Multicellular 3D Models for the Study of Cardiac Fibrosis
Abstract
1. Introduction
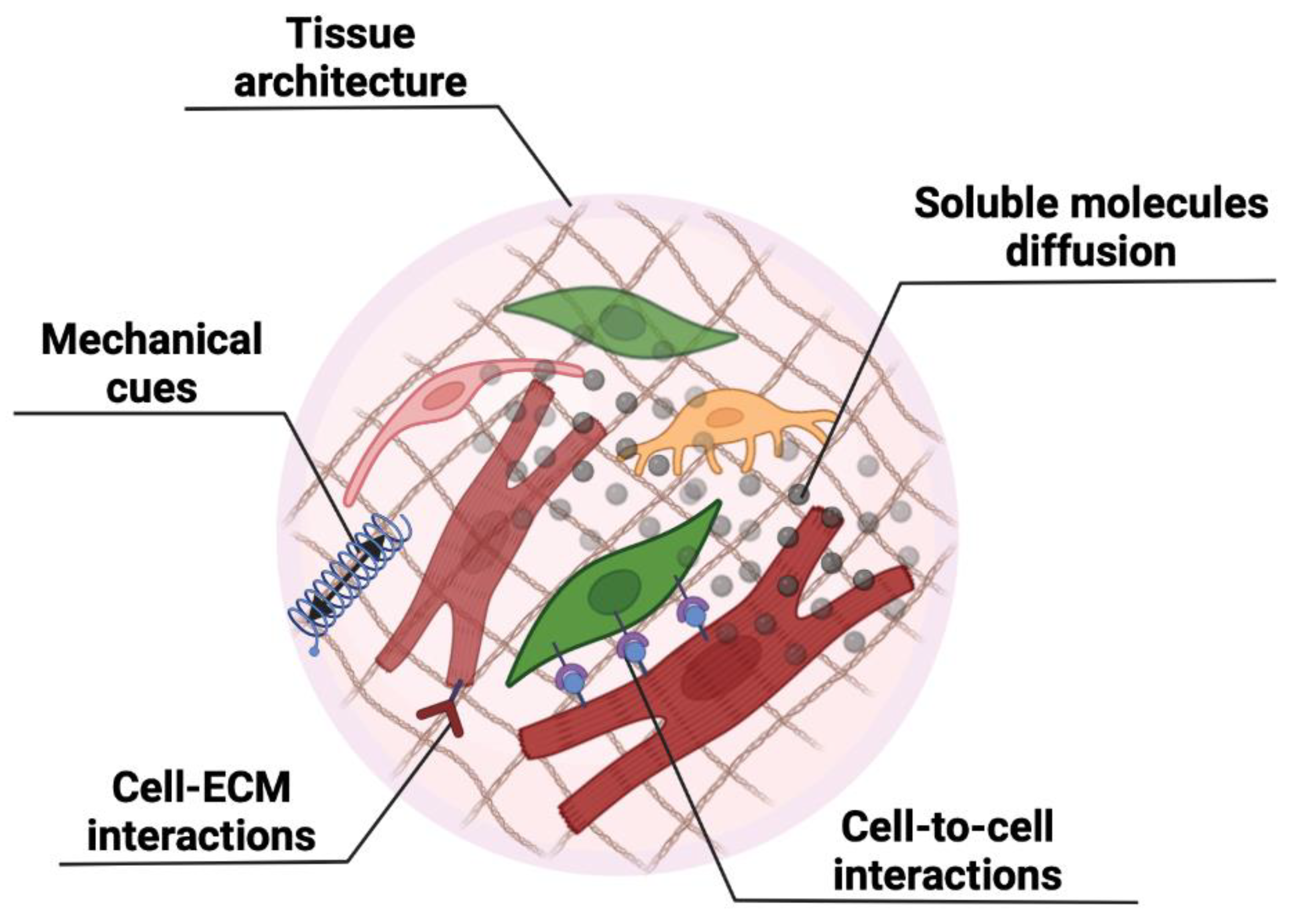
2. Sometimes Three Dimensions Are Better than Two
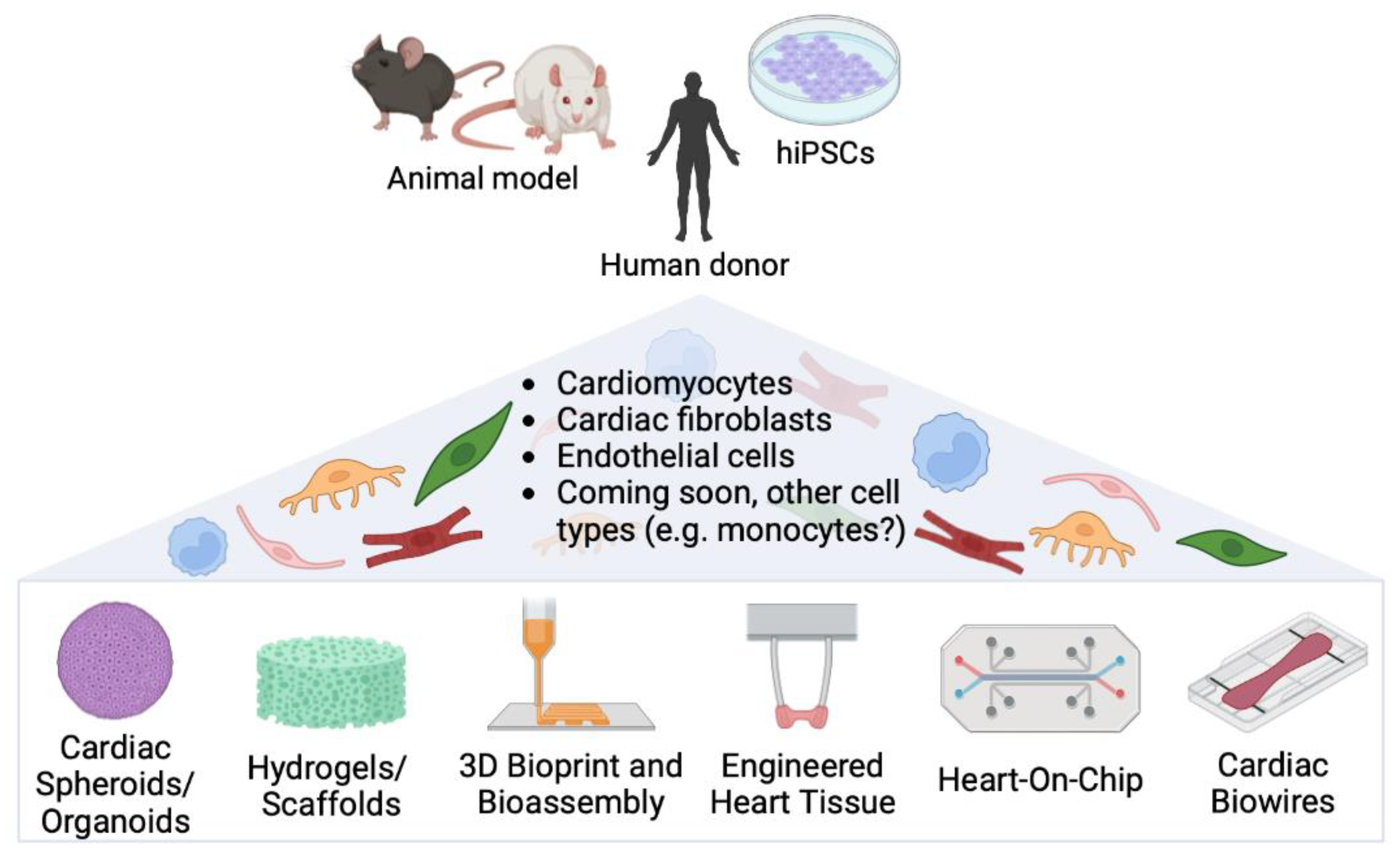
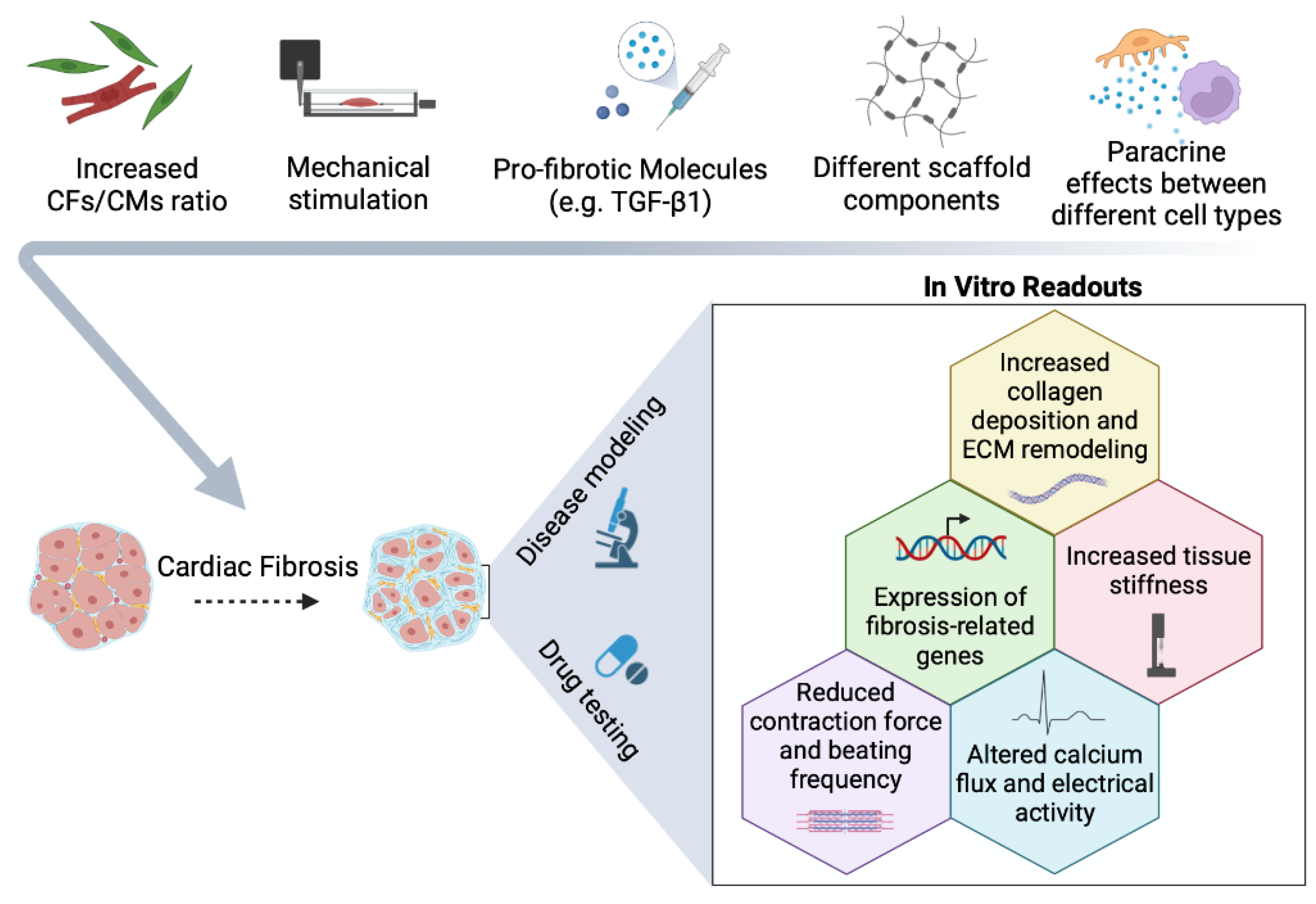
3. The More Cell Types, the Merrier
4. Microtissues: Cardiac Spheroids and Organoids
5. Bioprinting and Bioassembling
6. Engineered Heart Tissues
7. Heart-on-Chips and Cardiac Biowires
8. 3D Systems for Advanced Electromechanical Assessment
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Frangogiannis, N.G. Cardiac Fibrosis. Cardiovasc. Res. 2021, 117, 1450–1488. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, S.H.; Mouton, A.J.; DeLeon-Pennell, K.Y.; Genovese, F.; Karsdal, M.; Lindsey, M.L. Understanding Cardiac Extracellular Matrix Remodeling to Develop Biomarkers of Myocardial Infarction Outcomes. Matrix Biol. 2019, 75–76, 43–57. [Google Scholar] [CrossRef] [PubMed]
