De Novo Transcriptome Analysis Reveals Flowering-Related Genes That Potentially Contribute to Flowering-Time Control in the Japanese Cultivated Gentian Gentiana triflora
Abstract
1. Introduction
2. Results
2.1. Identification of Reference Genes Expressed in Gentian Leaves, and Gene Annotation via BLAST Analysis and Protein Motif Analysis
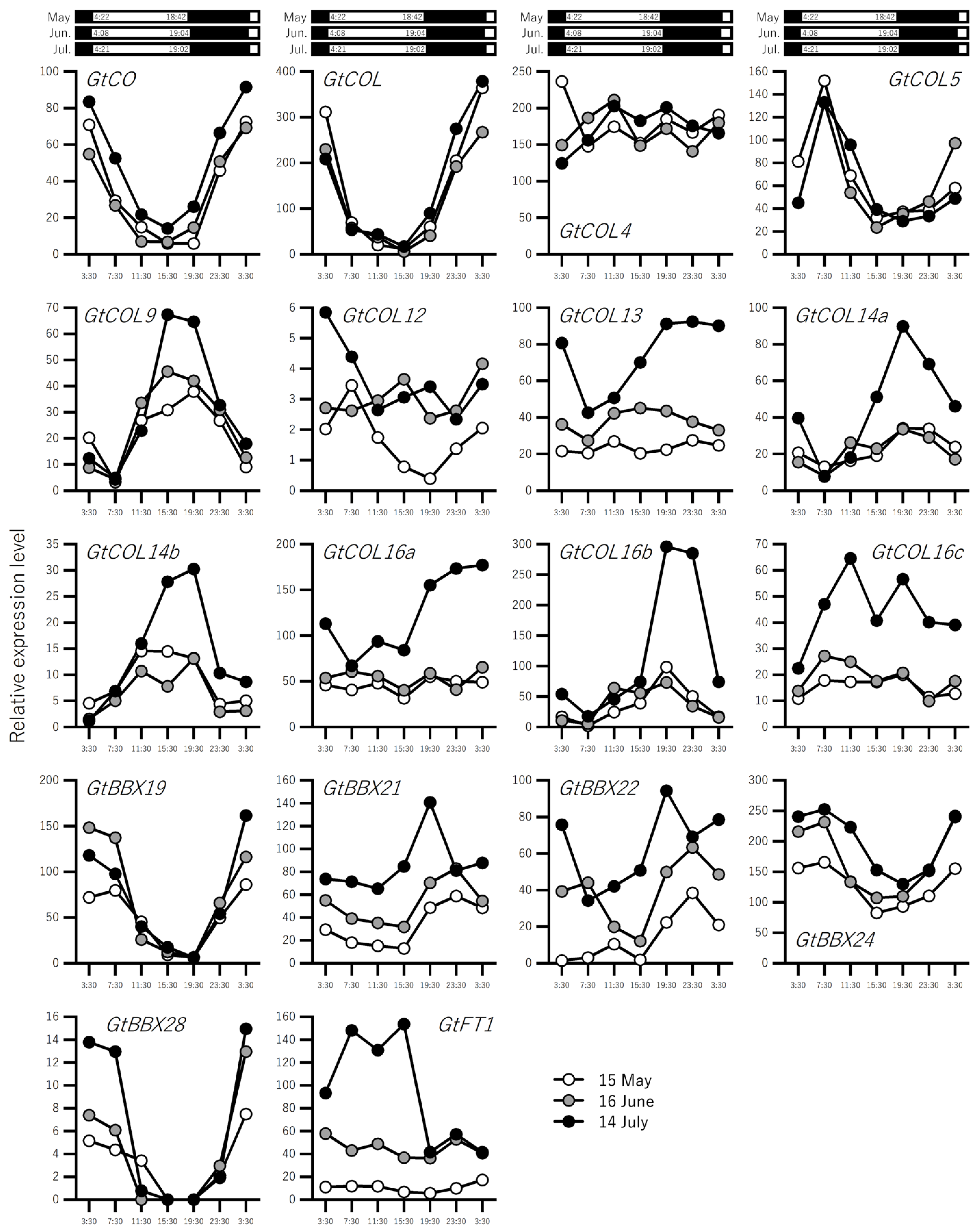
2.2. Expression Profiling of Flowering-Related Genes in Leaves over a Three-Month Period Prior to Flowering
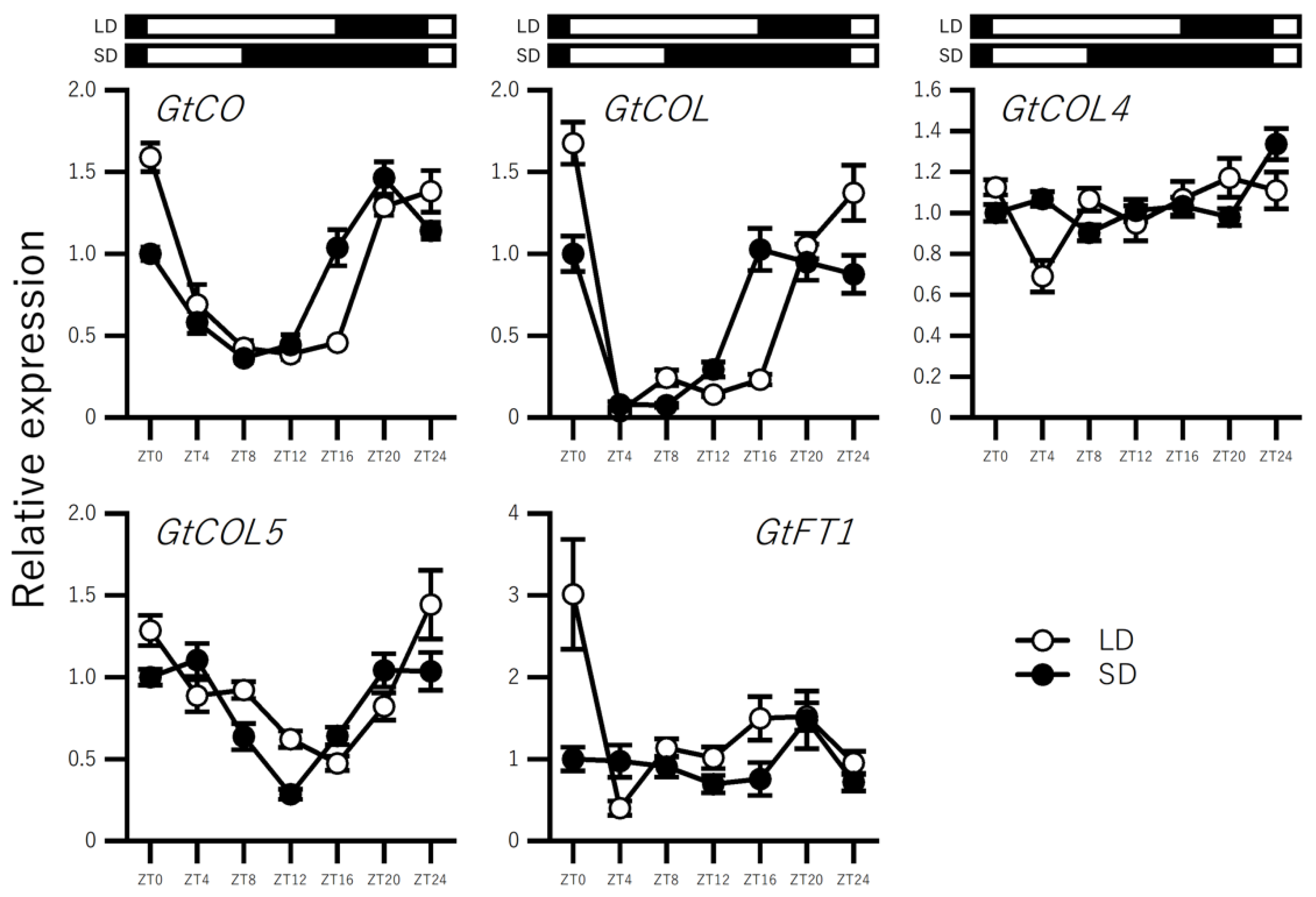
2.3. Effect of Photoperiod on the Expression of BBX Genes in In Vitro–Grown Seedlings
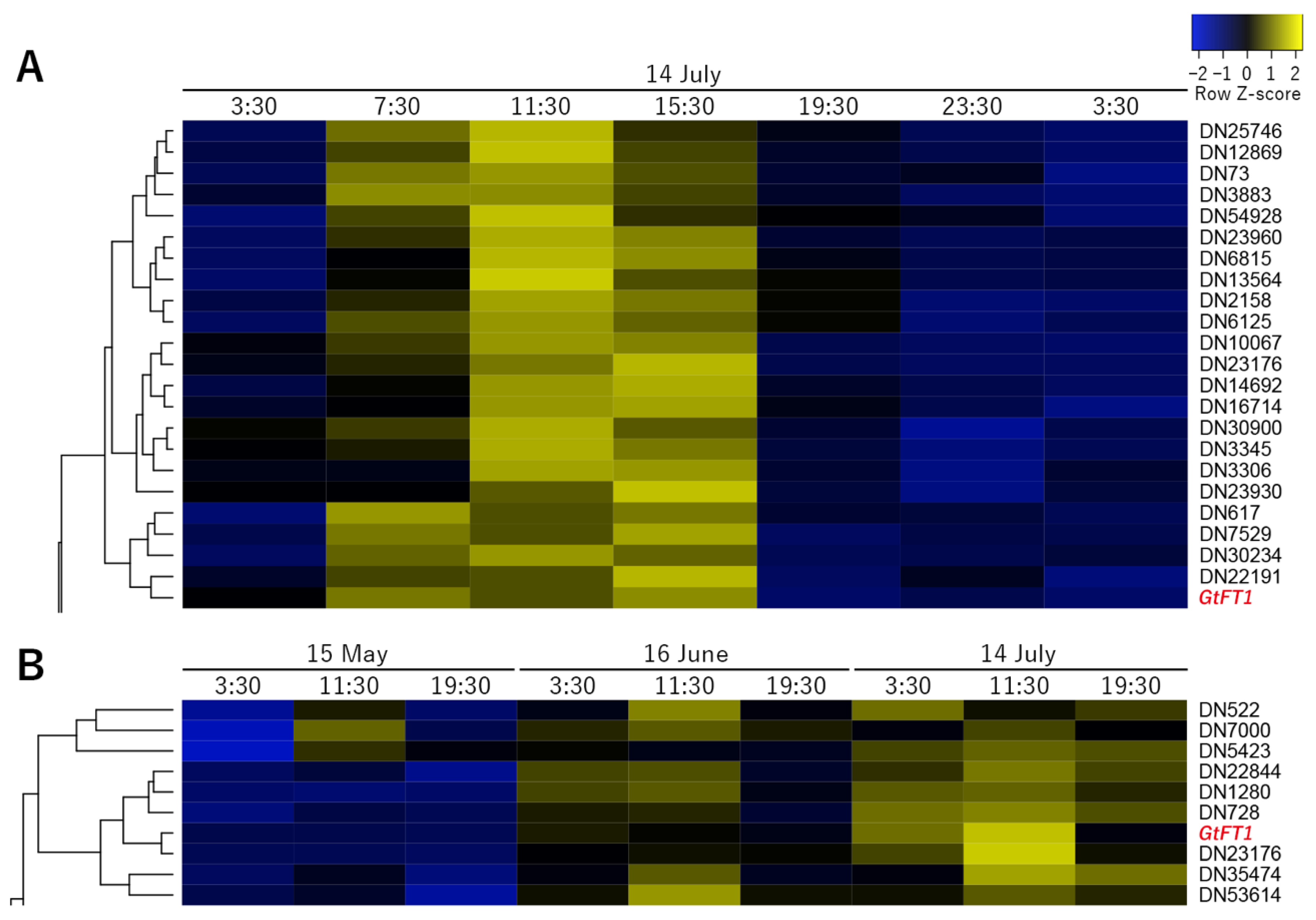
2.4. Coexpression Cluster Analysis
3. Discussion
4. Materials and Methods
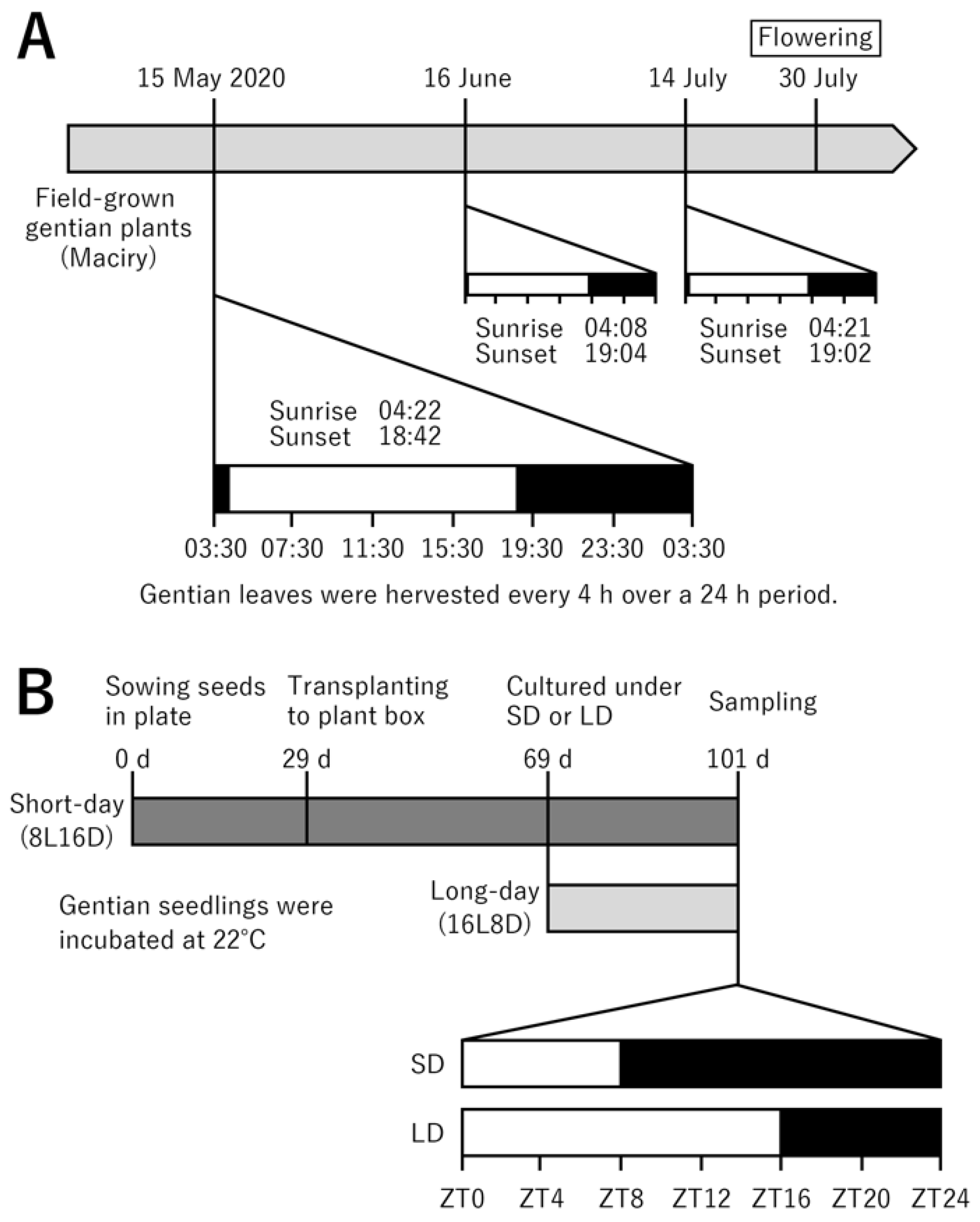
4.1. Plant Materials
4.2. RNA-Seq Analysis
4.3. Validation via qRT-PCR Analysis
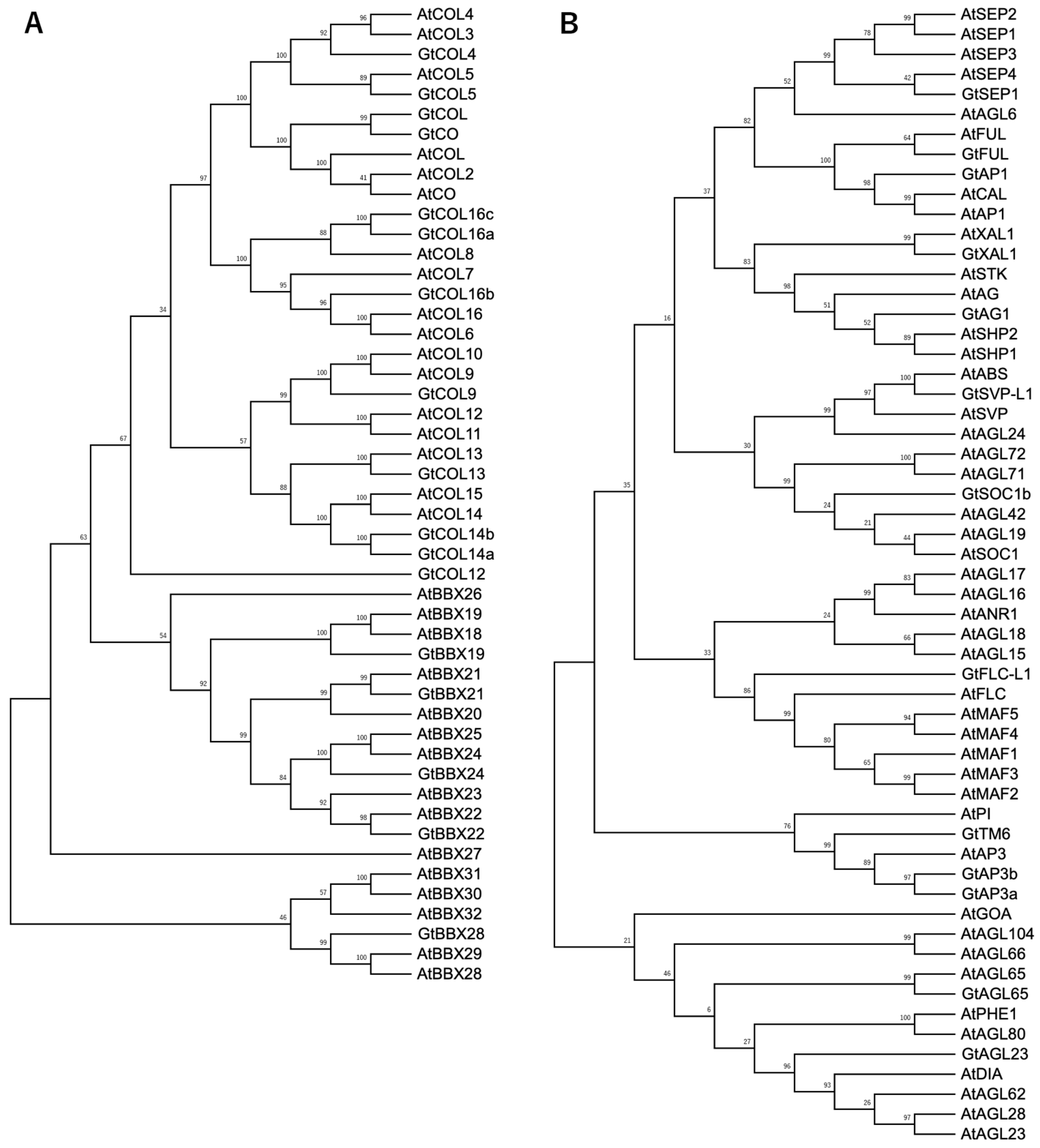
4.4. Phylogenetic Analysis of the BBX and MADS-Box Family Proteins
4.5. Analysis of BBX Gene Expression in Response to Photoperiod in In-Vitro-Grown Seedlings
4.6. Gene Coexpression Analysis in Field-Grown Gentian Plants
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kodama, K. IV-9, Gentian. In Horticulture in Japan 2006; The Japanese Society of Hortuculture Science, Ed.; Shoukadoh Publication: Kyoto, Japan, 2006; pp. 248–253. [Google Scholar]
- Eason, J.R.; Morgan, E.R.; Mullan, A.C.; Burge, G.K. Display life of Gentiana flowers is cultivar specific and influenced by sucrose, gibberellin, fluoride, and postharvest storage. N. Z. J. Crop Hortic. Sci. 2004, 32, 217–226. [Google Scholar] [CrossRef]
- Shimizu-Yumoto, H.; Ichimura, K. Effects of ethylene, pollination, and ethylene inhibitor treatments on flower senescence of gentians. Postharvest Biol. Technol. 2012, 63, 111–115. [Google Scholar] [CrossRef]
- Çelikel, F.G. Postharvest physiology of flowers from the family Gentianaceae. In The Gentianaceae—Volume 2: Biotechnology and Applications; Rybczyński, J.J., Davey, M.R., Mikuła, A., Eds.; Springer: Berlin/Heidelberg, Germany, 2015; pp. 287–305. [Google Scholar]
- Higuchi, Y.; Narumi, T.; Oda, A.; Nakano, Y.; Sumitomo, K.; Fukai, S.; Hisamatsu, T. The gated induction system of a systemic floral inhibitor, antiflorigen, determines obligate short-day flowering in chrysanthemums. Proc. Natl. Acad. Sci. USA 2013, 110, 17137–17142. [Google Scholar] [CrossRef]
