, R.F.; Zhang, H.W. OsERF2 controls rice root growth and hormone responses through tuning expression of key genes involved in hormone signaling and sucrose metabolism. Plant Mol. Biol. 2016, 90, 293–302. [Google Scholar] [CrossRef]
- Qi, W.W.; Sun, F.; Wang, Q.J.; Chen, M.L.; Huang, Y.Q.; Feng, Y.Q.; Luo, X.J.; Yang, J.S. Rice ethylene-response AP2/ERF factor OsEATB restricts internode elongation by down-regulating a gibberellin biosynthetic gene. Plant Physiol. 2011, 157, 216–228. [Google Scholar] [CrossRef]
- Ma, Z.M.; Wu, T.; Huang, K.; Jin, Y.M.; Li, Z.; Chen, M.J.; Yun, S.; Zhang, H.J.; Yang, X.; Chen, H.Y.; et al. A Novel AP2/ERF Transcription Factor, OsRPH1, Negatively Regulates Plant Height in Rice. Front. Plant Sci. 2020, 11, 709. [Google Scholar] [CrossRef] [PubMed]
- Yaish, M.W.; El-Kereamy, A.; Zhu, T.; Beatty, P.H.; Good, A.G.; Bi, Y.M.; Rothstein, S.J. The APETALA-2-like transcription factor OsAP2-39 controls key interactions between abscisic acid and gibberellin in rice. PLoS Genet. 2010, 6, e1001098. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; Wang, S.S.; Song, W.Z.; Zhang, J.Q.; Wang, Y.; Liu, Q.; Yu, J.P.; Ye, Y.F.; Li, S.; Chen, J.F.; et al. Enhanced sustainable green revolution yield via nitrogen-responsive chromatin modulation in rice. Science 2020, 367, eaaz2046. [Google Scholar] [CrossRef] [PubMed]
- Qiao, S.L.; Sun, S.Y.; Wang, L.L.; Wu, Z.H.; Li, C.X.; Li, X.M.; Wang, T.; Leng, L.N.; Tian, W.S.; Lu, T.G.; et al. The RLA1/SMOS1 Transcription Factor Functions with OsBZR1 to Regulate Brassinosteroid Signaling and Rice Architecture. Plant Cell 2017, 29, 292–309. [Google Scholar] [CrossRef] [PubMed]
- Hirano, K.; Yoshida, H.; Aya, K.; Kawamura, M.; Hayashi, M.; Hobo, T.; Sato-Izawa, K.; Kitano, H.; Ueguchi-Tanaka, M.; Matsuoka, M. SMALL ORGAN SIZE 1 and SMALL ORGAN SIZE 2/DWARF AND LOW-TILLERING Form a Complex to Integrate Auxin and Brassinosteroid Signaling in Rice. Mol. Plant 2017, 10, 590–604. [Google Scholar] [CrossRef]
- Sun, W.Q.; Gao, D.W.; Xiong, Y.; Tang, X.X.; Xiao, X.F.; Wang, C.R.; Yu, S.B. Hairy Leaf 6, an AP2/ERF Transcription Factor, Interacts with OsWOX3B and Regulates Trichome Formation in Rice. Mol. Plant 2017, 10, 1417–1433. [Google Scholar] [CrossRef]
- Wei, S.B.; Li, X.; Lu, Z.F.; Zhang, H.; Ye, X.Y.; Zhou, Y.J.; Li, J.; Yan, Y.Y.; Pei, H.C.; Duan, F.Y.; et al. A transcriptional regulator that boosts grain yields and shortens the growth duration of rice. Science 2022, 377, eabi8455. [Google Scholar] [CrossRef]
- Wang, J.C.; Ren, Y.L.; Liu, X.; Luo, S.; Zhang, X.; Liu, X.; Lin, Q.B.; Zhu, S.S.; Wan, H.; Yang, Y.; et al. Transcriptional activation and phosphorylation of OsCNGC9 confer enhanced chilling tolerance in rice. Mol. Plant 2021, 14, 315–329. [Google Scholar] [CrossRef]
- Qin, Q.L.; Liu, J.G.; Zhang, Z.; Peng, R.H.; Xiong, A.S.; Yao, Q.H.; Chen, J.M. Isolation, optimization, and functional analysis of the cDNA encoding transcription factor OsDREB1B in Oryza Sativa L. Mol. Breed. 2007, 19, 329–340. [Google Scholar] [CrossRef]
