Abstract
ATP-binding cassette subfamily G (ABCG) proteins play important roles in plant growth and development by transporting metabolites across cell membranes. To date, the genetic characteristics and potential functions of pomegranate ABCG proteins (PgrABCGs) have remained largely unknown. In this study, we found that 47 PgrABCGs were divided into five groups according to a phylogenetic analysis; groups I, II, III, and IV members are half-size proteins, and group V members are full-size proteins. PgrABCG14, PgrABCG21, and PgrABCG47 were highly expressed in the inner seed coat but had very low expression levels in the outer seed coat, and the expression levels of these three PgrABCG genes in the inner seed coats of hard-seeded pomegranate ‘Dabenzi’ were higher than those of soft-seeded pomegranate ‘Tunisia’. In addition, the expression of these three PgrABCG genes was highly correlated with the expression of genes involved in lignin biosynthesis and hormone signaling pathways. The evolution of PgrABCG14 presents a highly similar trend to the origin and evolution of lignin biosynthesis during land plant evolution. Ectopic expression of PgrABCG14 in Arabidopsis promoted plant growth and lignin accumulation compared to wild type plants; meanwhile, the expression levels of lignin biosynthesis-related genes (CAD5, C4H, and Prx71) and cytokinin response marker genes (ARR5 and ARR15) were significantly upregulated in transgenic plants, which suggests the potential role of PgrABCG14 in promoting plant growth and lignin accumulation. Taken together, these findings not only provide insight into the characteristics and evolution of PgrABCGs, but also shed a light on the potential functions of PgrABCGs in seed hardness development.
1. Introduction
Pomegranate (Punica granatum L.) is a perennial fruit tree grown in regions with Mediterranean climates. It grows in countries such as Tunisia, Turkey, Spain, Egypt, Morocco, the United States, China, India, Argentina, Israel, and South Africa [1]. The nutritional and medicinal pomegranate fruits attract much interest from consumers [2,3]. The soft and juicy outer seed coat is the edible part of the pomegranate, while the lignified inner seed coat is hard, which affects the feeling of swallowing fresh fruits. The lignin accumulation in the inner seed coat of hard-seed pomegranate is significantly higher than that of soft-seed pomegranate [4]. Thus, genetic studies of inner seed coat development are appealing for facilitation of breeding and development of pomegranate cultivars with low seed hardness.
In pomegranates, the differentiation and expansion of seed coat cells occurs in the early stages of fruit development [5]. Secondary cell wall thickening and lignin accumulation presents in the inner seed coat at the later stage of fruit development, leading to the formation of seed hardness [4]. In plants, the staple biosynthetic pathway of lignin is well understood: at the first step, monolignols including p-coumaryl alcohol (H-type monolignol), coniferyl alcohol (G-type monolignol), and sinapyl alcohol (S-type monolignol), are produced by phenylpropanoid pathways within the plant cell; at the second step, the monolignols are transported across the plasma membrane to the cell wall; and at the last step, the monolignols are polymerized to form lignin under the action of peroxidase and laccase [6,7,8]. To date, many genes encoding key enzymes and transcription factors that are involved in the biosynthesis and polymerization of monolignols have been characterized [9,10,11,12,13]. Recently, studies have demonstrated that monolignols can be passively diffused from inside the cell out to the cell wall through lipid bilayers [14]. However, transporters responsible for active transport of monolignols remains elusive.
Phytohormones play important roles in regulating cell differentiation and lignin accumulation. Indole acetic acid (IAA) and gibberellic acid (GA) were confirmed to control the lignin formation in primary phloem fibers and in xylem of Coleus blumei stems [15]. In Arabidopsis, Abscisic acid (ABA) regulates secondary cell wall formation and lignin deposition [16]. The IAA and ABA responsive transcription factor CgMYB58 upregulates lignin biosynthesis and triggers juice sac granulation in pummelo [17]. The differential distribution of IAA affects the lignin composition during the tilting response in Pinus radiata seedlings [18]. In carrots, exogenous cytokinin significantly upregulates most of the expression levels of lignin biosynthesis genes, causing elevated lignin accumulation in taproots [19]. Similarly, cytokinin elevated lignification and upregulate the expression level of several key genes related to lignin biosynthesis in bamboo [20,21]. These studies suggested that phytohormones may play important roles in regulating seed coat development, especially for lignification.
ABCG proteins are the G type of ATP-binding cassette (ABC) transporters, which can transport a variety of metabolites, including monolignols, hormones, and sterol glycosides. ABCG genes have been identified in many plants such as Arabidopsis thaliana [22], rice (Oryza sativa) [23], corn (Zea mays) [24], alfalfa (Medicago truncatula) [25], grapes (Vitis vinifera) [26], pineapple (Ananas comosus) [27], and tomato (Solanum lycopersicum) [28]. AtABCG14 plays an important role in delivering cytokinin from the root to the shoot. Knocking out AtABCG14 strongly impairs the plant’s growth and development, lignin synthesis and deposition are considerably delayed, and the numbers of lignified cells are substantially reduced in the interfascicular fibers and xylem bundles of the mutant plants [29,30]. In addition, AtABCG16, 25, 30, 31, 36, 37, and 40 are involved in hormone transport [31,32,33,34,35]. AtABCG9 functions in sterol glycoside transport in Arabidopsis [36]. In addition, AtABCG9, AtABCG11, and AtABCG14 were reportedly required for vascular development in Arabidopsis [37]. AtABCG29 functions as a p-coumaryl alcohol exporter involved in lignin biosynthesis in Arabidopsis [38]. The expression levels of AtABCG29, 30, 33, 34, and 37 were co-expressed with reference genes involved in lignification [39]. These studies imply the multifunctionality of ABCG transporters involving the translocation of diverse substrates.
In pomegranate genome, 47 ABCG genes were identified [40], but their genetic characteristics and potential functions remained unclear. In this study, integrated analyses of phylogenetic classification, gene and protein structures, expression patterns, and evolution were conducted to investigate the characteristics of the PgrABCG genes. We cloned one candidate gene, called PgrABCG14, to investigate its biological function by ectopic expression in Arabidopsis. This study not only provides insight into PgrABCG characteristics, but also provides valuable references for further studies on the potential function of PgrABCG genes involved in inner seed coat development, especially in lignification.
2. Results
2.1. Characterization of PgrABCG Genes and the Encoded Proteins
The PgrABCG genes were named according to their phylogenetic relationships with ABCG genes in Arabidopsis. The PgrABCG protein varied in length from 592 to 1492 amino acids, their molecular weight varied from 64.64 to 169.51 kDa, and the isoelectric points of these proteins ranged from 6.27 to 9.84 (Table S1). The number of exons ranged from 1 to 25 (Table S1). The structure prediction results showed that PgrABCGs contained four to fifteen transmembrane helices (TMHs) (Table S1). Subcellular localization prediction results showed that most PgrABCGs were localized to the PM (Table S1). However, PgrABCG7 was localized to the PM, chloroplasts, and cytoplasm; PgrABCG39 was localized to the PM and cytoplasm; and PgrABCG24 was localized to the PM, mitochondria and nucleus, implying that these proteins may have diverse functions.