- Hisamatsu, T. Denshosaibai no Kiso to Jissen; Hisamatsu, T., Ed.; Seibundo-Shinkosha: Tokyo, Japan, 2014; p. 225. (In Japanese) [Google Scholar]
- Song, J.; Zhang, S.; Wang, X.; Sun, S.; Liu, Z.; Wang, K.; Wan, H.; Zhou, G.; Li, R.; Yu, H.; et al. Variations in both FTL1 and SP5G, two tomato FT paralogs, control day-neutral flowering. Mol. Plant 2020, 13, 939–942. [Google Scholar] [CrossRef]
- Yang, A.; Xu, Q.; Hong, Z.; Wang, X.; Zeng, K.; Yan, L.; Liu, Y.; Zhu, Z.; Wang, H.; Xu, Y. Modified photoperiod response of CsFT promotes day neutrality and early flowering in cultivated cucumber. Theor. Appl. Genet. 2022, 135, 2735–2746. [Google Scholar] [CrossRef] [PubMed]
- Imamura, T.; Nakatsuka, T.; Higuchi, A.; Nishihara, M.; Takahashi, H. The gentian orthologs of the FT/TFL1 gene family control floral initiation in Gentiana. Plant Cell Physiol. 2011, 52, 1031–1041. [Google Scholar] [CrossRef]
- Higuchi, Y. Florigen and anti-florigen: Flowering regulation in horticultural crops. Breed. Sci. 2018, 68, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Yamagishi, N.; Kume, K.; Hikage, T.; Takahashi, Y.; Bidadi, H.; Wakameda, K.; Saitoh, Y.; Yoshikawa, N.; Tsutsumi, K. Identification and functional analysis of SVP ortholog in herbaceous perennial plant Gentiana triflora: Implication for its multifunctional roles. Plant Sci. 2016, 248, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Nakatsuka, T.; Saito, M.; Yamada, E.; Fujita, K.; Yamagishi, N.; Yoshikawa, N.; Nishihara, M. Isolation and characterization of the C-class MADS-box gene involved in the formation of double flowers in Japanese gentian. BMC Plant Biol. 2015, 15, 182. [Google Scholar] [CrossRef]
- Nakatsuka, T.; Saito, M.; Nishihara, M. Functional characterization of duplicated B-class MADS-box genes in Japanese gentian. Plant Cell Rep. 2016, 35, 895–904. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; Imamura, T.; Konno, N.; Takeda, T.; Fujita, K.; Konishi, T.; Nishihara, M.; Uchimiya, H. The gentio-oligosaccharide gentiobiose functions in the modulation of bud dormancy in the herbaceous perennial Gentiana. Plant Cell 2014, 26, 3949–3963. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; Nishihara, M.; Yoshida, C.; Itoh, K. Gentian FLOWERING LOCUS T orthologs regulate phase transitions: Floral induction and endodormancy release. Plant Physiol. 2022, 188, 1887–1899. [Google Scholar] [CrossRef] [PubMed]
