Advances in Fungal Elicitor-Triggered Plant Immunity
Abstract
1. Introduction
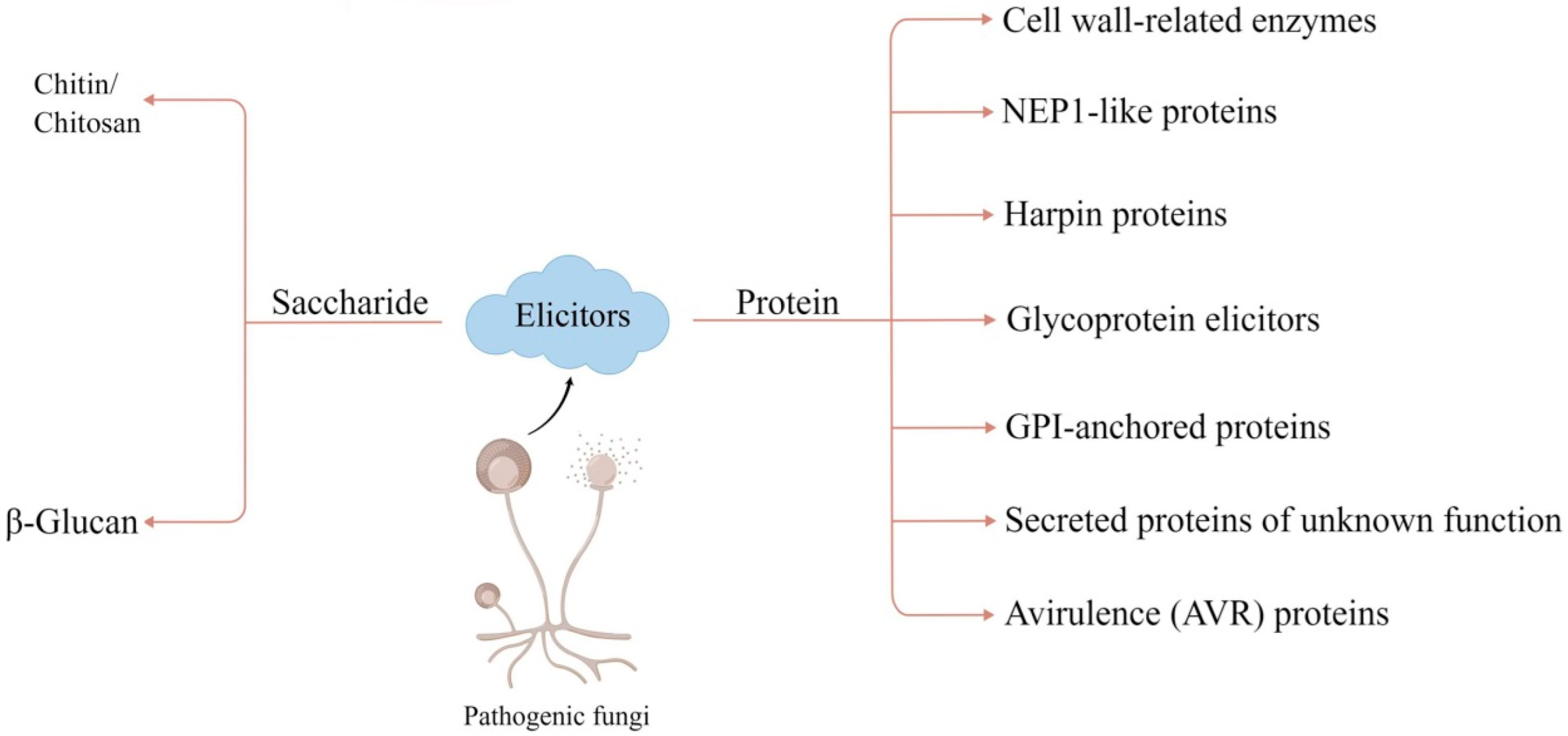
2. Classification of Fungal Elicitors
2.1. Saccharide Elicitors
2.2. Protein Elicitors
2.2.1. Cell Wall-Related Enzymes
2.2.2. NEP1-like Proteins (NLPs)
2.2.3. Harpin Proteins
2.2.4. Glycoprotein Elicitors
2.2.5. GPI-Anchored Proteins
2.2.6. Secreted Proteins of Unknown Function
2.2.7. Avirulence (AVR) Proteins
| Type | Origin | Elicitor Name | Receptor | Receptor Type | Co-Receptor | Ref. |
|---|---|---|---|---|---|---|
| Saccharide | fungi cell wall | chitin/chitosan | OsCEBiP, LYK5 | LysM-RLP, LysM-RLK | OsCERK1, CERK1 | [73,74,75,76] |
| β-Glucan | - | - | - | [27] | ||
| GH11 | Trichoderma viride | TvEIX | LeEIX2 | LRR-RLP | BAK1 | [31,32,77] |
| Botrytis cinerea | BcXyn11A | - | - | - | [34] | |
| Fusarium graminearum | FGSG_03624 | - | - | - | [35] | |
| Verticillium dahliae | VdEIX3 | NbEIX2 | LRR-RLP | - | [36] | |
| GH10 | Rhizoctonia solani | RSAG8_07159, FGSG_11487 | - | - | - | [78] |
| GH12 | B. cinerea | BcXYG1 | - | - | - | [79] |
| F. oysporum | FoEG1 | - | - | - | [80] | |
| V. dahliae | VdEG1 | - | - | - | [81] | |
| VdEG3 | - | - | - | |||
| GH16 | B. cinerea | BcCrh1 | - | - | - | [39] |
| GH18 | Magnaporthe oryza | MoChia1/MoChi | OsTPR1 | Tetratricopeptide repeat protein | - | [40] |
| MoChi/MoChia1 | OsMBL1 | Jacalin-related mannose-binding lectin | - | [41] | ||
| GH28 | B. cinerea | BcPG1 to BcPG4, BcPG6 | RLP42/RBPG1 | LRR-RLP | - | [82] |
| GH45 | R. solani | EG1 | - | - | - | [83] |
| CE | Sclerotinia sclerotiorum | SsCut1 | - | - | - | [42] |
| V. dahliae | VdCUT11 | - | - | - | [43] | |
| PL | V. dahliae | VdPEL1 | - | - | - | [44] |
| NLP | B. cinerea | BcNEP1, BcNEP2 | RLP23 | LRR-RLP | - | [84,85] |
| Harpin | Alternaria tenuissima | Hrip1 | - | - | - | [51] |
| M. oryzae | MoHrip1, MoHrip2 | - | - | - | [52,53] | |
| Glycoprotein | M. oryzae | GP66 | - | - | - | [56] |
| GPI-anchored protein | Ustilaginoidea virens | SGP1 | - | - | - | [63] |
| Secreted protein of unknown function | Rhynchosporium commune | RcCDI1 | - | - | - | [64] |
| Valsa mali | VmE02 | RE02 | LRR-RLP | - | [67] | |
| F. graminearum | Fg02685 | - | - | - | [66] |
| AVR | Receptor | Ref. | |||
|---|---|---|---|---|---|
| Species | Name | Species | Name | Type | |
| Albugo candida | CCG28, CCG30, CCG33, CCG40, CCG67, CCG71, CCG79 and CCG104 | Arabidopsis | WRR4A | TNL | [86] |
| CCG45, CCG57, CCG61 and CCG70 | Arabidopsis | WRR4B | TNL | ||
| Blumeria graminis f. sp. hordei | AVRa1, AVRa6, AVRa7, AVRa9, AVRa10, AVRa13 and AVRa22 | barley | MLA1, MLA6, MLA7, MLA9, MLA10, MLA13 and MLA22 | CNL | [87,88,89] |
| B. graminis f. sp. tritici (Bgt), B. graminis f. sp. Secalis, and B. graminis f. sp. Triticale | AvrPm2, BgsE-5845 and BgtriticaleE-5845 | wheat | Pm2 | CNL | [90] |
| Bgt | AvrPm3A2/F2, AVRPM3B2/C2 and AVRPM3D3 | wheat | PM3A, PM3F, PM3B, PM3C and PM3D | CNL | [91,92] |
| AvrPm17 | rye | Pm17 | CNL | [93] | |
| Cladosporium fulvum | apoplastic effectors, including Avr2, Avr4, Avr4E and Avr9 | tomato | Cf-2, Cf-4, Hcr9-4E and Cf-9 | PRR | [94,95,96,97,98] |
| Fusarium oxysporum f. sp. lycopersici | FoAvr2 | tomato | I2 | CNL | [99] |
| F. oxysporum f. sp. melonis | AvrFom2 | melon | Fom-2 | CNL | [100] |
| Leptosphaeria maculans | apoplastic AvrLm1 | oilseed rape | LepR3 | PRR | [101] |
| Magnaporthe oryzae | AvrPi9, Avr-Pi54, AvrPib, Avr-Pik, Avr-Pita and AvrPiz-t | rice | Pi9, Pi54, Pib, Pik, Pi-ta and Piz-t | CNL | [102,103,104,105,106,107] |
| Avr-Pia and Avr1-CO39 | rice | Pia-2/RGA5 | CNL | [108] | |
| AvrPii | rice | Pi5-1, Pi5-2, Pii-1 and Pii-2 | CNL | [109] | |
| Melampsora lini | AvrL2-A, AvrM, AvrP and AvrP123 | flax | L2, M, P and P2 | TNL | [110,111] |
| AvrL567 effectors | flax | L5, L6 and L7 | TNL | [112] | |
| Puccinia graminis f. sp. tritici | AvrSr27 and AvrSr35 | wheat | Sr27 and Sr35 | CNL | [72] |
| AvrSr50 | rye | Sr50 | CNL | [71] | |
| P. polysora | AvrRppC | maize | RppC | CNL | [113] |
| AvrRppK | maize | RppK | CNL | [114] | |
3. The Receptors of Fungal Elicitors
3.1. Types of Receptors
3.1.1. PRRs
3.1.2. NLRs
3.2. The Important Role of Immune Receptors Recognizing Fungal Elicitors in Disease Resistance Breeding
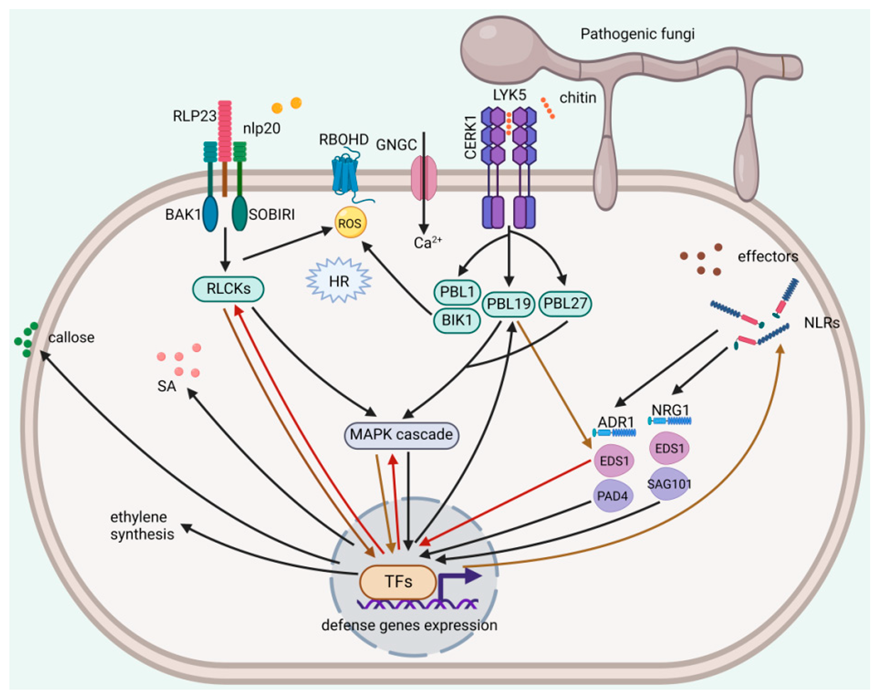
4. Signaling Pathways of Fungal Elicitor-Triggered Plant Immunity
4.1. Fungal PAMPs-Triggered PTI
4.2. Fungal Effector-Triggered ETI
4.3. Convergent Pathways between Fungal Elicitor-Triggered PTI and ETI
5. Conclusions and Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Savary, S.; Willocquet, L.; Pethybridge, S.J.; Esker, P.; McRoberts, N.; Nelson, A. The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 2019, 3, 430–439. [Google Scholar] [CrossRef] [PubMed]
- Fisher, M.C.; Henk, D.A.; Briggs, C.J.; Brownstein, J.S.; Madoff, L.C.; McCraw, S.L.; Gurr, S.J. Emerging fungal threats to animal, plant and ecosystem health. Nature 2012, 484, 186–194. [Google Scholar] [CrossRef] [PubMed]
- Fones, H.N.; Bebber, D.P.; Chaloner, T.M.; Kay, W.T.; Steinberg, G.; Gurr, S.J. Threats to global food security from emerging fungal and oomycete crop pathogens. Nat. Food 2020, 1, 332–342. [Google Scholar] [CrossRef]
- Mehta, A.; Brasileiro, A.C.M.; Souza, D.S.L.; Romano, E.; Campos, M.A.; Grossi-De-Sa, M.F.; Silva, M.S.; Franco, O.L.; Fragoso, R.R.; Bevitori, R.; et al. Plant-pathogen interactions: What is proteomics telling us? FEBS J. 2008, 275, 3731–3746. [Google Scholar] [CrossRef]
- Bi, K.; Liang, Y.; Mengiste, T.; Sharon, A. Killing softly: A roadmap of Botrytis cinerea pathogenicity. Trends Plant Sci. 2022. [Google Scholar] [CrossRef]
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef]
- de Wit, P.J.G.M. How plants recognize pathogens and defend themselves. Cell. Mol. Life. Sci. 2007, 64, 2726–2732. [Google Scholar] [CrossRef]
- Fang, Y.; Gu, Y. Regulation of plant immunity by nuclear membrane-associated mechanisms. Front. Immunol. 2021, 12, 771065. [Google Scholar] [CrossRef]