2.2. Phylogeny and Structure Analysis of PgrABCG Genes
In order to reveal the phylogenetic relationship between ABCG genes, a total of 208 ABCG proteins from four species, including 47 from pomegranate (Punica granatum), 43 from Arabidopsis thaliana, 55 from eucalyptus (Eucalyptus grandis), and 63 from grape (Vitis vinifera) were used to construct a phylogenetic tree. All ABCG genes were divided into five clusters (Figure 1). Cluster III contained the lowest number of ABCG genes (10 genes) and Cluster V contained the greatest number of ABCG genes (115 genes). ABCG genes can be divided into full- and half-size molecular proteins: half-size ABCG genes comprise only one nucleotide-binding domain (NBD) and one transmembrane domain (TMD), while full-size ABCG contain two NBDs and two TMDs [25]. According to the Hidden Markov Model profiles, PgrABCG proteins consist of 17 full-size proteins and 30 half-size proteins. Interestingly, all the full-size PgrABCG proteins were distributed in cluster V, while the half-size proteins were randomly distributed in the other four clusters. Most of the ABCG genes between different species in the same branches had one or more homologous genes, while some had no homologous genes in other species.
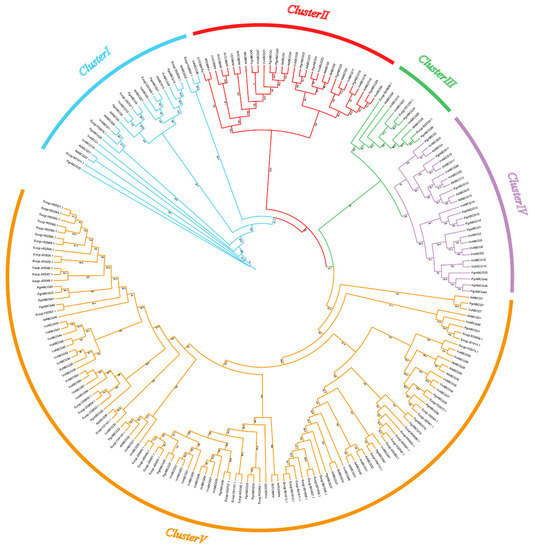
Figure 1.
Phylogenetic analysis of ABCG proteins from pomegranate, grape, Arabidopsis, and eucalyptus. The tree was generated with MEGA X using the neighbor-joining method based on 1000 bootstrap replicates.
The distribution of conserved motifs in PgrABCG proteins showed that most PgrABCG proteins contained motifs 1, 4, 7, and 15, and the PgrABCG proteins clustered on the same branch generally contained similar numbers and types of motifs (Figure S1A). Full-size ABCG proteins contained more motifs, and the unique motifs 8, 9, 10, 11, 12, 13, and 14 were generally distributed among the full-size ABCG proteins. The sequence information for each motif is provided in Table S2. The exon–intron structure of PgrABCG genes showed that the full size PgrABCG genes in cluster V contained more exons compared to the half-size PgrABCG genes in the other four clusters (Figure S1B).
2.3. Expression Patterns of PgrABCG Genes in Pomegranate Tissues
In order to obtain more information about PgrABCG genes, their transcription patterns in pomegranate tissues were determined by analyzing RNA-seq data. As shown in Figure 2, several PgrABCG genes were specifically expressed in roots (PgrABCG6, PgrABCG40, PgrABCG36, PgrABCG2, and PgrABCG20), flowers (PgrABCG31, PgrABCG1, PgrABCG28, and PgrABCG8), and peel (PgrABCG22). Interestingly, PgrABCG5, PgrABCG9, PgrABCG14, PgrABCG21, PgrABCG24, and PgrABCG47 were highly expressed in the inner seed coat, while they had very low expression levels in the outer seed coat. This suggests that these genes may function in the inner seed coat. Previous transcriptomic data showed that the relative expression levels of PgrABCG14, PgrABCG21, and PgrABCG47 in the inner seed coat of hard-seeded pomegranate ‘Dabenzi’ were higher than those in soft-seeded pomegranate ‘Tunisia’ [40]. In this study, a qRT-PCR analysis further proved the different expression levels of these three genes in ‘Dabenzi’ and ‘Tunisia’ (Figure 3). Accordingly, these three genes were selected as candidate genes involved in seed hardness development to be further studied.
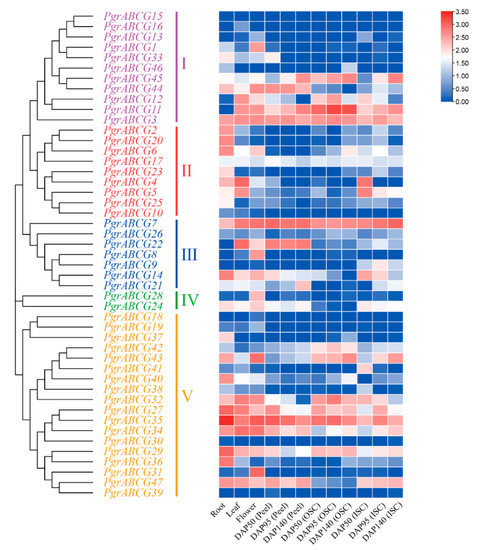
Figure 2.
Expression profile of PgrABCG genes in different tissues of pomegranate based on RNA-Seq data. The transcript data were calculated with log2 normalization based on FPKM values. “DAP” = days after pollination, “ISC” = Inner Seed Coat, “OSC” = Outer Seed Coat.
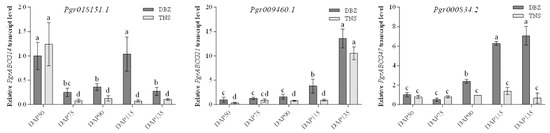
Figure 3.
Relative expression levels of the candidate PgrABCG genes in the inner seed coats of hard-seeded pomegranate ‘Dabenzi’ and soft-seeded pomegranate ‘Tunisia’ at five developmental stages. “DBZ” = ‘Dabenzi;’ “TNS” = ‘Tunisia;’ “DAP” = days after pollination. Error bars indicate standard deviation (n = 3). Different letters indicate a significant difference at p < 0.05 using Tukey’s test.
2.4. Co-Expression, and Evolution Analysis of the Candidate Genes
In order to further investigate the potential functions of ABCG genes in seed hardness development, we conducted WGCNA of protein-coded genes having transcriptional expression in inner seed coats with PgrABCG genes. The filtered genes were divided into 20 modules; the largest module contained 3641 genes and the smallest module only contained 75 genes (Figure S2A). Further, we associated the gene expression profiles in each module with all of the samples to generate a heat map of the module–sample matrix (Figure S2B). We found that the gene expression profiles in three modules, including light yellow, green, and red modules, were highly correlative to the inner seed coat (Figure S2C). Genes in these three modules were mapped in the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway. Among them, 27 and 41 genes were enriched in phenylpropanoid biosynthesis pathway and hormone signaling pathway, respectively. These genes were used to construct a co-expression network with the three candidate PgrABCG genes. Results showed that 16 genes were co-expressed with these PgrABCG genes (Figure 4A), the functional annotation of these co-expression genes was showed in Table S3. Among them, five genes are involved in lignin biosynthesis, including Pgr021671.1 encoding Caffeic acid 3-O-methyltransferase and Pgr014360.1 encoding shikimate O-hydroxy cinnamoyl transferase. Notably, PgrABCG14 was highly correlated with eight genes involved in hormone signaling pathways. Among them were Pgr011139.1 encoding isopentenyl transferase (cytokinin synthase), Pgr015797.1 encoding cytokinin response marker protein, Pgr000815.1 encoding auxin-responsive protein, Pgr003031.1 encoding ABA responsive element binding factor, and Pg022934.2 encoding brassinosteroid insensitive protein.