- Gutha, L.R.; Reddy, A.R. Rice DREB1B promoter shows distinct stress-specific responses, and the overexpression of cDNA in tobacco confers improved abiotic and biotic stress tolerance. Plant Mol. Biol. 2008, 68, 533–555. [Google Scholar] [CrossRef]
- Figueiredo, D.D.; Barros, P.M.; Cordeiro, A.M.; Serra, T.S.; Lourenço, T.; Chander, S.; Oliveira, M.M.; Saibo, N.J. Seven zinc-finger transcription factors are novel regulators of the stress responsive gene OsDREB1B. J. Exp. Bot. 2012, 63, 3643–3656. [Google Scholar] [CrossRef] [PubMed]
- Mao, D.; Chen, C. Colinearity and similar expression pattern of rice DREB1s reveal their functional conservation in the cold-responsive pathway. PLoS ONE 2012, 7, e47275. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.Q.; Meng, X.P.; Zhang, Y.; Xia, M.; Wang, X.P. Over-expression of OsDREB genes lead to enhanced drought tolerance in rice. Biotechnol. Lett. 2008, 30, 2191–2198. [Google Scholar] [CrossRef]
- Wang, Q.Y.; Guan, Y.C.; Wu, Y.R.; Chen, H.L.; Chen, F.; Chu, C.C. Overexpression of a rice OsDREB1F gene increases salt, drought, and low temperature tolerance in both Arabidopsis and rice. Plant Mol. Biol. 2008, 67, 589–602. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.H.; Li, X.P.; Zhou, H.L.; Zhang, J.S.; Gong, Z.Z.; Chen, S.Y. OsDREB4 Genes in Rice Encode AP2-Containing Proteins that Bind Specifically to the Dehydration-Responsive Element. J. Integr. Plant Biol. 2005, 47, 467–476. [Google Scholar] [CrossRef]
- Zhou, X.Y.; Jenks, M.A.; Liu, J.; Liu, A.L.; Zhang, X.W.; Xiang, J.H.; Zou, J.; Peng, Y.; Chen, X.B. Overexpression of Transcription Factor OsWR2 Regulates Wax and Cutin Biosynthesis in Rice and Enhances its Tolerance to Water Deficit. Plant Mol. Biol. Rep. 2014, 32, 719–731. [Google Scholar] [CrossRef]
- Kan, Y.; Mu, X.R.; Zhang, H.; Gao, J.; Shan, J.X.; Ye, W.W.; Lin, H.X. TT2 controls rice thermotolerance through SCT1-dependent alteration of wax biosynthesis. Nat. Plants 2022, 8, 53–67. [Google Scholar] [CrossRef]
- Lee, D.K.; Jung, H.; Jang, G.; Jeong, J.S.; Kim, Y.S.; Ha, S.H.; Do Choi, Y.; Kim, J.K. Overexpression of the OsERF71 Transcription Factor Alters Rice Root Structure and Drought Resistance. Plant Physiol. 2016, 172, 575–588. [Google Scholar] [CrossRef] [PubMed]
- Xiong, H.Y.; Yu, J.P.; Miao, J.L.; Li, J.J.; Zhang, H.L.; Wang, X.; Liu, P.L.; Zhao, Y.; Jiang, C.H.; Yin, Z.G.; et al. Natural Variation in OsLG3 Increases Drought Tolerance in Rice by Inducing ROS Scavenging. Plant Physiol. 2018, 178, 451–467. [Google Scholar] [CrossRef]
- Gao, F.; Chen, J.M.; Xiong, A.S.; Peng, R.H.; Liu, J.G.; Cai, B.; Yao, Q.H. Isolation and characterization of a novel AP2/EREBP-type transcription factor OsAP211 in Oryza sativa. Biol. Plantarum 2009, 53, 643–649. [Google Scholar] [CrossRef]