- Jin, Q.; Yin, S.; Li, G.; Guo, T.; Wan, M.; Li, H.; Li, J.; Ge, X.; King, G.J.; Li, Z.; et al. Functional homoeologous alleles of CONSTANS contribute to seasonal crop type in rapeseed. Theor. Appl. Genet. 2021, 134, 3287–3303. [Google Scholar] [CrossRef] [PubMed]
- Shalmani, A.; Fan, S.; Jia, P.; Li, G.; Muhammad, I.; Li, Y.; Sharif, R.; Dong, F.; Zuo, X.; Li, K.; et al. Genome identification of B-BOX gene family members in seven Rosaceae species and their expression analysis in response to flower induction in Malus domestica. Molecules 2018, 23, 1763. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Sun, J.; Jiang, A.; Bai, M.; Fan, C.; Liu, J.; Ning, G.; Wang, C. Alternate expression of CONSTANS-LIKE 4 in short days and CONSTANS in long days facilitates day-neutral response in Rosa chinensis. J. Exp. Bot. 2020, 71, 4057–4068. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Weers, B.D.; Morishige, D.T.; Mullet, J.E. CONSTANS is a photoperiod regulated activator of flowering in sorghum. BMC Plant Biol. 2014, 14, 148. [Google Scholar] [CrossRef]
- Zhou, R.; Liu, P.; Li, D.; Zhang, X.; Wei, X. Photoperiod response-related gene SiCOL1 contributes to flowering in sesame. BMC Plant Biol. 2018, 18, 343. [Google Scholar] [CrossRef] [PubMed]
- González-Schain, N.D.; Díaz-Mendoza, M.; Zurczak, M.; Suárez-López, P. Potato CONSTANS is involved in photoperiodic tuberization in a graft-transmissible manner. Plant J. 2012, 70, 678–690. [Google Scholar] [CrossRef]
- Chiang, C.; Viejo, M.; Aas, O.T.; Hobrak, K.T.; Strømme, C.B.; Fløistad, I.S.; Olsen, J.E. Interactive effects of light quality during day extension and temperature on bud set, bud burst and PaFTL2, PaCOL1-2 and PaSOC1 expression in Norway spruce (Picea abies (L.) Karst.). Forests 2021, 12, 337. [Google Scholar] [CrossRef]
- Böhlenius, H.; Huang, T.; Charbonnel-Campaa, L.; Brunner, A.M.; Jansson, S.; Strauss, S.H.; Nilsson, O. CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science 2006, 312, 1040–1043. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; He, Y.; Niu, S.; Yan, S.; Zhang, Y. Identification and characterization of the CONSTANS (CO)/CONSTANS-like (COL) genes related to photoperiodic signaling and flowering in tomato. Plant Sci. 2020, 301, 110653. [Google Scholar] [CrossRef] [PubMed]
- Martin, L.B.B.; Fei, Z.; Giovannoni, J.J.; Rose, J.K.C. Catalyzing plant science research with RNA-seq. Front. Plant Sci. 2013, 4, 66. [Google Scholar] [CrossRef] [PubMed]