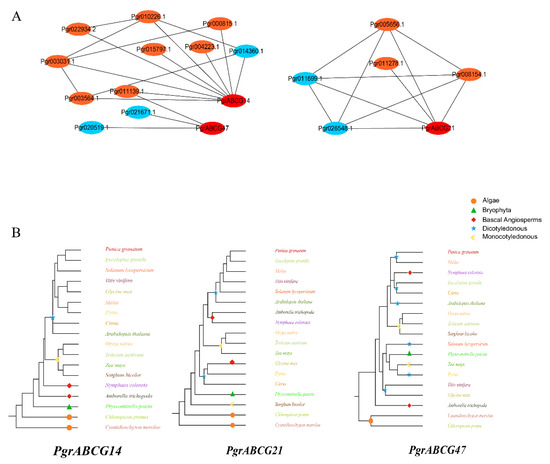
Figure 4.
Co-expression network and evolution of the candidate PgrABCG genes. (A) The red dots represent PgrABCG genes, the blue dots represent genes involved in phenylpropanoid biosynthesis pathways, and the orange dots represent genes involved in hormone signaling pathways. (B) Rooted evolutionary trees of the candidate PgrABCG genes and the homologous genes in typical plant species. Different colors represent different plant species.
In order to further understand the potential functions of the three candidate PgrABCG genes through a plant evolutional perspective, we constructed a rooted evolutionary tree of ABCG genes from typical species during plant evolution. The evolution of PgrABCG14 occurred from a sequence that originated in algae, then bryophytes, then basal angiosperms, and finally monocotyledonous and dicotyledonous plant species. This presents a highly similar trend to the origin and evolution of lignin biosynthesis during land plant evolution, wherein plant evolution took place from aquatic to terrestrial plants and from lower to higher plants (Figure 4B).
2.5. Functional Analysis of PgrABCG14
We cloned the candidate gene PgrABCG14 and transiently expressed PgrABCG14-GFP in tobacco leaves to investigate its subcellular localization. The results showed that PgrABCG14 was localized to the PM (Figure 5A). Consistent with the results, the localization of PgrABCG14 on PM was also observed in PgrABCG14-GFP transgenic Arabidopsis (Figure 5B). In order to analyze the tissue-specific expression of PgrABCG14, we generated transgenic Arabidopsis plants expressing the ProPgrABCG14:GUS fusion construct. GUS staining of seedlings showed that GUS activity could be observed in almost all tissue, including roots, leaves, flowers, and silique (Figure 6A–E), which was consistent with the mRNA expression results in pomegranate tissues (Figure 2). Interestingly, strong GUS activity was observed in the vascular tissues of roots (mature zones), old leaf, and petal.
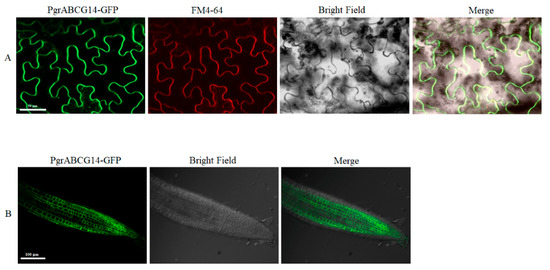
Figure 5.
Subcellular localization of PgrABCG14-GFP fusion protein. (A) Subcellular localization of PgrABCG14-GFP in tobacco epidermal cells. Scale bar, 50 μm. (B) Subcellular localization of PgrABCG14-GFP in transgenic Arabidopsis plants. Scale bar, 100 μm.

Figure 6.
GUS activity in transgenic Arabidopsis plants expressing the GUS reporter gene under the control of PgrABCG14 promoter. (A) 10-day rosette leaves, (B) root, (C) 45-day-old leaf, (D) inflorescences, (E) silique.
In order to examine the function of PgrABCG14 in plant, we ectopically expressed PgrABCG14-GFP in Arabidopsis. Semi-quantitative PCR showed that PgrABCG14 was successfully expressed in Arabidopsis (Figure S3). Two independent overexpression (OE) lines, OE-1 and OE-2, were selected for further functional analyses. Overexpression of PgrABCG14 was able to promote Arabidopsis plant growth and development (Figure 7A), and shoot biomass was increased significantly in overexpressed plants (Figure 7B). Meanwhile, the lignin content was increased significantly in OE plants (Figure 7C). Notably, the deeper lignin staining in vascular tissue of stems and ventral suture of siliques were observed in OE plants compared to wild plants (Figure 7D). The transcript levels of three lignin biosynthesis related genes (CAD5, C4H, and Prx71) were significantly higher in the shoots of OE lines than in those of the wild type (Figure 7E).
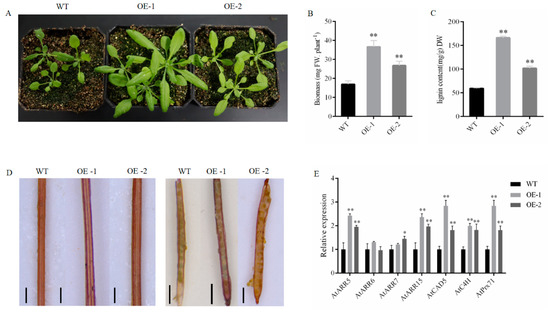
Figure 7.
Phenotypes of PgrABCG14 OE Arabidopsis plants under normal growth conditions. (A) Phenotypes of the wild type and PgrABCG14 OE lines grown in soil for 4 weeks. (B) Biomass production of plants grown as in (A). (C) Lignin content in the shoots of plants grown as in (A). (D) Lignin staining of the stems and siliques of wild-type and PgrABCG14 OE lines. (E) Expression levels of lignin biosynthesis-related genes and cytokinin response marker genes in the shoots of wild-type and PgrABCG14 OE plants. Data are means ± SD (n = 3). Asterisks indicate values significantly different from those of the wild type (* p < 0.05, ** p < 0.01, Student’s t test). Bars: (D) 200 mm.
As PgrABCG14 is orthologous to AtABCG14, a gene having a clear cytokinin transport function in Arabidopsis, we hypothesized that PgrABCG14 also has a role in cytokinin transport. In order to investigate the relationship between PgrABCG14 and cytokinin, we analyzed the expression levels of four cytokinin response marker genes in the shoots of wild type and OE plants. Transcript levels of ARR5 and ARR15 were significantly higher in the OE plants than those in the wild type plants (Figure 7E). In addition, we exogenously applied cytokinin to the leaves of wild and OE plants. Indeed, daily spraying with 1 μM tZ accelerated both the WT and OE plants development, and the OE plants exhibited early bolting and stem elongation phenotypes (Figure S4).