- Ramegowda, V.; Basu, S.; Krishnan, A.; Pereira, A. Rice GROWTH UNDER DROUGHT KINASE is required for drought tolerance and grain yield under normal and drought stress conditions. Plant Physiol. 2014, 166, 1634–1645. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.S.; Kim, J.K. Rice transcription factor AP37 involved in grain yield increase under drought stress. Plant Signal. Behav. 2009, 4, 735–736. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.J.; Kim, Y.S.; Kwon, C.W.; Park, H.K.; Jeong, J.S.; Kim, J.K. Overexpression of the transcription factor AP37 in rice improves grain yield under drought conditions. Plant Physiol. 2009, 150, 1368–1379. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.H.; Liu, K.; Zhang, J.; Li, X.; Xu, K.D.; Zhang, Y.; Qi, J.; Yu, D.S.; Wang, J.; Li, C.W. JcDREB2, a Physic Nut AP2/ERF Gene, Alters Plant Growth and Salinity Stress Responses in Transgenic Rice. Front. Plant Sci. 2017, 8, 306. [Google Scholar] [CrossRef] [PubMed]
- Serra, T.S.; Figueiredo, D.D.; Cordeiro, A.M.; Almeida, D.M.; Lourenço, T.; Abreu, I.A.; Sebastián, A.; Fernandes, L.; Contreras-Moreira, B.; Oliveira, M.M.; et al. OsRMC, a negative regulator of salt stress response in rice, is regulated by two AP2/ERF transcription factors. Plant Mol. Biol. 2013, 82, 439–455. [Google Scholar] [CrossRef]
- Cheng, X.L.; Zhang, S.X.; Tao, W.C.; Zhang, X.X.; Liu, J.; Sun, J.Q.; Zhang, H.W.; Pu, L.; Huang, R.F.; Chen, T. INDETERMINATE SPIKELET1 Recruits Histone Deacetylase and a Transcriptional Repression Complex to Regulate Rice Salt Tolerance. Plant Physiol. 2018, 178, 824–837. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.F.; Chen, X.J.; Liu, J.Q.; Ye, J.C.; Guo, Z.J. The rice ERF transcription factor OsERF922 negatively regulates resistance to Magnaporthe oryzae and salt tolerance. J. Exp. Bot. 2012, 63, 3899–3911. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.W.; Liao, F.X.; Wang, F.M.; Wen, W.W.; Li, J.J.; Mei, H.W.; Luo, L.J. Identification of rice transcription factors associated with drought tolerance using the Ecotilling method. PLoS ONE 2012, 7, e30765. [Google Scholar] [CrossRef]
- Tran, T.T.; Pérez-Quintero, A.L.; Wonni, I.; Carpenter, S.; Yu, Y.H.; Wang, L.; Leach, J.E.; Verdier, V.; Cunnac, S.; Bogdanove, A.J.; et al. Functional analysis of African Xanthomonas oryzae pv. oryzae TALomes reveals a new susceptibility gene in bacterial leaf blight of rice. PLoS Pathog. 2018, 14, e1007092. [Google Scholar] [CrossRef]
- Wamaitha, M.J.; Yamamoto, R.; Wong, H.L.; Kawasaki, T.; Kawano, Y.; Shimamoto, K. OsRap2.6 transcription factor contributes to rice innate immunity through its interaction with Receptor for Activated Kinase-C 1 (RACK1). Rice 2012, 5, 35. [Google Scholar] [CrossRef]
- Cao, Y.F.; Song, F.M.; Goodman, R.M.; Zheng, Z. Molecular characterization of four rice genes encoding ethylene-responsive transcriptional factors and their expressions in response to biotic and abiotic stress. J. Plant Physiol. 2006, 163, 1167–1178. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Ju, H.P.; Zhou, G.X.; Zhu, C.S.; Erb, M.; Wang, X.P.; Wang, P.; Lou, Y.G. An EAR-motif-containing ERF transcription factor affects herbivore-induced signaling, defense and resistance in rice. Plant J. 2011, 68, 583–596. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, D.D.; Yang, Z.H.; Jiang, S.; Qu, J.N.; He, W.; Liu, Z.M.; Xing, J.J.; Ma, Y.C.; Lin, Q.L.; et al. Roles of FERONIA-like receptor genes in regulating grain size and quality in rice. Sci. China Life Sci. 2021, 64, 294–310. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.K.; Yu, X.Q.; Cui, Y.J.; Xia, S.S.; Zeng, D.L.; Qian, Q.; Ren, D.Y. LRG1 maintains sterile lemma identity by regulating OsMADS6 expression in rice. Sci. China Life Sci. 2021, 64, 1190–1192. [Google Scholar] [CrossRef] [PubMed]