- Sripathi, V.R.; Anche, V.C.; Gossett, Z.B.; Walker, L.T. Recent applications of RNA sequencing in food and agriculture. In Applications of RNA-Seq in Biology and Medicine; Louis, I.V.-S., Ed.; IntechOpen: London, UK, 2021; p. 367. [Google Scholar]
- Hua, W.; Zheng, P.; He, Y.; Cui, L.; Kong, W.; Wang, Z. An insight into the genes involved in secoiridoid biosynthesis in Gentiana macrophylla by RNA-seq. Mol. Biol. Rep. 2014, 41, 4817–4825. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Guo, X.; Yang, X.; Wang, H.; Hua, W.; He, Y.; Kang, J.; Wang, Z. Transcriptional responses and gentiopicroside biosynthesis in methyl jasmonate-treated Gentiana macrophylla seedlings. PLoS ONE 2016, 11, e0166493. [Google Scholar] [CrossRef]
- Zhang, X.; Allan, A.C.; Li, C.; Wang, Y.; Yao, Q. De novo assembly and characterization of the transcriptome of the Chinese medicinal herb, Gentiana rigescens. Int. J. Mol. Sci. 2015, 16, 11550–11573. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.; Gao, S.; Wang, H.; Lei, T.; Shen, J.; Gao, J.; Chen, S.; Yin, J.; Liu, J. De novo sequencing transcriptome of endemic Gentiana straminea (Gentianaceae) to identify genes involved in the biosynthesis of active ingredients. Gene 2016, 575, 160–170. [Google Scholar] [CrossRef]
- Sasaki, N.; Nemoto, K.; Nishizaki, Y.; Sugimoto, N.; Tasaki, K.; Watanabe, A.; Goto, F.; Higuchi, A.; Morgan, E.; Hikage, T.; et al. Identification and characterization of xanthone biosynthetic genes contributing to the vivid red coloration of red-flowered gentian. Plant J. 2021, 107, 1711–1723. [Google Scholar] [CrossRef]
- Tasaki, K.; Watanabe, A.; Nemoto, K.; Takahashi, S.; Goto, F.; Sasaki, N.; Hikage, T.; Nishihara, M. Identification of candidate genes responsible for flower colour intensity in Gentiana triflora. Front. Plant Sci. 2022, 13, 906879. [Google Scholar] [CrossRef]
- Nemoto, K.; Niinae, T.; Goto, F.; Sugiyama, N.; Watanabe, A.; Shimizu, M.; Shiratake, K.; Nishihara, M. Calcium-dependent protein kinase 16 phosphorylates and activates the aquaporin PIP2;2 to regulate reversible flower opening in Gentiana scabra. Plant Cell 2022, 34, 2652–2670. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef]
- Bouché, F.; Lobet, G.; Tocquin, P.; Périlleux, C. FLOR-ID: An interactive database of flowering-time gene networks in Arabidopsis thaliana. Nucleic Acids Res. 2016, 44, D1167–D1171. [Google Scholar] [CrossRef]