- Picchio, V.; Bordin, A.; Floris, E.; Cozzolino, C.; Dhori, X.; Peruzzi, M.; Frati, G.; De Falco, E.; Pagano, F.; Chimenti, I. The Dynamic Facets of the Cardiac Stroma: From Classical Markers to Omics and Translational Perspectives. Am. J. Transl. Res. 2022, 14, 1172–1187. [Google Scholar] [PubMed]
- Farbehi, N.; Patrick, R.; Dorison, A.; Xaymardan, M.; Janbandhu, V.; Wystub-Lis, K.; Ho, J.W.K.; Nordon, R.E.; Harvey, R.P. Single-Cell Expression Profiling Reveals Dynamic Flux of Cardiac Stromal, Vascular and Immune Cells in Health and Injury. Elife 2019, 8, e43882. [Google Scholar] [CrossRef] [PubMed]
- Schirone, L.; Forte, M.; Palmerio, S.; Yee, D.; Nocella, C.; Angelini, F.; Pagano, F.; Schiavon, S.; Bordin, A.; Carrizzo, A.; et al. A Review of the Molecular Mechanisms Underlying the Development and Progression of Cardiac Remodeling. Oxid. Med. Cell. Longev. 2017, 2017, 3920195. [Google Scholar] [CrossRef] [PubMed]