3. Discussion
ABCG protein is a branch of the ABC transporter family, which was divided into ABCA-F subfamilies [41]. ABC transporters comprise four core domains: two nucleotide-binding domains (NBDs) and two transmembrane domains (TMDs). The ABC transporters are composed of two types of proteins: (i) half-size proteins, comprising only one NBD and TMD in so-called reverse orientation; and (ii) full-size proteins, containing two NBDs and two TMDs [25,42]. Half-size proteins have the capability to homodimerize and at the same time heterodimerize with another half-size proteins, thereby forming the functional ABC transporter that translocates substrates across a membrane [43]. According to the phylogenetic tree and Pfam domains of PgrABCGs, 30 full-size PgrABCGs belonged to cluster V, while 17 half-size PgrABCGs were randomly distributed to clusters I–IV, which is consistent with the phyletic evolution of ABCGs in Arabidopsis and grape [26,44]. Sequence analysis of ABC genes from different organisms suggests that half- and full-size transporters have a common ancestor and that the full-size transporters could derive from gene duplication of a half-size transporter gene [22,42,45,46]. In Arabidopsis, the G subfamily consists of 28 half-size transporters and 15 full-size transporters [44,45]. Some AtABCG genes have multiple homologs in pomegranate, while some AtABCG genes have no homologs in pomegranate, suggesting that gene duplication or loss may have occurred in the ABCG genes during evolution. We also found that the same type of PgrABCGs had similar conserved motifs, while some difference existed among conserved motifs between the two types of proteins. Full-size PgrABCG genes had similar numbers of exons and introns, while half-size PgrABCG genes contained diverse numbers of exons and introns. This is also reflected by the sequence identities among half- and full-size transporters from Arabidopsis, respectively.
Cell division in the seed coat accompanies the proliferation of endosperm during seed development in early stages [47]. At the early stage of pomegranate seed development, it is difficult to visually distinguish the inner and outer seed coats, and abundant cell division in the seed coat may facilitate the differentiation and expansion of the seed coat. During later stages of seed development, the outer seed coat develops into soft and juicy tissue, while the inner seed coat develops into lignified tissue. Among the PgrABCG genes, PgrABCG14, 21, and 47 showed high expression levels in the inner seed coat, but had very low expression levels in the outer seed coat. These three PgrABCG genes showed higher expression levels in the inner seed coat of the hard-seeded pomegranate ‘Dabenzi’ compared to that of the soft-seeded pomegranate ‘Tunisia’. In addition, these three PgrABCG genes were co-expressed with the genes involved in phenylpropanoid biosynthesis pathways and hormone signaling pathways. Phytohormones, including IAA, ABA, GA, and cytokinin, have been confirmed to play important roles in regulating cell differentiation and lignin accumulation [15,16,17,18,19,20,21]. Numerous studies have shown that ABCG proteins are involved in the transport of hormones and monolignols [29,30,31,32,33,34,35]. Seed hardness formation is the result of lignin accumulation in the inner seed coat. A model was proposed that monolignols can either be transported to the cell wall to polymerize into lignin, or be glycosylated into monolignol glucosides and then stored in the vacuole [48]. Therefore, it is reasonable to presume that PgrABCG14, 21, and 47 may be involved in seed hardness development through different biological processes, most likely through the lignin biosynthesis pathway and/or hormone signaling pathway.
ABCG proteins, such as AtABCG9, 14, 36, etc., are localized to the PM and are responsible for transporting specific substrates from intracellular to extracellular. Constitutive overexpression of PM-localized PgrABCG14 in Arabidopsis promoted plant growth and development. Its phenotype was similar to the VviABCG14 OE plants [49]. Interestingly, we found that the lignin content in PgrABCG14 OE plants was higher than that in WT, and the expression levels of three lignin biosynthesis-related genes (CAD5, C4H, and Prx71) were significantly higher in OE plants than those in the WT plants. In Arabidopsis, AtABCG14 is involved in the transport of tZ-type cytokinin from root to shoot. Mutation of AtABCG14 results in a significant decrease in xylem and phloem cells, considerably smaller rosette leaves, shorter and thinner stems, and reduced lignin content [29,30]. In our study, we found that the expression levels of two cytokinin response marker genes (ARR5 and ARR15) in PgrABCG14 OE plants were significantly higher than that in WT plants. In addition, exogenous application of tZ promoted early bolting in PgrABCG14 OE plants. The PgrABCG14 OE plants also have longer and thicker stems compared to WT plants. It was speculated that the PgrABCG14 gene may participate in plant development by involvement in cytokinin transport. In pomegranates, PgrABCG14 was highly expressed in the inner seed coat, and had a higher expression level in the inner seed coats of hard-seeded pomegranate ‘Dabenzi’ than in those of soft-seeded pomegranate ‘Tunisia’. Its expression was highly correlated with that of genes which functioned in phenylpropanoid biosynthesis and hormone signaling. We proposed that PgrABCG14 is a key candidate that is involved in seed hardness development. However, its detailed mechanisms require further study.
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
Pomegranate cultivars ‘Dabenzi’ (a hard-seeded cultivar) and ‘Tunisia’ (a soft-seeded cultivar) are two of the main cultivars in China. The fruit was harvested from these two cultivars, which were grown in the Gangji Eco-agricultural Demonstration site (31°51′9.05″ N, 117°06′34.33″ E), Academy of Agricultural Sciences, Hefei, China. For gene expression, inner seed coats from ‘Dabenzi’ and ‘Tunisia’ were obtained from fruits at 50, 75, 90, 115, and 135 days after pollination (DAP) in three biological replicates. All materials were quickly frozen in liquid nitrogen and stored at −80 °C for RNA extraction.
Arabidopsis plants (ecotype Col-0 and transgenic Arabidopsis plants) were grown in soil pots in an incubator under 16 h of light (150 μmol s−1 m−2) at 22 °C, 8 h of darkness at 20 °C, and 60% relative humidity. Nicotiana benthamiana was grown in soil pots in an incubator under 16 h of light (500 μmol s−1 m−2) at 25 °C, 8 h of darkness at 23 °C, and 60% relative humidity.
4.2. Sequence Analysis of PgrABCG Genes and the Encoded Proteins
The gene and protein sequences of 47 PgrABCG genes were obtained according to a previous study [40]. ExPASy (https://web.expasy.org/protparam/ (accessed on 5 September 2020)) was used to calculate the basic properties of ABCG proteins. The numbers of transmembrane helices (TMHs) of the PgrABCG proteins were identified using the TMHMM Server v.2.0 (http://www.cbs.dtu.dk/services/TMHMM/ (accessed on 6 September 2020)). The subcellular localization of the PgrABCG proteins was predicted by WOLF PSORT (https://www.psort.org/ (accessed on 6 September 2020)) [50].
4.3. Conserved Motifs and Gene Structure Analysis
The conserved motifs of the PgrABCG proteins were identified on the MEME (http://meme.nbcr.net/meme/cgi-bin/meme.cgi (accessed on 8 October 2020)) website, with up to 15 motifs [51]. The exon–intron structures of the PgrABCG genes were determined by comparing of the coding sequence of each PgrABCG gene with its genomic sequence using TBtools v0.655 [10].
4.4. Phylogenetic Analysis of ABCG Proteins in Four Species
The ABCG protein sequences of grape (a species with a recent genome triplication) [52], eucalyptus (a species as the same family of pomegranate), Arabidopsis, and pomegranate were used to construct an unrooted tree using MEGA 7.0, based on the neighbor-joining method with bootstrap parameters of 1000 replicates [53].
4.5. Expression Analysis of PgrABCG Genes in Different Tissues
The abundance of PgrABCG transcripts in the root, flower, leaf, and the three developmental stages of the peel and seed coat (inner and outer seed coat) of ‘Dabenzi’ were collected from the RNA-seq data deposited in NCBI Sequence Read Archive (SRA) (accession SRP100581) [54]. The transcriptional abundances of ABCG genes were estimated using the fragments per kilobase per million (FPKM) method, and illustrated with heat map generation by TBtools v0.655 [10]. The detailed FRKM values are listed in Table S4.