- Sák, M.; Dokupilová, I.; Kaňuková, a.; Mrkvová, M.; Kraic, J. Biotic and abiotic elicitors of stilbenes production in Vitis vinifera L. cell culture. Plants 2021, 10, 490. [Google Scholar] [CrossRef]
- Patel, Z.M.; Mahapatra, R.; Jampala, S.S.M. Role of fungal elicitors in plant defense mechanism. Mol. Asp. Plant Benef. Microbes Agric. 2020, 143–158. [Google Scholar] [CrossRef]
- Sun, L.; Zhang, J. Regulatory role of receptor-like cytoplasmic kinases in early immune signaling events in plants. Fems. Microbiol. Rev. 2020, 44, 845–856. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.H.; Lee, H.S.; Belkhadir, Y. Coding of plant immune signals by surface receptors. Curr. Opin. Plant Biol. 2021, 62, 102044. [Google Scholar] [CrossRef] [PubMed]
- Geoghegan, I.A.; Gurr, S.J. Investigating chitin deacetylation and chitosan hydrolysis during vegetative growth in Magnaporthe oryzae. Cell. Microbiol. 2017, 19, e12743. [Google Scholar] [CrossRef] [PubMed]
- Fesel, P.H.; Zuccaro, A. beta-glucan: Crucial component of the fungal cell wall and elusive MAMP in plants. Fungal Genet. Biol. 2016, 90, 53–60. [Google Scholar] [CrossRef]
- Iriti, M.; Faoro, F. Chitosan as a MAMP, searching for a PRR. Plant Signal. Behav. 2009, 4, 66–68. [Google Scholar] [CrossRef]
- Pusztahelyi, T. Chitin and chitin-related compounds in plant–fungal interactions. Mycology 2018, 9, 189–201. [Google Scholar] [CrossRef]
- Miya, A.; Albert, P.; Shinya, T.; Desaki, Y.; Ichimura, K.; Shirasu, K.; Narusaka, Y.; Kawakami, N.; Kaku, H.; Shibuya, N. CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 2007, 104, 19613–19618. [Google Scholar] [CrossRef]
- Shimizu, T.; Nakano, T.; Takamizawa, D.; Desaki, Y.; Ishii-Minami, N.; Nishizawa, Y.; Minami, E.; Okada, K.; Yamane, H.; Kaku, H.; et al. Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J. 2010, 64, 204–214. [Google Scholar] [CrossRef]
- Yamaguchi, T.; Yamada, A.; Hong, N.; Ogawa, T.; Shibuya, I.N. Differences in the recognition of glucan elicitor signals between rice and soybean: Beta-glucan fragments from the rice blast disease fungus Pyricularia oryzae that elicit phytoalexin biosynthesis in suspension-cultured rice cells. Plant Cell 2000, 12, 817–826. [Google Scholar]
- Sun, C. Induction Effect of Yeast Cell Wall on Resistance of Postharvest Pathogenic Fungi in Pear and Tomato Fruits and Related Mechanism. Ph.D. Thesis, Zhejiang University, Hangzhou, China, 2019. [Google Scholar]
- Bowman, S.M.; Free, S.J. The structure and synthesis of the fungal cell wall. BioEssays 2006, 28, 799–808. [Google Scholar] [CrossRef]
- Shahinian, S.; Bussey, H. β-1,6-Glucan synthesis in Saccharomyces cerevisiae. Mol. Microbiol. 2000, 35, 477–489. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Herrera, J.; Ortiz-Castellanos, L. Cell wall glucans of fungi. A review. Cell Surface 2019, 5, 100022. [Google Scholar] [CrossRef]
- Ayers, A.R.; Ebel, J.; Valent, B.; Albersheim, P. Host-pathogen interactions: X. fractionation and biological activity of an elicitor isolated from the mycelial walls of Phytophthora megasperma var. sojae. Plant Physiol. 1976, 57, 760–765. [Google Scholar] [CrossRef] [PubMed]
- Cosio, E.G.; Feger, M.; Miller, C.J.; Antelo, L.; Ebel, J. High-affinity binding of fungal β-glucan elicitors to cell membranes of species of the plant family Fabaceae. Planta 1996, 200, 92–99. [Google Scholar] [CrossRef]
- Côté, F.; Roberts, K.A.; Hahn, M.G. Identification of high-affinity binding sites for the hepta-beta-glucoside elicitor in membranes of the model legumes Medicago truncatula and Lotus japonicus. Planta 2000, 211, 596–605. [Google Scholar]
- Rebaque, D.; Hierro, I.D.; López, G.; Bacete, L.; Vilaplana, F.; Dallabernardina, P.; Pfrengle, F.; Jordá, L.; Sánchez-Vallet, A.; Pérez, R. Cell wall-derived mixed-linked β-1,3/1,4-glucans trigger immune responses and disease resistance in plants. Plant J. 2021, 106, 601–615. [Google Scholar] [CrossRef]
- Rotblat, B.; Enshell-Seijffers, D.; Gershoni, J.M.; Schuster, S.; Avni, A. Identification of an essential component of the elicitation active site of the EIX protein elicitor. Plant J. 2002, 32, 1049–1055. [Google Scholar] [CrossRef]
- Yang, C.; Liu, R.; Pang, J.; Ren, B.; Zhou, H.; Wang, G.; Wang, E.; Liu, J. Poaceae-specific cell wall-derived oligosaccharides activate plant immunity via OsCERK1 during Magnaporthe oryzae infection in rice. Nat. Commun. 2021, 12, 2178. [Google Scholar] [CrossRef]
- Bradley, E.L.; Ökmen, B.; Doehlemann, G.; Henrissat, B.; Bradshaw, R.E.; Mesarich, C.H. Secreted glycoside hydrolase proteins as effectors and invasion patterns of plant-associated fungi and oomycetes. Front. Plant Sci. 2022, 13, 853106. [Google Scholar] [CrossRef]
- Fuchs, Y.; Saxena, A.; Gamble, H.R.; Anderson, J.D. Ethylene biosynthesis-inducing protein from cellulysin is an endoxylanase. Plant Physiol. 1989, 89, 138–143. [Google Scholar] [CrossRef]
- Enkerli, J.; Felix, G.; Boller, T. The enzymatic activity of fungal xylanase is not necessary for its elicitor activity. Plant Physiol. 1999, 121, 391–398. [Google Scholar] [CrossRef] [PubMed]
- Sella, L.; Gazzetti, K.; Faoro, F.; Odorizzi, S.; D’Ovidio, R.; Schäfer, W.; Favaron, F. A Fusarium graminearum xylanase expressed during wheat infection is a necrotizing factor but is not essential for virulence. Plant Physiol. Bioch. 2013, 64, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Frías, M.; González, M.; González, C.; Brito, N. A 25-residue peptide from Botrytis cinerea xylanase bcxyn11a elicits plant defenses. Front. Plant Sci. 2019, 10, 474. [Google Scholar] [CrossRef] [PubMed]
- Tundo, S.; Paccanaro, M.C.; Bigini, V.; Savatin, D.V.; Faoro, F.; Favaron, F.; Sella, L. The Fusarium graminearum FGSG_03624 xylanase enhances plant immunity and increases resistance against bacterial and fungal pathogens. Int. J. Mol. Sci. 2021, 22, 10811. [Google Scholar] [CrossRef]
- Yin, Z.; Wang, N.; Pi, L.; Li, L.; Duan, W.; Wang, X.; Dou, D. Nicotiana benthamiana LRR-RLP NbEIX2 mediates the perception of an EIX-like protein from Verticillium dahliae. J. Integr. Plant Biol. 2021, 63, 949–960. [Google Scholar] [CrossRef]