- Talar, U.; Kiełbowicz-Matuk, A. Beyond Arabidopsis: BBX regulators in crop plants. Int. J. Mol. Sci. 2021, 22, 2906. [Google Scholar] [CrossRef]
- Valverde, F. CONSTANS and the evolutionary origin of photoperiodic timing of flowering. J. Exp. Bot. 2011, 62, 2453–2463. [Google Scholar] [CrossRef]
- Song, Y.H.; Estrada, D.A.; Johnson, R.S.; Kim, S.K.; Lee, S.Y.; MacCoss, M.J.; Imaizumi, T. Distinct roles of FKF1, GIGANTEA, and ZEITLUPE proteins in the regulation of CONSTANS stability in Arabidopsis photoperiodic flowering. Proc. Natl. Acad. Sci. USA 2014, 111, 17672–17677. [Google Scholar] [CrossRef] [PubMed]
- Irish, V.F. The flowering of Arabidopsis flower development. Plant J. 2010, 61, 1014–1028. [Google Scholar] [CrossRef]
- Ditta, G.; Pinyopich, A.; Robles, P.; Pelaz, S.; Yanofsky, M.F. The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr. Biol. 2004, 14, 1935–1940. [Google Scholar] [CrossRef]
- Huang, H.; Tudor, M.; Weiss, C.A.; Hu, Y.; Ma, H. The Arabidopsis MADS-box gene AGL3 is widely expressed and encodes a sequence-specific DNA-binding protein. Plant Mol. Biol. 1995, 28, 549–567. [Google Scholar] [CrossRef]
- Ma, Y.Q.; Pu, Z.Q.; Tan, X.M.; Meng, Q.; Zhang, K.L.; Yang, L.; Ma, Y.Y.; Huang, X.; Xu, Z.Q. SEPALLATA-like genes of Isatis indigotica can affect the architecture of the inflorescences and the development of the floral organs. PeerJ 2022, 10, e13034. [Google Scholar] [CrossRef] [PubMed]
- Pu, Z.Q.; Ma, Y.Y.; Lu, M.X.; Ma, Y.Q.; Xu, Z.Q. Cloning of a SEPALLATA4-like gene (IiSEP4) in Isatis indigotica Fortune and characterization of its function in Arabidopsis thaliana. Plant Physiol. Biochem. 2020, 154, 229–237. [Google Scholar] [CrossRef]
- Liu, Y.; Li, X.; Ma, D.; Chen, Z.; Wang, J.W.; Liu, H. CIB1 and CO interact to mediate CRY2-dependent regulation of flowering. EMBO Rep. 2018, 19, e45762. [Google Scholar] [CrossRef] [PubMed]
- Fernández, V.; Takahashi, Y.; Le Gourrierec, J.; Coupland, G. Photoperiodic and thermosensory pathways interact through CONSTANS to promote flowering at high temperature under short days. Plant J. 2016, 86, 426–440. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.H.; Lee, H.J.; Ryu, J.Y.; Park, C.M. SPL3/4/5 Integrate developmental aging and photoperiodic signals into the FT-FD module in Arabidopsis flowering. Mol. Plant 2016, 9, 1647–1659. [Google Scholar] [CrossRef]