4.6. Semi-Quantitative PCR and Quantitative Real Time Polymerase Chain Reaction (qRT-PCR) Analysis
The total RNA from pomegranate and Arabidopsis tissues was extracted using the RNA prep Pure Plant Kit (TianGen Bio-chemical Technology Co., Ltd., Beijing, China). First, 1 μg of RNA was used for cDNA synthesis using the Prime ScriptTM RT Reagent Kit (TaKaRa Biomedical Technology Co., Ltd., Tokyo, Japan). The RNA samples were treated with DNase in the process of RNA extraction and cDNA synthesis. The procedures were carried out strictly following the manufacturer’s instructions. The expression levels of PgrABCG14 in the transgenic Arabidopsis lines were tested by semi-quantitative PCR. The PCR program was performed as follows: 1 cycle of 95 °C for 5 min, followed by 25 cycles of 95 °C for 30 s, 59 °C for 30 s, 72 °C for 20 s, and, finally, 1 cycle of 72 °C for 5 min. qRT-PCR was carried out using SYBR Premix Ex Taq II (TaKaRa Biomedical Technology Co., Ltd., Tokyo, Japan) with an AB 7500 Fast Real-Time PCR System (Thermo Fisher Instruments Co., Ltd., Shanghai, China). The gene-specific primers were designed online using NCBI Primer-BLAST. PgrActin7 and AtActin2 were used as reference genes for pomegranate and Arabidopsis, respectively. The gene-specific primers and product sizes were listed in Table S5. The relative expression of specific genes was compared using the 2−ΔΔCt method and calculated by normalizing the PCR threshold cycle (Ct) value to the expression of the reference gene. Each analysis was carried out in triplicate. Prism 6.0 was used to draw a histogram [55].
4.7. Co-Expression Analysis
The initial transcriptome data were collected from the RNA-seq data, which have been described above. Firstly, the genes having transcriptional expression in the inner seed coat were selected for the weighted gene co-expression network analysis (WGCNA). The parameters were as following: the weighted network was unsigned, the soft threshold power was 6, and Pearson’s correlation values (≥0.8) were used to identify the correlation between the modules and the PgrABCG genes. Then, the inner seed coat was used as a respective input file for the detection of significant relationships (p < 0.05) among the inner seed coat and the eigengene of each module. Genes were annotated to be involved in phenylpropanoid biosynthesis and hormone signaling pathways of these modules. All PgrABCG genes were used to construct co-expression networks using the Pearson’s correlation coefficient. Then, PCC values lower than 0.99 were removed. Networks were visualized by Cytoscape v3.7.1 [56].
4.8. Evolutionary Analysis of Candidate PgrABCG Genes
Algae plants including Cyanidioschyzon merolae and Chloropicon primus; bryophytes including Physcomitrella patens; basal angiosperms including Amborella and Nymphaea; monocotyledonous plants including O. sativa, T. aestivum, Z. mays, and Sorghum bicolor; and dicotyledonous plants including E. grandis, V. vinifera, Solanum lycopersicum, Malus, A. thaliana, P. granatum, Glycine max, citrus, and Pyrus, adding up to a total of 18 typical species, were selected in order to study the evolution of ABCG genes in the process of plant evolution. The protein sequence of each candidate PgrABCG gene was used for BLASTP search (with >90% coverage) to obtain the homologous protein sequences of each species using the NCBI species database (accessed on 10 November 2020). A rooted evolutionary tree of homologous ABCG proteins was constructed using DNAMAN v6.0.3.99 [57].
4.9. Vector Construction and Plant Transformation
The coding region of PgrABCG14 was amplified using two primers (F, 5′-CACGGGGGACGAGCTCGGTACCATGCCTCTCCACTCCATAGCACC-3′; R, 5′-CCATGTCGACTCTAGAGGATCCCCTCAGCACCCGATGCAGTG-3′), then inserted into the pCAMBIA1300GFP vector [58]. The constructed expression vector was then used for wild type (WT) Arabidopsis transformation. WT transformed with the pCAMBIA1300GFP vector being used as the control plant. The T3 homozygous plants were used for phenotypic observation and functional experiment. The fragment 2.0 kb upstream of the PgrABCG14 start codon was amplified using two primers (F, 5′- CGAGCTCGGTACCCGGGGATCCAAATGTAGAGTACTCGGAACCGATC-3′; R, 5′-ATTTACCCTCAGATCTACCATGGGGCAAAAGTAGTGGCAGTACAATT-3′) and inserted into the pCAMBIA1305 vector [59]. The constructed expression vector was transformed into WT Arabidopsis. The T3 homozygous plants were used for GUS histochemical analysis. All the Arabidopsis transformation was carried out by floral dipping [60]. The coding region of PgrABCG14 was amplified using two primers (F, 5′-TTAAGTCCGGAGCTAGCTCTAGAATGCCTCTCCACTCCATAGCACC-3′; R, 5′-TCGCCCTTGCTCACCATGGATCCCCTCAGCACCCGATGCAGTG-3′) and inserted into the pCAMBIA1305GFP vector [59]. Then, the constructed expression vector was injected into tobacco epidermal cells by agrobacterium tumefaciens infection, and cultured in the dark for 3 days [61]. The expression of GFP fusion protein in cells was observed by laser confocal microscopy (Olympus Co., Ltd., Beijing, China). All of the constructed expression vectors described above were confirmed by sequencing.
4.10. GUS Histochemical Analysis
Histochemical staining was performed as described [62]. Arabidopsis plants were grown in soil pots under normal conditions. The Arabidopsis tissues were collected and washed twice with staining buffer [50 mM sodium phosphate buffer (pH 7.2), 0.2% Triton X-100, 2 mM potassium ferrocyanide, and 2 mM potassium ferricyanide]. Then, staining solution [50 mM sodium phosphate buffer (pH 7.2), 0.2% Triton X-100, 2 mM potassium ferrocyanide, 2 mM potassium ferricyanide, and 2 mM X-Gluc] was added to the samples. The samples were evacuated for 20 min by vacuum pump in the dark, followed by overnight incubation at 37 °C. Stained tissues were washed in increasing ethanol gradients from 50% to 100% to remove chlorophyll, and then observed with a stereomicroscope (Motic Instruments Co., Ltd., Xiamen, China).
4.11. Exogenous Cytokinin Treatment
The seeds were sown on Murashige and Skoog (MS) solid medium. After four leaves grew, the seedings were transferred into 1/2 Hoaglang nutrient solution (5 mM CaNO3, 5 mM KNO3, 2 mM MgSO4, 1 mM NH4H2PO4, 0.185 mM H3BO3, 2 μM [NH4]6Mo7O24, 36.6 μM MnSO4, 3 μM ZnSO4, 1.28 μM CuSO4, 40 μM Fe (III)-EDTA) and grown in a phytotron. After 1 week, the plants were sprayed with 1uM trans-zeatin (tZ) or 0.1% Dimethyl sulfoxide (DMSO) solution (mock) every day for the following 21 days, as Zhang et al. described [30], and then the plant phenotype was observed and photographed.