- Rafiei, V.; Vélëz, H.; Tzelepis, G. The role of glycoside hydrolases in phytopathogenic fungi and oomycetes virulence. Int. J. Mol. Sci. 2021, 22, 9359. [Google Scholar] [CrossRef]
- Li, Y.; Han, Y.-J.; Qu, M.-Y.; Chen, J.; Chen, X.-F.; Geng, X.-Q.; Wang, Z.-H.; Chen, S.-B. Apoplastic cell death-inducing proteins of filamentous plant pathogens: Roles in plant-pathogen interactions. Front. Genet. 2020, 11, 661. [Google Scholar] [CrossRef]
- Bi, K.; Scalschi, L.; Jaiswal, N.; Mengiste, T.; Fried, R.; Sanz, A.B.; Arroyo, J.; Zhu, W.; Masrati, G.; Sharon, A. The Botrytis cinerea Crh1 transglycosylase is a cytoplasmic effector triggering plant cell death and defense response. Nat. Commun. 2021, 12, 2166. [Google Scholar] [CrossRef]
- Yang, C.; Yu, Y.; Huang, J.; Meng, F.; Pang, J.; Zhao, Q.; Islam, M.A.; Xu, N.; Tian, Y.; Liu, J. Binding of the Magnaporthe oryzae chitinase MoChia1 by a rice tetratricopeptide repeat protein allows free chitin to trigger immune responses. Plant Cell 2019, 31, 172–188. [Google Scholar] [CrossRef]
- Han, Y.; Song, L.; Peng, C.; Liu, X.; Liu, L.; Zhang, Y.; Wang, W.; Zhou, J.; Wang, S.; Ebbole, D.; et al. A Magnaporthe chitinase interacts with a rice jacalin-related lectin to promote host colonization. Plant Physiol. 2019, 179, 1416–1430. [Google Scholar] [CrossRef]
- Zhang, H.; Wu, Q.; Cao, S.; Zhao, T.; Chen, L.; Zhuang, P.; Zhou, X.; Gao, Z. A novel protein elicitor (SsCut) from Sclerotinia sclerotiorum induces multiple defense responses in plants. Plant Mol. Biol. 2014, 86, 495–511. [Google Scholar] [CrossRef] [PubMed]
- Gui, Y.-J.; Zhang, W.-Q.; Zhang, D.-D.; Zhou, L.; Short, D.P.G.; Wang, J.; Ma, X.-F.; Li, T.-G.; Kong, Z.-Q.; Wang, B.-L.; et al. A Verticillium dahliae extracellular cutinase modulates plant immune responses. Mol. Plant Microbe Interact. 2017, 31, 260–273. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhang, Y.; Li, B.; Yang, X.; Dong, Y.; Qiu, D. A Verticillium dahliae pectate lyase induces plant immune responses and contributes to virulence. Front. Plant Sci. 2018, 9, 1271. [Google Scholar] [CrossRef]
- Van den Ackerveken, G. How plants differ in toxin-sensitivity. Science 2017, 358, 1383–1384. [Google Scholar] [CrossRef] [PubMed]
- Seidl, M.F.; Ackerveken, G. Activity and phylogenetics of the broadly occurring family of microbial Nep1-Like proteins. Annu. Rev. Phytopathol. 2019, 57, 367–386. [Google Scholar] [CrossRef] [PubMed]
- Böhm, H.; Albert, I.; Oome, S.; Raaymakers, T.M.; Van den Ackerveken, G.; Nürnberger, T. A conserved peptide pattern from a widespread microbial virulence factor triggers pattern-induced immunity in Arabidopsis. PloS Pathog. 2014, 10, e1004491. [Google Scholar] [CrossRef]
- Albert, I.; Bohm, H.; Albert, M.; Feiler, C.E.; Imkampe, J.; Wallmeroth, N.; Brancato, C.; Raaymakers, T.M.; Oome, S.; Zhang, H.; et al. An RLP23-SOBIR1-BAK1 complex mediates NLP-triggered immunity. Nat. Plants 2015, 1, 15140. [Google Scholar] [CrossRef]
- Lenarcic, T.; Pirc, K.; Hodnik, V.; Albert, I.; Borisek, J.; Magistrato, A.; Nurnberger, T.; Podobnik, M.; Anderluh, G. Molecular basis for functional diversity among microbial Nep1-like proteins. PLoS Pathog. 2019, 15, e1007951. [Google Scholar] [CrossRef]
- Wei, Z.M.; Laby, R.J.; Zumoff, C.H.; Bauer, D.W.; He, S.Y.; Collmer, A.; Beer, S.V. Harpin, elicitor of the hypersensitive response produced by the plant pathogen Erwinia amylovora. Science 1992, 257, 85–88. [Google Scholar] [CrossRef]
- Kulye, M.; Liu, H.; Zhang, Y.; Zeng, H.; Yang, X.; Qiu, D. Hrip1, a novel protein elicitor from necrotrophic fungus, Alternaria tenuissima, elicits cell death, expression of defence-related genes and systemic acquired resistance in tobacco. Plant Cell Environ. 2012, 35, 2104–2120. [Google Scholar] [CrossRef]
- Chen, M.; Zeng, H.; Qiu, D.; Guo, L.; Yang, X.; Shi, H.; Zhou, T.; Zhao, J. Purification and characterization of a novel hypersensitive response-inducing elicitor from Magnaporthe oryzae that triggers defense response in rice. PLoS ONE 2012, 7, e37654. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Zhang, C.; Zi, Q.; Qiu, D.; Liu, W.; Zeng, H. A novel elicitor identified from Magnaporthe oryzae triggers defense responses in tobacco and rice. Plant Cell Rep. 2014, 33, 1865–1879. [Google Scholar] [CrossRef] [PubMed]
- Miao, X.-Y.; Qu, H.-P.; Han, Y.-L.; He, C.-F.; Qiu, D.-W.; Cheng, Z.-W. The protein elicitor Hrip1 enhances resistance to insects and early bolting and flowering in Arabidopsis thaliana. PLoS ONE 2019, 14, e0216082. [Google Scholar] [CrossRef] [PubMed]
- Roby, D.; Toppan, A.; Esquerré-Tugayé, M.-T. Cell surfaces in plant-microorganism interactions. Plant Physiol. 1985, 77, 700–704. [Google Scholar] [CrossRef]
- Li, Y.-F.; Wang, Z.-Z.; Jia, X.-L. Membrane lipid peroxidation and hypersensitive reaction induced by a glycoprotein elicitor from Magnaporthe grisea in rice leaves. J. Plant Physiol. Mol. Biol. 2004, 30, 147–152. [Google Scholar]
- Yang, X.; Qiu, D.; Zeng, H.; Yuan, J.; Mao, J. Purification and characterization of a glycoprotein elicitor from Alternaria tenuissima. World J. Microb. Biot. 2009, 25, 2035–2042. [Google Scholar] [CrossRef]
- Radhajeyalakshmi, R.; Velazhahan, R.; Samiyappan, R.; Doraiswamy, S. Systemic induction of pathogenesis related proteins (PRs) in Alternaria solani elicitor sensitized tomato cells as resistance response. Sci. Res. Essays 2009, 4, 685–689. [Google Scholar]
- Zurzolo, C.; Simons, K. Glycosylphosphatidylinositol-anchored proteins: Membrane organization and transport. Biochim. Biophys. Acta. 2016, 1858, 632–639. [Google Scholar] [CrossRef]
- Fujita, M.; Kinoshita, T. GPI-anchor remodeling: Potential functions of GPI-anchors in intracellular trafficking and membrane dynamics. Biochim. Biophys. Acta. 2012, 1821, 1050–1058. [Google Scholar] [CrossRef]
- Oliveira-Garcia, E.; Deising, H.B. The glycosylphosphatidylinositol anchor biosynthesis genes GPI12, GAA1, and GPI8 are essential for cell-wall integrity and pathogenicity of the maize anthracnose fungus Colletotrichum graminicola. Mol. Plant-Microbe Interact. 2016, 29, 889–901. [Google Scholar] [CrossRef]