- Nishihara, M.; Tasaki, K.; Sasaki, N.; Takahashi, H. Development of basic technologies for improvement of breeding and cultivation of Japanese gentian. Breed. Sci. 2018, 68, 14–24. [Google Scholar] [CrossRef]
- Fekih, R.; Yamagishi, N.; Yoshikawa, N. Apple latent spherical virus vector-induced flowering for shortening the juvenile phase in Japanese gentian and lisianthus plants. Planta 2016, 244, 203–214. [Google Scholar] [CrossRef]
- Li, T.; Yu, X.; Ren, Y.; Kang, M.; Yang, W.; Feng, L.; Hu, Q. The chromosome-level genome assembly of Gentiana dahurica (Gentianaceae) provides insights into gentiopicroside biosynthesis. DNA Res. 2022, 29, dsac008. [Google Scholar] [CrossRef] [PubMed]
- Mishiba, K.; Yamane, K.; Nakatsuka, T.; Nakano, Y.; Yamamura, S.; Abe, J.; Kawamura, H.; Takahata, Y.; Nishihara, M. Genetic relationships in the genus Gentiana based on chloroplast DNA sequence data and nuclear DNA content. Breed. Sci. 2009, 59, 119–127. [Google Scholar] [CrossRef]
- Lo, C.C.; Chain, P.S. Rapid evaluation and quality control of next generation sequencing data with FaQCs. BMC Bioinform. 2014, 15, 366. [Google Scholar] [CrossRef]
- Tian, F.; Yang, D.C.; Meng, Y.Q.; Jin, J.; Gao, G. PlantRegMap: Charting functional regulatory maps in plants. Nucleic Acids Res. 2020, 48, D1104–D1113. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Tian, F.; Yang, D.C.; Meng, Y.Q.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017, 45, D1040–D1045. [Google Scholar] [CrossRef]
- Jin, J.; Zhang, H.; Kong, L.; Gao, G.; Luo, J. PlantTFDB 3.0: A portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res. 2014, 42, D1182–D1187. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; He, K.; Tang, X.; Li, Z.; Lv, L.; Zhao, Y.; Luo, J.; Gao, G. An Arabidopsis transcriptional regulatory map reveals distinct functional and evolutionary features of novel transcription factors. Mol. Biol. Evol. 2015, 32, 1767–1773. [Google Scholar] [CrossRef] [PubMed]
- Babicki, S.; Arndt, D.; Marcu, A.; Liang, Y.; Grant, J.R.; Maciejewski, A.; Wishart, D.S. Heatmapper: Web-enabled heat mapping for all. Nucleic Acids Res. 2016, 44, W147–W153. [Google Scholar] [CrossRef] [PubMed]

| Trinity Status | ||
|---|---|---|
| Contigs | Total number (>179 bp) | 521,292 |
| Total assembled bases (bp) | 411,975,909 | |
| Average of contig length (bp) | 790 | |
| N50 (bp) | 1360 | |
| Unigenes | Total number | 37,919 |
| Total assembled bases (bp) | 39,296,556 | |
| Average of contig length (bp) | 1036 | |