4.12. Lignin Histochemical Staining and Content Determination
The main stem at 1 cm above the ground and the siliques were used for staining as described [63], with the following steps: firstly, dissolve phloroglucinol in 95% absolute ethanol and 30% HCL solution (volume ratio = 1:1), the final concentration of phloroglucinol is 10%; then soak the samples to be stained in the prepared staining solution for 5 min; finally, the stained tissues were observed and photographed by stereomicroscope (Motic Instruments Co., Ltd., Xiamen, China). The deeper color in staining tissues indicated higher lignin deposition.
The determination of the plants’ lignin content was conducted as described [64], the steps as follows: firstly, 20 mg of the dry sample was added into a 15 mL centrifuge tube; then 2 mL of acetyl bromine and acetic acid was added (volume ratio = 1:1). This was kept in a constant temperature water bath at 70 °C for 30 min, and shaken every 8 min to fully mix the solution. Then, 3 mL of 2 mol/L NaOH were added, and the reaction was terminated after about 20 min. Next, 0.2 mL supernatant was placed into a new 2 mL centrifuge tube, and 3.8 mL of acetic acid was added and mixed fully. Finally, the absorbance value at 280 nm was determined.
5. Conclusions
PgrABCG genes were divided into five groups according to phylogenetic analysis. Members of Group I, II, III, and IV were half-size ABCG proteins, while group V members were full-size ABCG proteins. PgrABCG14, PgrABCG21, and PgrABCG47 were selected as candidate genes involved in the seed hardness development according to the expression patterns, co-expression, and evolutionary analysis. Overexpression of PgrABCG14 in Arabidopsis promoted plant development and lignin accumulation. This study provides a reference for understanding the characteristics and potential functions of PgrABCG genes.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms231911661/s1.
Author Contributions
Conceptualization, J.L. and G.Q.; data curation, Q.Y., J.L. and Y.Y.; methodology, Q.Y., J.L. and Y.Y.; resources, B.J., G.L. and Y.S.; supervision, Z.C. and Y.X.; writing—original draft, Q.Y. and J.L.; writing—review and editing, G.Q., C.L. and H.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by Natural Science Foundation of Anhui Province (1708085MC85, 2108085QC129), Major Science and Technology Project of Anhui Province (201903b06020017), and Major Science and Technology Project of Huaibei City (HK2021014).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All datasets presented in this study are included in the article/Supplementary Material.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Stover, E.; Mercure, E.W. The pomegranate: A new look at the fruit of paradise. Hortscience 2007, 42, 1088–1092. [Google Scholar] [CrossRef]
- Shahamirian, M.; Eskandari, M.H.; Niakousari, M.; Esteghlal, S.; Hashemi Gahruie, H.; Mousavi Khaneghah, A. Incorporation of pomegranate rind powder extract and pomegranate juice into frozen burgers: Oxidative stability, sensorial and microbiological characteristics. J. Food Sci. Technol. 2019, 56, 1174–1183. [Google Scholar] [CrossRef] [PubMed]
- Van Nieuwenhove, C.P.; Moyano, A.; Castro-Gómez, P.; Fontecha, J.; Sáez, G.; Zárate, G.; Pizarro, P.L. Comparative study of pomegranate and jacaranda seeds as functional components for the conjugated linolenic acid enrichment of yogurt. LWT 2019, 111, 401–407. [Google Scholar] [CrossRef]
- Xie, X.; Huang, Y.; Tian, S.; Li, G.; Cao, S. Study on the relationship between seed hardness development of soft-seeded pomegranate and the microstructure of seed coat cell wall. Acta Hortic. Sin. 2017, 44, 1174–1180. [Google Scholar]
- Figueiredo, D.D.; Batista, R.A.; Roszak, P.J.; Hennig, L.; Kohler, C. Auxin production in the endosperm drives seed coat development in Arabidopsis. Elife 2016, 5, e20542. [Google Scholar] [CrossRef]
- Barakate, A.; Stephens, J.; Goldie, A.; Hunter, W.N.; Marshall, D.; Hancock, R.D.; Lapierre, C.; Morreel, K.; Boerjan, W.; Halpin, C. Syringyl lignin is unaltered by severe sinapyl alcohol dehydrogenase suppression in tobacco. Plant Cell 2011, 23, 4492–4506. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.B.; Ma, Q.H.; Xu, Y. Lignin biosynthesis and its molecular regulation. Prog. Nat. Sci. -Mater. Int. 2003, 13, 321–328. [Google Scholar]
- Orella, M.J.; Gani, T.Z.H.; Vermaas, J.V.; Stone, M.L.; Anderson, E.M.; Beckham, G.T.; Brushett, F.R.; Román-Leshkov, Y. Lignin-KMC: A Toolkit for Simulating Lignin Biosynthesis. ACS Sustain. Chem. Eng. 2019, 7, 18313–18322. [Google Scholar] [CrossRef]
- Cao, Y.; Meng, D.; Li, X.; Wang, L.; Cai, Y.; Jiang, L. A Chinese White Pear (Pyrus bretschneideri) BZR Gene PbBZR1 Act as a Transcriptional Repressor of Lignin Biosynthetic Genes in Fruits. Front Plant Sci. 2020, 11, 1087. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant. 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Li, L.; Yang, K.; Wang, S.; Lou, Y.; Zhu, C.; Gao, Z. Genome-wide analysis of laccase genes in moso bamboo highlights PeLAC10 involved in lignin biosynthesis and in response to abiotic stresses. Plant Cell Rep. 2020, 39, 751–763. [Google Scholar] [CrossRef]
- Sakamoto, S.; Kamimura, N.; Tokue, Y.; Nakata, M.T.; Yamamoto, M.; Hu, S.; Masai, E.; Mitsuda, N.; Kajita, S. Identification of enzymatic genes with the potential to reduce biomass recalcitrance through lignin manipulation in Arabidopsis. Biotechnol. Biofuels 2020, 13, 97. [Google Scholar] [CrossRef]
- Tu, M.; Wang, X.; Yin, W.; Wang, Y.; Li, Y.; Zhang, G.; Li, Z.; Song, J.; Wang, X. Grapevine VlbZIP30 improves drought resistance by directly activating VvNAC17 and promoting lignin biosynthesis through the regulation of three peroxidase genes. Hortic. Res. 2020, 7, 150. [Google Scholar] [CrossRef] [PubMed]
- Perkins, M.L.; Schuetz, M.; Unda, F.; Chen, K.T.; Bally, M.B.; Kulkarni, J.A.; Yan, Y.; Pico, J.; Castellarin, S.D.; Mansfield, S.D.; et al. Monolignol export by diffusion down a polymerization-induced concentration gradient. Plant Cell 2022, 34, 2080–2095. [Google Scholar] [CrossRef] [PubMed]