- Liu, C.; Xing, J.; Cai, X.; Hendy, A.; He, W.; Yang, J.; Huang, J.; Peng, Y.L.; Ryder, L.; Chen, X.L. GPI7-mediated glycosylphosphatidylinositol anchoring regulates appressorial penetration and immune evasion during infection of Magnaporthe oryzae. Environ. Microbiol. 2020, 22, 2581–2595. [Google Scholar] [CrossRef]
- Song, T.; Zhang, Y.; Zhang, Q.; Zhang, X.; Shen, D.; Yu, J.; Yu, M.; Pan, X.; Cao, H.; Yong, M.; et al. The N-terminus of an Ustilaginoidea virens Ser-Thr-rich glycosylphosphatidylinositol-anchored protein elicits plant immunity as a MAMP. Nat. Commun. 2021, 12, 2451. [Google Scholar] [CrossRef]
- Franco-Orozco, B.; Berepiki, A.; Ruiz, O.; Gamble, L.; Griffe, L.L.; Wang, S.; Birch, P.R.J.; Kanyuka, K.; Avrova, A. A new proteinaceous pathogen-associated molecular pattern PAMP identified in Ascomycete fungi induces cell death in Solanaceae. New Phytol. 2017, 214, 1657–1672. [Google Scholar] [CrossRef] [PubMed]
- Nie, J.; Yin, Z.; Li, Z.; Wu, Y.; Huang, L. A small cysteine-rich protein from two kingdoms of microbes is recognized as a novel pathogen-associated molecular pattern. New Phytol. 2019, 222, 995–1011. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Hu, S.; Jin, M.; Xu, Y.; Jiang, Q.; Ma, J.; Zhang, Y.; Qi, P.; Chen, G.; Jiang, Y.; et al. The N-terminus of a Fusarium graminearum-secreted protein enhances broad-spectrum disease resistance in plants. Mol. Plant Pathol. 2022. [Google Scholar] [CrossRef]
- Nie, J.; Zhou, W.; Liu, J.; Tan, N.; Zhou, J.M.; Huang, L. A receptor-like protein from Nicotiana benthamiana mediates VmE02 PAMP-triggered immunity. New Phytol. 2021, 229, 2260–2272. [Google Scholar] [CrossRef] [PubMed]
- Petit-Houdenot, Y.; Fudal, I. Complex interactions between fungal avirulence genes and their corresponding plant resistance genes and consequences for disease resistance management. Front. Plant Sci. 2017, 8, 1072. [Google Scholar] [CrossRef]
- van Kan, J.A.; van den Ackerveken, G.F.; de Wit, P.J. Cloning and characterization of cDNA of avirulence gene avr9 of the fungal pathogen Cladosporium fulvum, causal agent of tomato leaf mold. Mol. Plant-Microbe Interact. 1991, 4, 52–59. [Google Scholar] [CrossRef]
- Zhang, S.; Li, C.; Si, J.; Han, Z.; Chen, D. Action mechanisms of effectors in plant-pathogen interaction. Int. J. Mol. Sci. 2022, 23, 6758. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Upadhyaya, N.M.; Ortiz, D.; Sperschneider, J.; Li, F.; Bouton, C.; Breen, S.; Dong, C.; Xu, B.; Zhang, X. Loss of AvrSr50 by somatic exchange in stem rust leads to virulence for Sr50 resistance in wheat. Science 2017, 358, 1607–1610. [Google Scholar] [CrossRef]
- Salcedo, A.; Rutter, W.; Wang, S.; Akhunova, A.; Bolus, S.; Chao, S.; Anderson, N.; Soto, M.D.; Rouse, M.; Szabo, L. Variation in the AvrSr35 gene determines Sr35 resistance against wheat stem rust race Ug99. Science 2017, 358, 1604–1606. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Yan, L.; Tanaka, K.; Nguyen, C.T.; Stacey, G. The kinase LYK5 is a major chitin receptor in Arabidopsis and forms a chitin-induced complex with related kinase CERK1. Elife 2014, 3, e03766. [Google Scholar] [CrossRef] [PubMed]
- Erwig, J.; Ghareeb, H.; Kopischke, M.; Hacke, R.; Matei, A.; Petutschnig, E.; Lipka, V. Chitin-induced and CHITIN ELICITOR RECEPTOR KINASE1 (CERK1) phosphorylation-dependent endocytosis of Arabidopsis thaliana LYSIN MOTIF-CONTAINING RECEPTOR-LIKE KINASE5 (LYK5). New Phytol. 2017, 215, 382–396. [Google Scholar] [CrossRef] [PubMed]
- Kaku, H.; Nishizawa, Y.; Ishii-Minami, N.; Akimoto-Tomiyama, C.; Dohmae, N.; Takio, K.; Minami, E.; Shibuya, N. Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc. Natl. Acad. Sci. USA 2006, 103, 11086–11091. [Google Scholar] [CrossRef] [PubMed]
- Kouzai, Y.; Nakajima, K.; Hayafune, M.; Ozawa, K.; Kaku, H.; Shibuya, N.; Minami, E.; Nishizawa, Y. CEBiP is the major chitin oligomer-binding protein in rice and plays a main role in the perception of chitin oligomers. Plant Mol. Biol. 2014, 84, 519–528. [Google Scholar] [CrossRef]
- Bar, M.; Sharfman, M.; Ron, M.; Avni, A. BAK1 is required for the attenuation of ethylene-inducing xylanase (Eix)-induced defense responses by the decoy receptor LeEix1. Plant J. 2010, 63, 791–800. [Google Scholar] [CrossRef]
- Anderson, J.P.; Sperschneider, J.; Win, J.; Kidd, B.; Yoshida, K.; Hane, J.; Saunders, D.G.O.; Singh, K.B. Comparative secretome analysis of Rhizoctonia solani isolates with different host ranges reveals unique secretomes and cell death inducing effectors. Sci. Rep. 2017, 7, 10410. [Google Scholar] [CrossRef]
- Zhu, W.J.; Ronen, M.; Gur, Y.; Minz-Dub, A.; Masrati, G.; Ben-Tal, N.; Savidor, A.; Sharon, I.; Eizner, E.; Valerius, O.; et al. BcXYG1, a secreted xyloglucanase from Botrytis cinerea, triggers both cell death and plant immune responses. Plant Physiol. 2017, 175, 438–456. [Google Scholar] [CrossRef]
- Zhang, L.; Yan, J.P.; Fu, Z.C.; Shi, W.J.; Ninkuu, V.; Li, G.Y.; Yang, X.F.; Zeng, H.M. FoEG1, a secreted glycoside hydrolase family 12 protein from Fusarium oxysporum, triggers cell death and modulates plant immunity. Mol. Plant Pathol. 2021, 22, 522–538. [Google Scholar] [CrossRef]
- Gui, Y.J.; Chen, J.Y.; Zhang, D.D.; Li, N.Y.; Li, T.G.; Zhang, W.Q.; Wang, X.Y.; Short, D.P.G.; Li, L.; Guo, W.; et al. Verticillium dahliae manipulates plant immunity by glycoside hydrolase 12 proteins in conjunction with carbohydrate-binding module 1. Environ. Microbiol. 2017, 19, 1914–1932. [Google Scholar] [CrossRef]
- Zhang, L.S.; Kars, I.; Essenstam, B.; Liebrand, T.W.H.; Wagemakers, L.; Elberse, J.; Tagkalaki, P.; Tjoitang, D.; van den Ackerveken, G.; van Kan, J.A.L. Fungal endopolygalacturonases are recognized as microbe-associated molecular patterns by the Arabidopsis receptor-like protein RESPONSIVENESS TO BOTRYTIS POLYGALACTURONASES1. Plant Physiol. 2014, 164, 352–364. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.A.; Han, C.; Chen, J.Y.; Li, H.Y.; He, K.; Liu, A.X.; Li, D.C. Fungal cellulase is an elicitor but its enzymatic activity is not required for its elicitor activity. Mol. Plant Pathol. 2015, 16, 14–26. [Google Scholar] [CrossRef] [PubMed]