| Classification | AP2 | ERF | RAV | Soloist |
|---|---|---|---|---|
| Number | 24 | 136 | 5 | 0 |
| Gene Name | Gene Family | Gene Locus | Gene Function | Reference(s) |
|---|---|---|---|---|
| FZP; BFL1; SGDP7 | ERF | LOC_Os07g47330 | spikelet development | [19,20] |
| OsSNB; SSH1 | AP2 | LOC_Os07g13170 | spikelet development | [21] |
| OsIDS1 | AP2 | LOC_Os03g60430 | spikelet development | [22] |
| MFS1 | ERF | LOC_Os05g41760 | spikelet development | [18] |
| Gene Name | Gene Family | Gene Locus | Gene Function | Reference |
|---|---|---|---|---|
| OsIDS1 | AP2 | LOC_Os03g60430 | flowering | [22] |
| OsSNB; SSH1 | AP2 | LOC_Os07g13170 | flowering | [23] |
| Gene Name | Gene Family | Gene Locus | Gene Function | Reference(s) |
|---|---|---|---|---|
| OsSNB; SSH1 | AP2 | LOC_Os07g13170 | grain size; grain shattering | [24,25] |
| SHAT1 | AP2 | LOC_Os04g55560 | grain shattering | [26] |
| OsERF115 | ERF | LOC_Os08g41030 | grain size | [27] |
| FZP; BFL1; SGDP7 | ERF | LOC_Os07g47330 | grain size, grain number | [19,28] |
| SERF1 | ERF | LOC_Os05g34730 | grain size; grain quality | [29] |
| RSR1 | AP2 | LOC_Os05g03040 | grain quality | [30] |
| Gene Name | Gene Family | Gene Locus | Gene Function | Reference |
|---|---|---|---|---|
| BBM1 | AP2 | LOC_Os11g19060 | embryonic development | [31] |
| BBM2 | AP2 | LOC_Os02g40070 | embryonic development | [31] |
| BBM3 | AP2 | LOC_Os01g67410 | embryonic development | [31] |
| Gene Name | Gene Family | Gene Locus | Gene Function | Reference(s) |
|---|---|---|---|---|
| Crl5 | AP2 | LOC_Os07g03250 | root initiation and formation | [33,34] |
| OsERF48; OsDRAP1 | ERF | LOC_Os08g31580 | root initiation and formation | [35] |
| OsERF3; OsAP37 | ERF | LOC_Os01g58420 | root initiation and formation | [36] |
| Gene Name | Gene Family | Gene Locus | Gene Function | Reference(s) |
|---|---|---|---|---|
| OsDERF1 | ERF | LOC_Os08g35240 | drought tolerance | [37] |
| OsEATB | ERF | LOC_Os09g28440 | internode development | [40] |
| HL6 | ERF | LOC_Os06g44750 | epidermis hair development | [46] |
| OsERF2; OsWR4 | ERF | LOC_Os06g08340 | root development and formation | [39] |
| OsRPH1 | ERF | LOC_Os05g49700 | internode development | [41] |
| SMOS1; SHB; RLA1; NGR5 | ERF | LOC_Os05g32270 | signal transduction | [43,44,45] |
| OsAP2-39 | ERF | LOC_Os04g52090 | signal transduction | [42] |
| SNORKEL1 | ERF | AB510478 | internode development | [38] |
| SNORKEL2 | ERF | AB510479 | internode development | [38] |
| Gene Name | Gene Family | Gene Locus | Gene Function | Reference |
|---|---|---|---|---|
| SMOS1; SHB; RLA1; NGR5 | ERF | LOC_Os05g32270 | nutrient use | [43] |