- Aloni, R.; Tollier, M.T.; Monties, B. The Role of Auxin and Gibberellin in Controlling Lignin Formation in Primary Phloem Fibers and in Xylem of Coleus blumei Stems. Plant Physiol. 1990, 94, 1743–1747. [Google Scholar] [CrossRef]
- Liu, C.; Yu, H.; Rao, X.L.; Li, L.G.; Dixon, R.A. Correction for Liu et al., Abscisic acid regulates secondary cell-wall formation and lignin deposition in Arabidopsis thaliana through phosphorylation of NST1. Proc. Natl. Acad. Sci. USA 2021, 118, e2106367118. [Google Scholar]
- Shi, M.; Liu, X.; Zhang, H.; He, Z.; Yang, H.; Chen, J.; Feng, J.; Yang, W.; Jiang, Y.; Yao, J.L.; et al. The IAA- and ABA-responsive transcription factor CgMYB58 upregulates lignin biosynthesis and triggers juice sac granulation in pummelo. Hortic. Res. 2020, 7, 139. [Google Scholar] [CrossRef]
- Salazar, R.; Pollmann, S.; Morales-Quintana, L.; Herrera, R.; Caparros-Ruiz, D.; Ramos, P. In seedlings of Pinus radiata, jasmonic acid and auxin are differentially distributed on opposite sides of tilted stems affecting lignin monomer biosynthesis and composition. Plant Physiol. Biochem. 2019, 135, 215–223. [Google Scholar] [CrossRef]
- Khadr, A.; Wang, Y.H.; Zhang, R.R.; Wang, X.R.; Xu, Z.S.; Xiong, A.S. Cytokinin (6-benzylaminopurine) elevates lignification and the expression of genes involved in lignin biosynthesis of carrot. Protoplasma 2020, 257, 1507–1517. [Google Scholar] [CrossRef] [PubMed]
- Ogita, S.; Nomura, T.; Kishimoto, T.; Kato, Y. A novel xylogenic suspension culture model for exploring lignification in Phyllostachys bamboo. Plant Methods 2012, 8, 40. [Google Scholar] [CrossRef]
- Ogita, S.; Nomura, T.; Kato, Y.; Uehara-Yamaguchi, Y.; Inoue, K.; Yoshida, T.; Sakurai, T.; Shinozaki, K.; Mochida, K. Transcriptional alterations during proliferation and lignification in Phyllostachys nigra cells. Sci. Rep. 2018, 8, 11347. [Google Scholar] [CrossRef] [PubMed]
- Cho, C.H.; Jang, S.; Choi, B.Y.; Hong, D.; Choi, D.S.; Choi, S.; Kim, H.; Han, S.K.; Kim, S.; Kim, M.S.; et al. Phylogenetic analysis of ABCG subfamily proteins in plants: Functional clustering and coevolution with ABCGs of pathogens. Physiol. Plant 2021, 172, 1422–1438. [Google Scholar] [CrossRef]
- Gupta, B.B.; Selter, L.L.; Baranwal, V.K.; Arora, D.; Mishra, S.K.; Sirohi, P.; Poonia, A.K.; Chaudhary, R.; Kumar, R.; Krattinger, S.G.; et al. Updated inventory, evolutionary and expression analyses of G (PDR) type ABC transporter genes of rice. Plant Physiol. Biochem. 2019, 142, 429–439. [Google Scholar] [CrossRef] [PubMed]
- Pang, K.; Li, Y.; Liu, M.; Meng, Z.; Yu, Y. Inventory and general analysis of the ATP-binding cassette (ABC) gene superfamily in maize (Zea mays L.). Gene 2013, 526, 411–428. [Google Scholar] [CrossRef]
- Jasinski, M.; Banasiak, J.; Radom, M.; Kalitkiewicz, A.; Figlerowicz, M. Full-size ABC transporters from the ABCG subfamily in medicago truncatula. Mol. Plant Microbe Interact 2009, 22, 921–931. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cakir, B.; Kilickaya, O. Whole-genome survey of the putative ATP-binding cassette transporter family genes in Vitis vinifera. PLoS ONE 2013, 8, e78860. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Li, Y.; Zhao, L.; Hou, Z.; Yan, M.; Hu, B.; Liu, Y.; Azam, S.M.; Zhang, Z.; Rahman, Z.U.; et al. Genome-Wide Identification and Expression Profiling of ATP-Binding Cassette (ABC) Transporter Gene Family in Pineapple (Ananas comosus (L.) Merr.) Reveal the Role of AcABCG38 in Pollen Development. Front Plant Sci. 2017, 8, 2150. [Google Scholar] [CrossRef]
- Ofori, P.A.; Mizuno, A.; Suzuki, M.; Martinoia, E.; Reuscher, S.; Aoki, K.; Shibata, D.; Otagaki, S.; Matsumoto, S.; Shiratake, K. Genome-wide analysis of ATP binding cassette (ABC) transporters in tomato. PLoS ONE 2018, 13, e0200854. [Google Scholar] [CrossRef]
- Ko, D.; Kang, J.; Kiba, T.; Park, J.; Kojima, M.; Do, J.; Kim, K.Y.; Kwon, M.; Endler, A.; Song, W.Y.; et al. Arabidopsis ABCG14 is essential for the root-to-shoot translocation of cytokinin. Proc. Natl. Acad. Sci. USA 2014, 111, 7150–7155. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Novak, O.; Wei, Z.; Gou, M.; Zhang, X.; Yu, Y.; Yang, H.; Cai, Y.; Strnad, M.; Liu, C.J. Arabidopsis ABCG14 protein controls the acropetal translocation of root-synthesized cytokinins. Nat. Commun. 2014, 5, 3274. [Google Scholar] [CrossRef]
- Strader, L.C.; Bartel, B. The Arabidopsis PLEIOTROPIC DRUG RESISTANCE8/ABCG36 ATP binding cassette transporter modulates sensitivity to the auxin precursor indole-3-butyric acid. Plant Cell 2009, 21, 1992–2007. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.; Hwang, J.U.; Lee, M.; Kim, Y.Y.; Assmann, S.M.; Martinoia, E.; Lee, Y. PDR-type ABC transporter mediates cellular uptake of the phytohormone abscisic acid. Proc. Natl. Acad. Sci. USA 2010, 107, 2355–2360. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.; Yim, S.; Choi, H.; Kim, A.; Lee, K.P.; Lopez-Molina, L.; Martinoia, E.; Lee, Y. Abscisic acid transporters cooperate to control seed germination. Nat. Commun. 2015, 6, 8113. [Google Scholar] [CrossRef]
- Kuromori, T.; Miyaji, T.; Yabuuchi, H.; Shimizu, H.; Sugimoto, E.; Kamiya, A.; Moriyama, Y.; Shinozaki, K. ABC transporter AtABCG25 is involved in abscisic acid transport and responses. Proc. Natl. Acad. Sci. USA 2010, 107, 2361–2366. [Google Scholar] [CrossRef]
- Ji, H.; Peng, Y.; Meckes, N.; Allen, S.; Stewart, C.N., Jr.; Traw, M.B. ATP-dependent binding cassette transporter G family member 16 increases plant tolerance to abscisic acid and assists in basal resistance against Pseudomonas syringae DC3000. Plant Physiol. 2014, 166, 879–888. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.; Ohyama, K.; Kim, Y.Y.; Jin, J.Y.; Lee, S.B.; Yamaoka, Y.; Muranaka, T.; Suh, M.C.; Fujioka, S.; Lee, Y. The Role of Arabidopsis ABCG9 and ABCG31 ATP Binding Cassette Transporters in Pollen Fitness and the Deposition of Steryl Glycosides on the Pollen Coat. Plant Cell. 2014, 26, 310–324. [Google Scholar] [CrossRef] [PubMed]