- Schouten, A.; van Baarlen, P.; van Kan, J.A.L. Phytotoxic Nep1-like proteins from the necrotrophic fungus Botrytis cinerea associate with membranes and the nucleus of plant cells. New Phytol. 2008, 177, 493–505. [Google Scholar] [CrossRef] [PubMed]
- Ono, E.; Mise, K.; Takano, Y. RLP23 is required for Arabidopsis immunity against the grey mould pathogen Botrytis cinerea. Sci. Rep. 2020, 10, 13798. [Google Scholar] [CrossRef]
- Redkar, A.; Cevik, V.; Bailey, K.; Zhao, H.; Kim, D.S.; Zou, Z.; Furzer, O.J.; Fairhead, S.; Borhan, M.H.; Holub, E.B.; et al. The Arabidopsis WRR4A and WRR4B paralogous NLR proteins both confer recognition of multiple Albugo candida effectors. New Phytol. 2022. [Google Scholar] [CrossRef]
- Lu, X.L.; Kracher, B.; Saur, I.M.L.; Bauer, S.; Ellwood, S.R.; Wise, R.; Yaeno, T.; Maekawa, T.; Schulze-Lefert, P. Allelic barley MLA immune receptors recognize sequence-unrelated avirulence effectors of the powdery mildew pathogen. Proc. Natl. Acad. Sci. USA 2016, 113, E6486–E6495. [Google Scholar] [CrossRef]
- Saur, I.M.L.; Bauer, S.; Kracher, B.; Lu, X.L.; Franzeskakis, L.; Muller, M.C.; Sabelleck, B.; Kummel, F.; Panstruga, R.; Maekawa, T.; et al. Multiple pairs of allelic MLA immune receptor-powdery mildew AVR(A) effectors argue for a direct recognition mechanism. Elife 2019, 8, e44471. [Google Scholar] [CrossRef]
- Bauer, S.; Yu, D.L.; Lawson, A.W.; Saur, I.M.L.; Frantzeskakis, L.; Kracher, B.; Logemann, E.; Chai, J.J.; Maekawa, T.; Schulze-Lefert, P. The leucine-rich repeats in allelic barley MLA immune receptors define specificity towards sequence-unrelated powdery mildew avirulence effectors with a predicted common RNase-like fold. PloS Pathog. 2021, 17, e1009223. [Google Scholar] [CrossRef]
- Praz, C.R.; Bourras, S.; Zeng, F.S.; Sanchez-Martin, J.; Menardo, F.; Xue, M.F.; Yang, L.J.; Roffler, S.; Boni, R.; Herren, G.; et al. AvrPm2 encodes an RNase-like avirulence effector which is conserved in the two different specialized forms of wheat and rye powdery mildew fungus. New Phytol. 2017, 213, 1301–1314. [Google Scholar] [CrossRef]
- Bourras, S.; McNally, K.E.; Ben-David, R.; Parlange, F.; Roffler, S.; Praz, C.R.; Oberhaensli, S.; Menardo, F.; Stirnweis, D.; Frenkel, Z.; et al. Multiple avirulence loci and allele-specific effector recognition control the Pm3 race-specific resistance of wheat to powdery mildew. Plant Cell 2015, 27, 2991–3012. [Google Scholar]
- Bourras, S.; Kunz, L.; Xue, M.F.; Praz, C.R.; Muller, M.C.; Kalin, C.; Schlafli, M.; Ackermann, P.; Fluckiger, S.; Parlange, F.; et al. The AvrPm3-Pm3 effector-NLR interactions control both race-specific resistance and host-specificity of cereal mildews on wheat. Nat. Commun. 2019, 10, 2292. [Google Scholar] [CrossRef] [PubMed]
- Muller, M.C.; Kunz, L.; Schudel, S.; Lawson, A.W.; Kammerecker, S.; Isaksson, J.; Wyler, M.; Graf, J.; Sotiropoulos, A.G.; Praz, C.R.; et al. Ancient variation of the AvrPm17 gene in powdery mildew limits the effectiveness of the introgressed rye Pm17 resistance gene in wheat. Proc. Natl. Acad. Sci. USA 2022, 119, e2108808119. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.A.; Thomas, C.M.; Hammondkosack, K.E.; Balintkurti, P.J.; Jones, J.D.G. Isolation of the tomato Cf-9 gene for resistance to Cladosporium Fulvum by transposon tagging. Science 1994, 266, 789–793. [Google Scholar] [CrossRef] [PubMed]
- Dixon, M.S.; Hatzixanthis, K.; Jones, D.A.; Harrison, K.; Jones, J.D.G. The tomato Cf-5 disease resistance gene and six homologs show pronounced allelic variation in leucine-rich repeat copy number. Plant Cell 1998, 10, 1915–1925. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.M.; Jones, D.A.; Parniske, M.; Harrison, K.; Balint-Kurti, P.J.; Hatzixanthis, K.; Jones, J.D.G. Characterization of the tomato Cf-4 gene for resistance to Cladosporium fulvum identifies sequences that determine recognitional specificity in Cf-4 and Cf-9. Plant Cell 1997, 9, 2209–2224. [Google Scholar]
- Dixon, M.S.; Jones, D.A.; Keddie, J.S.; Thomas, C.M.; Harrison, K.; Jones, J.D.G. The tomato Cf-2 disease resistance locus comprises two functional genes encoding leucine-rich repeat proteins. Cell 1996, 84, 451–459. [Google Scholar] [CrossRef]
- Takken, F.L.W.; Thomas, C.M.; Joosten, M.H.A.J.; Golstein, C.; Westerink, N.; Hille, J.; Nijkamp, H.J.J.; De Wit, P.J.G.M.; Jones, J.D.G. A second gene at the tomato Cf-4 locus confers resistance to Cladosporium fulvum through recognition of a novel avirulence determinant. Plant J. 1999, 20, 279–288. [Google Scholar] [CrossRef]
- Houterman, P.M.; Ma, L.; van Ooijen, G.; de Vroomen, M.J.; Cornelissen, B.J.C.; Takken, F.L.W.; Rep, M. The effector protein Avr2 of the xylem-colonizing fungus Fusarium oxysporum activates the tomato resistance protein I-2 intracellularly. Plant J. 2009, 58, 970–978. [Google Scholar] [CrossRef]
- Schmidt, S.M.; Lukasiewicz, J.; Farrer, R.; van Dam, P.; Bertoldo, C.; Rep, M. Comparative genomics of Fusarium oxysporum f. sp melonis reveals the secreted protein recognized by the Fom-2 resistance gene in melon. New Phytol. 2016, 209, 307–318. [Google Scholar] [CrossRef]
- Larkan, N.J.; Lydiate, D.J.; Parkin, I.A.P.; Nelson, M.N.; Epp, D.J.; Cowling, W.A.; Rimmer, S.R.; Borhan, M.H. The Brassica napus blackleg resistance gene LepR3 encodes a receptor-like protein triggered by the Leptosphaeria maculans effector AVRLM1. New Phytol. 2013, 197, 595–605. [Google Scholar] [CrossRef]