| N50 (bp) | 1206 | |
| Pathway | No. of Arabidopsis FLOR-ID Genes | No. of Hit Genes |
|---|---|---|
| Photoperiod | 139 | 82 |
| Vernalization | 90 | 63 |
| Aging | 53 | 26 |
| Hormones | 77 | 34 |
| Ambient temperature | 38 | 20 |
| Sugar | 46 | 21 |
| Autonomous | 116 | 86 |
| Circadian clock | 25 | 18 |
| Total 1 | 325 | 212 |
| Transcript ID | Gene Model | BlastP | Arabidopsis | Gentian |
|---|---|---|---|---|
| TRINITY_DN25746_c0_g1_i1.p1 | AT5G22290.1 | NAC domain-containing protein 89 | NAC089 | |
| TRINITY_DN12869_c1_g1_i7.p1 | AT4G34530.1 | Transcription factor bHLH63 | CIB1 | |
| TRINITY_DN73_c4_g1_i3.p1 | AT2G16770.1 | Basic leucine zipper 23 | bZIP23 | |
| TRINITY_DN3883_c0_g3_i1.p1 | AT5G48150.3 | Scarecrow-like transcription factor PAT1 | PAT1 | |
| TRINITY_DN54928_c0_g1_i3.p1 | AT1G29280.1 | Probable WRKY transcription factor 65 | WRKY65 | |
| TRINITY_DN23960_c0_g1_i5.p1 | AT2G28550.3 | Ethylene-responsive transcription factor RAP2-7 | TOE1 | |
| TRINITY_DN6815_c0_g2_i1.p1 | AT2G44940.1 | Ethylene-responsive transcription factor ERF034 | ||
| TRINITY_DN13564_c0_g1_i1.p1 | AT2G28350.1 | Auxin response factor 10 | ARF10 | |
| TRINITY_DN2158_c0_g1_i8.p1 | AT3G61150.1 | Homeobox-leucine zipper protein HDG1 | HDG1 | |
| TRINITY_DN6125_c0_g1_i1.p1 | AT5G60850.1 | Dof zinc finger protein DOF5.4 | OBP4 | |
| TRINITY_DN10067_c0_g2_i3.p1 | AT4G06598.1 | Uncharacterized protein At4g06598 | ||
| TRINITY_DN23176_c0_g1_i1.p1 | AT2G03710.2 | Agamous-like MADS-box protein AGL3 | SEP4 | GtSEP1 |
| TRINITY_DN14692_c0_g1_i4.p1 | AT1G74650.1 | MYB31 | ||
| TRINITY_DN16714_c0_g1_i3.p1 | AT3G11450.1 | |||
| TRINITY_DN30900_c0_g1_i2.p1 | AT3G54340.1 | Floral homeotic protein APETALA 3 | AP3 | GtAP3b |
| TRINITY_DN3345_c0_g3_i1.p1 | AT3G23690.1 | Transcription factor bHLH77 | CIL2 | |
| TRINITY_DN3306_c0_g1_i10.p1 | AT2G30590.1 | Probable WRKY transcription factor 21 | WRKY21 | |
| TRINITY_DN23930_c0_g2_i8.p1 | AT3G53310.1 | B3 domain-containing protein REM20 | ||
| TRINITY_DN617_c0_g2_i3.p1 | AT3G10760.1 | |||
| TRINITY_DN7529_c0_g1_i1.p1 | AT4G34590.1 | bZIP transcription factor 11 | ATB2 | |
| TRINITY_DN30234_c0_g1_i2.p1 | AT5G26930.1 | GATA transcription factor 23 | GATA23 | |
| TRINITY_DN22191_c0_g1_i5.p1 | AT3G54340.1 | Floral homeotic protein APETALA 3 | AP3 | GtAP3a |
| TRINITY_DN2269_c0_g4_i2.p1 | AT1G65480.2 | Protein FLOWERING LOCUS T | FT | GtFT1 |