- Le Hir, R.; Sorin, C.; Chakraborti, D.; Moritz, T.; Schaller, H.; Tellier, F.; Robert, S.; Morin, H.; Bako, L.; Bellini, C. ABCG9, ABCG11 and ABCG14 ABC transporters are required for vascular development in Arabidopsis. Plant J. 2013, 76, 811–824. [Google Scholar] [CrossRef] [PubMed]
- Alejandro, S.; Lee, Y.; Tohge, T.; Sudre, D.; Osorio, S.; Park, J.; Bovet, L.; Lee, Y.; Geldner, N.; Fernie, A.R.; et al. AtABCG29 is a monolignol transporter involved in lignin biosynthesis. Curr. Biol. 2012, 22, 1207–1212. [Google Scholar] [CrossRef]
- Takeuchi, M.; Kegasa, T.; Watanabe, A.; Tamura, M.; Tsutsumi, Y. Expression analysis of transporter genes for screening candidate monolignol transporters using Arabidopsis thaliana cell suspensions during tracheary element differentiation. J. Plant Res. 2018, 131, 297–305. [Google Scholar] [CrossRef]
- Qin, G.; Liu, C.; Li, J.; Qi, Y.; Gao, Z.; Zhang, X.; Yi, X.; Pan, H.; Ming, R.; Xu, Y. Diversity of metabolite accumulation patterns in inner and outer seed coats of pomegranate: Exploring their relationship with genetic mechanisms of seed coat development. Hortic. Res. 2020, 7, 10. [Google Scholar] [CrossRef]
- Andolfo, G.; Ruocco, M.; Di Donato, A.; Frusciante, L.; Lorito, M.; Scala, F.; Ercolano, M.R. Genetic variability and evolutionary diversification of membrane ABC transporters in plants. BMC Plant. Biol. 2015, 15, 51. [Google Scholar] [CrossRef] [PubMed]
- Crouzet, J.; Trombik, T.; Fraysse, A.S.; Boutry, M. Organization and function of the plant pleiotropic drug resistance ABC transporter family. FEBS Lett. 2006, 580, 1123–1130. [Google Scholar] [CrossRef] [PubMed]
- McFarlane, H.E.; Shin, J.J.; Bird, D.A.; Samuels, A.L. Arabidopsis ABCG transporters, which are required for export of diverse cuticular lipids, dimerize in different combinations. Plant Cell 2010, 22, 3066–3075. [Google Scholar] [CrossRef]
- Sanchez-Fernandez, R.; Davies, T.G.; Coleman, J.O.; Rea, P.A. The Arabidopsis thaliana ABC protein superfamily, a complete inventory. J. Biol. Chem. 2001, 276, 30231–30244. [Google Scholar] [CrossRef] [PubMed]
- Verrier, P.J.; Bird, D.; Burla, B.; Dassa, E.; Forestier, C.; Geisler, M.; Klein, M.; Kolukisaoglu, Ü.; Lee, Y.; Martinoia, E. Plant ABC proteins—A unified nomenclature and updated inventory. Trends Plant Sci. 2008, 13, 151–159. [Google Scholar] [CrossRef]
- Theodoulou, F.L. Plant ABC transporters. Biochim. Biophys. Acta 2000, 1465, 79–103. [Google Scholar] [CrossRef]
- Sun, X.; Shantharaj, D.; Kang, X.; Ni, M. Transcriptional and hormonal signaling control of Arabidopsis seed development. Curr. Opin. Plant Biol. 2010, 13, 611–620. [Google Scholar] [CrossRef]
- Miao, Y.C.; Liu, C.J. ATP-binding cassette-like transporters are involved in the transport of lignin precursors across plasma and vacuolar membranes. Proc. Natl. Acad. Sci. USA 2010, 107, 22728–22733. [Google Scholar] [CrossRef]
- Wang, L.; Xue, J.; Yan, J.; Liu, M.; Tang, Y.; Wang, Y.; Zhang, C. Expression and functional analysis of VviABCG14 from Vitis vinifera suggest the role in cytokinin transport and the interaction with VviABCG7. Plant Physiol. Biochem. 2020, 153, 1–10. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Jiao, Y.; Leebens-Mack, J.; Ayyampalayam, S.; Bowers, J.E.; McKain, M.R.; McNeal, J.; Rolf, M.; Ruzicka, D.R.; Wafula, E.; Wickett, N.J.; et al. A genome triplication associated with early diversification of the core eudicots. Genome Biol. 2012, 13, R3. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Tamura, K.; Nei, M. MEGA: Molecular Evolutionary Genetics Analysis software for microcomputers. Comput. Appl. Biosci. 1994, 10, 189–191. [Google Scholar] [CrossRef] [PubMed]
- Qin, G.; Xu, C.; Ming, R.; Tang, H.; Guyot, R.; Kramer, E.M.; Hu, Y.; Yi, X.; Qi, Y.; Xu, X.; et al. The pomegranate (Punica granatum L.) genome and the genomics of punicalagin biosynthesis. Plant J. 2017, 91, 1108–1128. [Google Scholar] [CrossRef]
- Bentley, H.A.; Belloni, D.R.; Tsongalis, G.J. Parameters involved in the conversion of real-time PCR assays from the ABI prism 7700 to the Cepheid SmartCycler® II. Clin. Biochem. 2005, 38, 183–186. [Google Scholar] [CrossRef]
- Smoot, M.E.; Ono, K.; Ruscheinski, J.; Wang, P.L.; Ideker, T. Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics 2011, 27, 431–432. [Google Scholar] [CrossRef]
- Xu, Y.-L.; Zhang, H.-G.; Shi, T.-Y.; Yao, Y.; Zhang, L.; Wang, Y.-H.; Liu, L. SNP Analysis of 4CL Gene in Larix olgensis Henry Populations. Bull. Bot. Res. 2013, 33, 208–213. [Google Scholar]
- Gu, D.; Zhou, X.; Yin, X.; Wu, M.; Chen, W.; Xu, E.; Liu, Y.; Gong, C.; Zhang, W.; Chen, X. Metal tolerance protein family members are involved in Mn homeostasis through internal compartmentation and exocytosis in Brassica napus. Environ. Exp. Bot. 2022, 195, 104785. [Google Scholar] [CrossRef]
- Ma, G.; Li, J.; Li, J.; Li, Y.; Gu, D.; Chen, C.; Cui, J.; Chen, X.; Zhang, W. OsMTP11, a trans-Golgi network localized transporter, is involved in manganese tolerance in rice. Plant Sci. 2018, 274, 59–69. [Google Scholar] [CrossRef]
- Bechtold, N.; Pelletier, G. In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol. 1998, 82, 259–266. [Google Scholar]
- Gallois, P.; Marinho, P. Leaf disk transformation using Agrobacterium tumefaciens-expression of heterologous genes in tobacco. Methods Mol. Biol. 1995, 49, 39–48. [Google Scholar]
- Pickardt, T.; Ziervogel, B.; Schade, V.; Ohl, L.; Bäumlein, H.; Meixner, M. Developmental-regulation and tissue-specific expression of two different seed promoter GUS-fusions in transgenic lines of Vicia narbonensis. J. Plant Physiol. 1998, 152, 621–629. [Google Scholar] [CrossRef]
- Zhong, R.; Ripperger, A.; Ye, Z.H. Ectopic deposition of lignin in the pith of stems of two Arabidopsis mutants. Plant Physiol. 2000, 123, 59–70. [Google Scholar] [CrossRef] [PubMed]
- Poke, F.S.; Wright, J.K.; Raymond, C.A. Predicting Extractives and Lignin Contents in Eucalyptus globulus Using Near Infrared Reflectance Analysis. J. Wood Chem. Technol. 2005, 24, 55–67. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).