- Devanna, N.B.; Vijayan, J.; Sharma, T.R. The blast resistance gene Pi54of cloned from Oryza officinalis interacts with Avr-Pi54 through its novel non-LRR domains. PLoS ONE 2014, 9, e104840. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Kou, Y.J.; Bao, J.D.; Li, Y.; Tang, M.Z.; Zhu, X.L.; Ponaya, A.; Xiao, G.; Li, J.B.; Li, C.Y.; et al. Comparative genomics identifies the Magnaporthe oryzae avirulence effector AvrPi9 that triggers Pi9-mediated blast resistance in rice. New Phytol. 2015, 206, 1463–1475. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.L.; Wang, L.; Wu, W.H.; He, L.Y.; Yang, X.F.; Pan, Q.H. Function and evolution of Magnaporthe oryzae avirulence gene AvrPib responding to the rice blast resistance gene Pib. Sci. Rep. 2015, 5, 11642. [Google Scholar] [CrossRef] [PubMed]
- Kanzaki, H.; Yoshida, K.; Saitoh, H.; Fujisaki, K.; Hirabuchi, A.; Alaux, L.; Fournier, E.; Tharreau, D.; Terauchi, R. Arms race co-evolution of Magnaporthe oryzae AVR-Pik and rice Pik genes driven by their physical interactions. Plant J. 2012, 72, 894–907. [Google Scholar] [CrossRef]
- Li, W.; Wang, B.H.; Wu, J.; Lu, G.D.; Hu, Y.J.; Zhang, X.; Zhang, Z.G.; Zhao, Q.; Feng, Q.Y.; Zhang, H.Y.; et al. The Magnaporthe oryzae avirulence gene AvrPiz-t encodes a predicted secreted protein that triggers the immunity in rice mediated by the blast resistance gene Piz-t. Mol. Plant-Microbe Interact. 2009, 22, 411–420. [Google Scholar] [CrossRef]
- Jia, Y.L.; Zhou, E.; Lee, S.; Bianco, T. Coevolutionary dynamics of rice blast resistance gene Pi-ta and Magnaporthe oryzae avirulence gene AVR-Pita 1. Phytopathology 2016, 106, 676–683. [Google Scholar] [CrossRef]
- Cesari, S.; Thilliez, G.; Ribot, C.; Chalvon, V.; Michel, C.; Jauneau, A.; Rivas, S.; Alaux, L.; Kanzaki, H.; Okuyama, Y.; et al. The rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae effectors AVR-Pia and AVR1-CO39 by direct binding. Plant Cell 2013, 25, 1463–1481. [Google Scholar] [CrossRef]
- Vo, K.T.X.; Lee, S.K.; Halane, M.K.; Song, M.Y.; Hoang, T.V.; Kim, C.Y.; Park, S.Y.; Jeon, J.; Kim, S.T.; Sohn, K.H.; et al. Pi5 and Pii paired NLRs are functionally exchangeable and confer similar disease resistance specificity. Mol. Cells 2019, 42, 637–645. [Google Scholar]
- Anderson, C.; Khan, M.A.; Catanzariti, A.M.; Jack, C.A.; Nemri, A.; Lawrence, G.J.; Upadhyaya, N.M.; Hardham, A.R.; Ellis, J.G.; Dodds, P.N.; et al. Genome analysis and avirulence gene cloning using a high-density RADseq linkage map of the flax rust fungus, Melampsora lini. BMC Genom. 2016, 17, 667. [Google Scholar] [CrossRef]
- Ve, T.; Williams, S.J.; Catanzariti, A.M.; Rafiqi, M.; Rahman, M.; Ellis, J.G.; Hardham, A.R.; Jones, D.A.; Anderson, P.A.; Dodds, P.N.; et al. Structures of the flax-rust effector AvrM reveal insights into the molecular basis of plant-cell entry and effector-triggered immunity. Proc. Natl. Acad. Sci. USA 2013, 110, 17594–17599. [Google Scholar] [CrossRef]
- Dodds, P.N.; Lawrence, G.J.; Catanzariti, A.M.; Ayliffe, M.A.; Ellis, J.G. The Melampsora lini AvrL567 avirulence genes are expressed in haustoria and their products are recognized inside plant cells. Plant Cell 2004, 16, 755–768. [Google Scholar] [CrossRef] [PubMed]
- Deng, C.; Leonard, A.; Cahill, J.; Lv, M.; Li, Y.R.; Thatcher, S.; Li, X.Y.; Zhao, X.D.; Du, W.J.; Li, Z.; et al. The RppC-AvrRppC NLR-effector interaction mediates the resistance to southern corn rust in maize. Mol. Plant 2022, 15, 904–912. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Zhang, B.; Ding, J.; Wang, H.; Deng, C.; Wang, J.; Yang, Q.; Pi, Q.; Zhang, R.; Zhai, H.; et al. Cloning southern corn rust resistant gene RppK and its cognate gene AvrRppK from Puccinia polysora. Nat. Commun. 2022, 13, 4392. [Google Scholar] [CrossRef]
- Wang, J.; Chai, J. Structural insights into the plant immune receptors PRRs and NLRs. Plant Physiol. 2020, 182, 1566–1581. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Xu, G.; He, P.; Shan, L. SERKing coreceptors for receptors. Trends Plant Sci. 2016, 21, 1017–1033. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Liu, Z.; Song, C.; Hu, Y.; Han, Z.; She, J.; Fan, F.; Wang, J.; Jin, C.; Chang, J. Chitin-Induced dimerization activates a plant immune receptor. Science 2012, 336, 1160–1164. [Google Scholar] [CrossRef]
- Ngou, B.P.M.; Jones, J.D.G.; Ding, P.T. Plant immune networks. Trends Plant Sci. 2022, 27, 255–273. [Google Scholar] [CrossRef]
- Jones, J.D.G.; Vance, R.E.; Dangl, J.L. Intracellular innate immune surveillance devices in plants and animals. Science 2016, 354, aaf6395. [Google Scholar] [CrossRef]
- Liu, X.X.; Wan, L. Molecular insights into the biochemical functions and signalling mechanisms of plant NLRs. Mol. Plant Pathol. 2022, 23, 772–780. [Google Scholar] [CrossRef]
- Rodriguez-Moreno, L.; Song, Y.; Thomma, B.P.H.J. Transfer and engineering of immune receptors to improve recognition capacities in crops. Curr. Opin. Plant Biol. 2017, 38, 42–49. [Google Scholar] [CrossRef]
- Schultink, A.; Steinbrenner, A.D. A playbook for developing disease-resistant crops through immune receptor identification and transfer. Curr. Opin. Plant Biol. 2021, 62, 102089. [Google Scholar] [CrossRef] [PubMed]
- Brunner, S.; Stirnweis, D.; Quijano, C.D.; Buesing, G.; Herren, G.; Parlange, F.; Barret, P.; Tassy, C.; Sautter, C.; Winzeler, M.; et al. Transgenic Pm3 multilines of wheat show increased powdery mildew resistance in the field. Plant Biotechnol. J. 2012, 10, 398–409. [Google Scholar] [CrossRef] [PubMed]
- Fradin, E.F.; Abd-El-Haliem, A.; Masini, L.; van den Berg, G.C.M.; Joosten, M.H.A.J.; Thomma, B.P.H.J. Interfamily transfer of tomato Ve1 mediates Verticillium resistance in Arabidopsis. Plant Physiol. 2011, 156, 2255–2265. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Deng, Y.W.; Ning, Y.S.; He, Z.H.; Wang, G.L. Exploiting broad-spectrum disease resistance in crops: From molecular dissection to breeding. Annu. Rev. Plant Biol. 2020, 71, 575–603. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Li, J.F.; Ao, Y.; Qu, J.W.; Li, Z.Q.; Su, J.B.; Zhang, Y.; Liu, J.; Feng, D.R.; Qi, K.B.; et al. Lysin motif-containing proteins LYP4 and LYP6 play dual roles in peptidoglycan and chitin perception in rice innate immunity. Plant Cell 2012, 24, 3406–3419. [Google Scholar] [CrossRef]