| OsDREB1C | ERF | LOC_Os06g03670 | photosynthetic efficiency; nitrogen use | [47] |
| Gene Name | Gene Family | Gene Locus | Gene Function | Reference(s) |
|---|---|---|---|---|
| OsDREB1A | ERF | LOC_Os09g35030 | cold tolerance | [48] |
| OsDREB1B | ERF | LOC_Os09g35010 | cold tolerance; heat tolerance; | [49,50,51,52] |
| OsDREB1D | ERF | LOC_Os06g06970 | cold tolerance; salinity tolerance | [52] |
| OsDREB1E | ERF | LOC_Os04g48350 | drought tolerance | [52] |
| OsDREB1G; OsDREB1I | ERF | LOC_Os08g43210 | drought tolerance | [53] |
| OsDREB2B | ERF | LOC_Os05g27930 | drought tolerance | [53] |
| OsDREB1E | ERF | LOC_Os04g48350 | drought tolerance | [53] |
| OsDREB1F; RCBF2 | ERF | LOC_Os01g73770 | salinity tolerance; drought tolerance; temperature tolerance | [54] |
| OsDREB1-1; CR350 | ERF | LOC_Os04g48350 | salinity tolerance; drought tolerance | [55] |
| OsDREB4-1; CR223 | ERF | LOC_Os02g43940 | salinity tolerance; drought tolerance | [55] |
| OsWR2 | ERF | LOC_Os06g40150 | temperature tolerance | [56,57] |
| OsERF71 | ERF | LOC_Os06g09390 | drought tolerance | [58] |
| OsLG3; OsERF62; OsRAF | ERF | LOC_Os03g08470 | drought tolerance | [59] |
| OsAP211; ARAG1 | ERF | LOC_Os02g43970 | drought tolerance | [60] |
| OsAP37 | ERF | LOC_Os01g58420 | drought tolerance | [61,62,63] |
| OsAP23 | ERF | LOC_Os03g05590 | salinity tolerance | [64] |
| OsEREBP2 | ERF | LOC_Os01g64790 | salinity tolerance; drought tolerance; temperature tolerance | [65] |
| SERF1 | ERF | LOC_Os05g34730 | salinity tolerance | [29] |
| OsIDS1 | AP2 | LOC_Os03g60430 | salinity tolerance | [66] |
| OsERF922 | ERF | LOC_Os01g54890 | salinity tolerance; drought tolerance | [67,68] |
| Gene Name | Gene Family | Gene Locus | Gene Function | Reference |
|---|---|---|---|---|
| OsERF123 | ERF | LOC_Os09g39810 | bacterial blight | [69] |
| OsERF922 | ERF | LOC_Os01g54890 | blast fungus | [67] |
| OsRap2.6; OsERF101 | ERF | LOC_Os04g32620 | blast fungus | [70] |
| OsBIERF1 | ERF | LOC_Os09g26420 | blast fungus | [71] |
| OsERF3; OsAP37 | ERF | LOC_Os01g58420 | chilo suppressalis; brown planthoppers | [72] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, W.; Ding, C.; Hu, H.; Dong, G.; Zhang, G.; Qian, Q.; Ren, D. Molecular Events of Rice AP2/ERF Transcription Factors. Int. J. Mol. Sci. 2022, 23, 12013. https://doi.org/10.3390/ijms231912013
Xie W, Ding C, Hu H, Dong G, Zhang G, Qian Q, Ren D. Molecular Events of Rice AP2/ERF Transcription Factors. International Journal of Molecular Sciences. 2022; 23(19):12013. https://doi.org/10.3390/ijms231912013
Chicago/Turabian StyleXie, Wei, Chaoqing Ding, Haitao Hu, Guojun Dong, Guangheng Zhang, Qian Qian, and Deyong Ren. 2022. "Molecular Events of Rice AP2/ERF Transcription Factors" International Journal of Molecular Sciences 23, no. 19: 12013. https://doi.org/10.3390/ijms231912013
APA StyleXie, W., Ding, C., Hu, H., Dong, G., Zhang, G., Qian, Q., & Ren, D. (2022). Molecular Events of Rice AP2/ERF Transcription Factors. International Journal of Molecular Sciences, 23(19), 12013. https://doi.org/10.3390/ijms231912013