| Transcript ID | Gene Model | BlastP | Arabidopsis | Gentian |
|---|---|---|---|---|
| TRINITY_DN522_c0_g1_i1.p1 | AT1G14920.1 | DELLA protein GAI | GAI | |
| TRINITY_DN7000_c1_g1_i1.p1 | AT4G00870.1 | Transcription factor bHLH14 | ||
| TRINITY_DN5423_c0_g1_i2.p1 | AT3G19860.1 | Transcription factor bHLH121 | bHLH121 | |
| TRINITY_DN22844_c0_g1_i6.p1 | AT5G01310.1 | APTX | ||
| TRINITY_DN1280_c1_g1_i2.p1 | AT1G58110.1 | |||
| TRINITY_DN728_c3_g1_i2.p1 | AT1G78600.1 | B-box zinc finger protein 22 | BBX22 | GtBBX22 |
| TRINITY_DN2269_c0_g4_i2.p1 | AT1G65480.2 | Protein FLOWERING LOCUS T | FT | GtFT1 |
| TRINITY_DN23176_c0_g1_i1.p1 | AT2G03710.2 | Agamous-like MADS-box protein AGL3 | SEP4 | GtSEP1 |
| TRINITY_DN35474_c0_g1_i1.p1 | AT5G51980.2 | |||
| TRINITY_DN53614_c0_g1_i5.p1 | AT1G67310.1 | Calmodulin-binding transcription activator 4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Takase, T.; Shimizu, M.; Takahashi, S.; Nemoto, K.; Goto, F.; Yoshida, C.; Abe, A.; Nishihara, M. De Novo Transcriptome Analysis Reveals Flowering-Related Genes That Potentially Contribute to Flowering-Time Control in the Japanese Cultivated Gentian Gentiana triflora. Int. J. Mol. Sci. 2022, 23, 11754. https://doi.org/10.3390/ijms231911754
Takase T, Shimizu M, Takahashi S, Nemoto K, Goto F, Yoshida C, Abe A, Nishihara M. De Novo Transcriptome Analysis Reveals Flowering-Related Genes That Potentially Contribute to Flowering-Time Control in the Japanese Cultivated Gentian Gentiana triflora. International Journal of Molecular Sciences. 2022; 23(19):11754. https://doi.org/10.3390/ijms231911754
Chicago/Turabian StyleTakase, Tomoyuki, Motoki Shimizu, Shigekazu Takahashi, Keiichirou Nemoto, Fumina Goto, Chiharu Yoshida, Akira Abe, and Masahiro Nishihara. 2022. "De Novo Transcriptome Analysis Reveals Flowering-Related Genes That Potentially Contribute to Flowering-Time Control in the Japanese Cultivated Gentian Gentiana triflora" International Journal of Molecular Sciences 23, no. 19: 11754. https://doi.org/10.3390/ijms231911754
APA StyleTakase, T., Shimizu, M., Takahashi, S., Nemoto, K., Goto, F., Yoshida, C., Abe, A., & Nishihara, M. (2022). De Novo Transcriptome Analysis Reveals Flowering-Related Genes That Potentially Contribute to Flowering-Time Control in the Japanese Cultivated Gentian Gentiana triflora. International Journal of Molecular Sciences, 23(19), 11754. https://doi.org/10.3390/ijms231911754