- Luo, M.; Xie, L.Q.; Chakraborty, S.; Wang, A.H.; Matny, O.; Jugovich, M.; Kolmer, J.A.; Richardson, T.; Bhatt, D.; Hoque, M.; et al. A five-transgene cassette confers broad-spectrum resistance to a fungal rust pathogen in wheat. Nat. Biotechnol. 2021, 39, 561–566. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, X.; Yuan, G.X.; Wang, D.L.; Zheng, Y.Y.; Ma, M.Q.; Guo, L.W.; Bhadauria, V.; Peng, Y.L.; Liu, J.F. A designer rice NLR immune receptor confers resistance to the rice blast fungus carrying noncorresponding avirulence effectors. Proc. Natl. Acad. Sci. USA 2021, 118, e2110751118. [Google Scholar] [CrossRef]
- Wang, N.; Tang, C.; Fan, X.; He, M.; Gan, P.; Zhang, S.; Hu, Z.; Wang, X.; Yan, T.; Shu, W.; et al. Inactivation of a wheat protein kinase gene confers broad-spectrum resistance to rust fungi. Cell 2022, 185, 2961–2974. [Google Scholar] [CrossRef]
- Ngou, B.P.M.; Ding, P.T.; Jones, J.D.G. Thirty years of resistance: Zig-zag through the plant immune system. Plant Cell 2022, 34, 1447–1478. [Google Scholar] [CrossRef]
- Wang, W.; Feng, B.; Zhou, J.-M.; Tang, D. Plant immune signaling: Advancing on two frontiers. J. Integr. Plant Biol. 2020, 62, 2–24. [Google Scholar] [CrossRef]
- Abdul Malik, N.A.; Kumar, I.S.; Nadarajah, K. Elicitor and receptor molecules: Orchestrators of plant defense and immunity. Int. J. Mol. Sci. 2020, 21, 963. [Google Scholar] [CrossRef] [PubMed]
- DeFalco, T.A.; Zipfel, C. Molecular mechanisms of early plant pattern-triggered immune signaling. Mol. Cell 2021, 81, 3449–3467. [Google Scholar] [CrossRef]
- Xue, D.-X.; Li, C.-L.; Xie, Z.-P.; Staehelin, C. LYK4 is a component of a tripartite chitin receptor complex in Arabidopsis thaliana. J. Exp. Bot. 2019, 70, 5507–5516. [Google Scholar] [CrossRef] [PubMed]
- Paparella, C.; Savatin, D.V.; Marti, L.; De Lorenzo, G.; Ferrari, S. The Arabidopsis LYSIN MOTIF-CONTAINING RECEPTOR-LIKE KINASE3 regulates the cross talk between immunity and abscisic acid responses. Plant Physiol. 2014, 165, 262–276. [Google Scholar] [CrossRef] [PubMed]
- Le, M.H.; Cao, Y.; Zhang, X.C.; Stacey, G. LIK1, a CERK1-interacting kinase, regulates plant immune responses in Arabidopsis. PLoS ONE 2014, 9, e102245. [Google Scholar] [CrossRef]
- Liang, X.; Zhou, J.M. Receptor-like cytoplasmic kinases: Central players in plant receptor kinase–mediated signaling. Annu. Rev. Plant Biol. 2018, 69, 267–299. [Google Scholar] [CrossRef]
- Tian, W.; Hou, C.; Ren, Z.; Wang, C.; Zhao, F.; Dahlbeck, D.; Hu, S.; Zhang, L.; Niu, Q.; Li, L.; et al. A calmodulin-gated calcium channel links pathogen patterns to plant immunity. Nature 2019, 572, 131–135. [Google Scholar] [CrossRef]
- Kadota, Y.; Sklenar, J.; Derbyshire, P.; Stransfeld, L.; Asai, S.; Ntoukakis, V.; Jonathan, D.G.J.; Shirasu, K.; Menke, F.; Jones, A.; et al. Direct regulation of the NADPH oxidase RBOHD by the PRR-associated kinase BIK1 during plant immunity. Mol. Cell 2014, 54, 43–55. [Google Scholar] [CrossRef]
- Rao, S.; Zhou, Z.; Miao, P.; Bi, G.; Hu, M.; Wu, Y.; Feng, F.; Zhang, X.; Zhou, J.-M. Roles of receptor-like cytoplasmic kinase VII members in pattern-triggered immune signaling. Plant Physiol. 2018, 177, 1679–1690. [Google Scholar] [CrossRef]
- Lv, S.; Zhang, Y.; Li, C.; Liu, Z.; Yang, N.; Pan, L.; Wu, J.; Wang, J.; Yang, J.; Lv, Y.; et al. Strigolactone-triggered stomatal closure requires hydrogen peroxide synthesis and nitric oxide production in an abscisic acid-independent manner. New Phytol. 2018, 217, 290–304. [Google Scholar] [CrossRef]
- Lin, W.; Tsuda, K.; Sato, M.; Cohen, J.D.; Katagiri, F.; Glazebrook, J. Arabidopsis CaM binding protein CBP60g contributes to MAMP-induced SA accumulation and is involved in disease resistance against Pseudomonas syringae. PLoS Pathog. 2009, 5, e1000301. [Google Scholar]
- Waszczak, C.; Carmody, M.; Kangasjärvi, J. Reactive oxygen species in plant signaling. Annu. Rev. Plant Biol. 2018, 69, 209–236. [Google Scholar] [CrossRef] [PubMed]
- Dvořák, P.; Krasylenko, Y.; Zeiner, A.; Šamaj, J.; Takáč, T. Signaling toward reactive oxygen species-scavenging enzymes in plants. Front. Plant Sci. 2021, 11, 2178. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Guleria, S.; Ghosh, D.; Dogra, V.; Kumar, S. Managing reactive oxygen species—Some learnings from high altitude extremophytes. Environ. Exp. Bot. 2021, 189, 104525. [Google Scholar] [CrossRef]
- Zhang, J.; Li, W.; Xiang, T.; Liu, Z.; Laluk, K.; Ding, X.; Zou, Y.; Gao, M.; Zhang, X.; Chen, S.; et al. Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe 2010, 7, 290–301. [Google Scholar] [CrossRef]
- Shinya, T.; Yamaguchi, K.; Desaki, Y.; Yamada, K.; Narisawa, T.; Kobayashi, Y.; Maeda, K.; Suzuki, M.; Tanimoto, T.; Takeda, J.; et al. Selective regulation of the chitin-induced defense response by the Arabidopsis receptor-like cytoplasmic kinase PBL27. Plant J. 2014, 79, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Yamada, K.; Yamaguchi, K.; Shirakawa, T.; Nakagami, H.; Mine, A.; Ishikawa, K.; Fujiwara, M.; Narusaka, M.; Narusaka, Y.; Ichimura, K.; et al. The Arabidopsis CERK1-associated kinase PBL27 connects chitin perception to MAPK activation. EMBO J. 2016, 35, 2468–2483. [Google Scholar] [CrossRef]
- Bi, G.; Zhou, Z.; Wang, W.; Li, L.; Rao, S.; Wu, Y.; Zhang, X.; Menke, F.L.H.; Chen, S.; Zhou, J.-M. Receptor-like cytoplasmic kinases directly link diverse pattern recognition receptors to the activation of mitogen-activated protein kinase cascades in Arabidopsis. Plant Cell 2018, 30, 1543–1561. [Google Scholar] [CrossRef]
- Li, G.; Meng, X.; Wang, R.; Mao, G.; Han, L.; Liu, Y.; Zhang, S. Dual-level regulation of ACC synthase activity by MPK3/MPK6 cascade and its downstream WRKY transcription factor during ethylene induction in Arabidopsis. PLoS Genet. 2012, 8, e1002767. [Google Scholar] [CrossRef]
- Martel, A.; Ruiz-Bedoya, T.; Breit-McNally, C.; Laflamme, B.; Desveaux, D.; Guttman, D.S. The ETS-ETI cycle: Evolutionary processes and metapopulation dynamics driving the diversification of pathogen effectors and host immune factors. Curr. Opin. Plant Biol. 2021, 62, 102011. [Google Scholar] [CrossRef]
- Fick, A.; Swart, V.; van den Berg, N. The ups and downs of plant NLR expression during pathogen infection. Front. Plant Sci. 2022, 13, 921148. [Google Scholar] [CrossRef] [PubMed]
- Dodds, P.N.; Rathjen, J.P. Plant immunity: Towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 2010, 11, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; M Ca Dams, S.A.; Bryan, G.T.; Hershey, H.P.; Valent, B. Direct interaction of resistance gene and avirulence gene products confers rice BLAST resistance. EMBO J. 2000, 19, 4004–4014. [Google Scholar] [CrossRef] [PubMed]
- Li, W. Molecular mechanisms of Cf-dependent ETI and Xoo-induced nonhost resistance in Nicotiana benthamiana. Ph.D. Thesis, Zhejiang University, Hangzhou, China, 2015. [Google Scholar]
- Kang, H.; Nguyen, Q.-M.; Iswanto, A.B.B.; Hong, J.C.; Bhattacharjee, S.; Gassmann, W.; Kim, S.H. Nuclear localization of HopA1Pss61 is required for effector-triggered immunity. Plants 2021, 10, 888. [Google Scholar] [CrossRef] [PubMed]
- Lapin, D.; Bhandari, D.D.; Parker, J.E. Origins and immunity networking functions of EDS1 family proteins. Annu. Rev. Phytopathol. 2020, 58, 253–276. [Google Scholar] [CrossRef] [PubMed]
- Bhandari, D.D.; Lapin, D.; Kracher, B.; von Born, P.; Bautor, J.; Niefind, K.; Parker, J.E. An EDS1 heterodimer signalling surface enforces timely reprogramming of immunity genes in Arabidopsis. Nat. Commun. 2019, 10, 772. [Google Scholar] [CrossRef]
- Lapin, D.; Kovacova, V.; Sun, X.; Dongus, J.A.; Parker, J.E. A coevolved EDS1-SAG101-NRG1 module mediates cell death signaling by TIR-domain immune receptors. Plant Cell 2019, 31, 2430–2455. [Google Scholar] [CrossRef]
- Zhou, J.-M.; Zhang, Y. Plant immunity: Danger perception and signaling. Cell 2020, 181, 978–989. [Google Scholar] [CrossRef]
- Xiao, S.; Calis, O.; Patrick, E.; Zhang, G.; Charoenwattana, P.; Muskett, P.; Parker, J.E.; Turner, J.G. The atypical resistance gene, RPW8, recruits components of basal defence for powdery mildew resistance in Arabidopsis. Plant J. 2005, 42, 95–110. [Google Scholar] [CrossRef]
- De Jong, C.F.; HonÉE, G.U.Y.; Joosten, M.H.A.J.; De Wit, P.J.G.M. Early defence responses induced by AVR9 and mutant analogues in tobacco cell suspensions expressing the Cf-9 resistance gene. Physiol. Mol. Plant P. 2000, 56, 169–177. [Google Scholar] [CrossRef]
- Heese, A.; Ludwig, A.A.; Jones, J.D.G. Rapid phosphorylation of a syntaxin during the Avr9/Cf-9-race-specific signaling pathway. Plant Physiol. 2005, 138, 2406–2416. [Google Scholar] [CrossRef] [PubMed]
- Dongus, J.A.; Parker, J.E. EDS1 signalling: At the nexus of intracellular and surface receptor immunity. Curr. Opin. Plant Biol. 2021, 62, 102039. [Google Scholar] [CrossRef] [PubMed]
- Stergiopoulos, I.; Wit, P.D. Fungal effector proteins. Annu. Rev. Phytopathol. 2009, 47, 233–263. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xue, J.; Wang, F.-Z.; Huang, X.; Gong, B.-Q.; Tao, Y.; Shen, W.; Tao, K.; Yao, N.; Xiao, S.; et al. Plasma membrane-nucleo-cytoplasmic coordination of a receptor-like cytoplasmic kinase promotes EDS1-dependent plant immunity. Nat. Plants 2022, 8, 802–816. [Google Scholar] [CrossRef]
- Pruitt, R.N.; Locci, F.; Wanke, F.; Zhang, L.; Saile, S.C.; Joe, A.; Karelina, D.; Hua, C.; Fröhlich, K.; Wan, W.-L.; et al. The EDS1–PAD4–ADR1 node mediates Arabidopsis pattern-triggered immunity. Nature 2021, 598, 495–499. [Google Scholar] [CrossRef]
- Wan, L.; Essuman, K.; Anderson, R.G.; Sasaki, Y.; Monteiro, F.; Chung, E.H.; Nishimura, E.O.; DiAntonio, A.; Milbrandt, J.; Dangl, J.L.; et al. TIR domains of plant immune receptors are NAD(+)-cleaving enzymes that promote cell death. Science 2019, 365, 799–803. [Google Scholar] [CrossRef]
- Horsefield, S.; Burdett, H.; Zhang, X.X.; Manik, M.K.; Shi, Y.; Chen, J.; Qi, T.C.; Gilley, J.; Lai, J.S.; Rank, M.X.; et al. NAD(+) cleavage activity by animal an plant TIR domains in cell death pathways. Science 2019, 365, 793–799. [Google Scholar] [CrossRef]
- Tian, H.; Wu, Z.; Chen, S.; Ao, K.; Huang, W.; Yaghmaiean, H.; Sun, T.; Xu, F.; Zhang, Y.; Wang, S.; et al. Activation of TIR signalling boosts pattern-triggered immunity. Nature 2021, 598, 500–503. [Google Scholar] [CrossRef]
- Perez-Hernandez, A.; Gonzalez, M.; Gonzalez, C.; Brito, N. The elicitor protein BcIEB1 and the derived peptide ieb35 provide long-term plant protection. Plant Pathol. 2020, 69, 807–817. [Google Scholar] [CrossRef]
- Cheng, Y.L.; Lin, Y.L.; Cao, H.H.; Li, Z.G. Citrus postharvest green mold: Recent advances in fungal pathogenicity and fruit resistance. Microorganisms 2020, 8, 449. [Google Scholar] [CrossRef]
- Tian, S.; Torres, R.; Ballester, A.R.; Li, B.; Vilanova, L.; Gonzalez-Candelas, L. Molecular aspects in pathogen-fruit interactions: Virulence and resistance. Postharvest Biol. Tec. 2016, 122, 11–21. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, Y.P.; Sun, Y.J.; Wang, H.B.; Qi, J.M.; Wan, B.W.; Ye, W.W.; Lin, Y.C.; Shao, Y.Y.; Dong, S.M.; et al. Leucine-rich repeat receptor-like gene screen reveals that Nicotiana RXEG1 regulates glycoside hydrolase 12 MAMP detection. Nat. Commun. 2018, 9, 594. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.H.; Ngou, B.P.M.; Ding, P.T.; Xiu-Fan, X. PTI-ETI crosstalk: An integrative view of plant immunity. Curr. Opin. Plant Biol. 2021, 62, 102030. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, J.; Cheng, Y. Advances in Fungal Elicitor-Triggered Plant Immunity. Int. J. Mol. Sci. 2022, 23, 12003. https://doi.org/10.3390/ijms231912003
Guo J, Cheng Y. Advances in Fungal Elicitor-Triggered Plant Immunity. International Journal of Molecular Sciences. 2022; 23(19):12003. https://doi.org/10.3390/ijms231912003
Chicago/Turabian StyleGuo, Jia, and Yulin Cheng. 2022. "Advances in Fungal Elicitor-Triggered Plant Immunity" International Journal of Molecular Sciences 23, no. 19: 12003. https://doi.org/10.3390/ijms231912003
APA StyleGuo, J., & Cheng, Y. (2022). Advances in Fungal Elicitor-Triggered Plant Immunity. International Journal of Molecular Sciences, 23(19), 12003. https://doi.org/10.3390/ijms231912003