- Henderson, N.C.; Rieder, F.; Wynn, T.A. Fibrosis: From Mechanisms to Medicines. Nature 2020, 587, 555–566. [Google Scholar] [CrossRef] [PubMed]
- Thomas, T.P.; Grisanti, L.A. The Dynamic Interplay between Cardiac Inflammation and Fibrosis. Front. Physiol. 2020, 11, 1–20. [Google Scholar] [CrossRef]
- Bansal, S.S.; Ismahil, M.A.; Goel, M.; Zhou, G.; Rokosh, G.; Hamid, T.; Prabhu, S.D. Dysfunctional and Proinflammatory Regulatory T-Lymphocytes Are Essential for Adverse Cardiac Remodeling in Ischemic Cardiomyopathy. Circulation 2019, 139, 206–221. [Google Scholar] [CrossRef]
- Boufenzer, A.; Lemarié, J.; Simon, T.; Derive, M.; Bouazza, Y.; Tran, N.; Maskali, F.; Groubatch, F.; Bonnin, P.; Bastien, C.; et al. TREM-1 Mediates Inflammatory Injury and Cardiac Remodeling Following Myocardial Infarction. Circ. Res. 2015, 116, 1772–1782. [Google Scholar] [CrossRef]
- Dewald, O.; Zymek, P.; Winkelmann, K.; Koerting, A.; Ren, G.; Abou-Khamis, T.; Michael, L.H.; Rollins, B.J.; Entman, M.L.; Frangogiannis, N.G. CCL2/Monocyte Chemoattractant Protein-1 Regulates Inflammatory Responses Critical to Healing Myocardial Infarcts. Circ. Res. 2005, 96, 881–889. [Google Scholar] [CrossRef]
- Nicin, L.; Wagner, J.U.G.; Luxán, G.; Dimmeler, S. Fibroblast-Mediated Intercellular Crosstalk in the Healthy and Diseased Heart. FEBS Lett. 2022, 596, 638–654. [Google Scholar] [CrossRef] [PubMed]
- Duval, K.; Grover, H.; Han, L.H.; Mou, Y.; Pegoraro, A.F.; Fredberg, J.; Chen, Z. Modeling Physiological Events in 2D vs. 3D Cell Culture. Physiology 2017, 32, 266. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Placone, J.K.; Engler, A.J. Understanding the Extracellular Forces That Determine Cell Fate and Maintenance. Development 2017, 144, 4261–4270. [Google Scholar] [CrossRef] [PubMed]
- Pagliarosi, O.; Picchio, V.; Chimenti, I.; Messina, E.; Gaetani, R. Building an Artificial Cardiac Microenvironment: A Focus on the Extracellular Matrix. Front. Cell Dev. Biol. 2020, 8, 559032. [Google Scholar] [CrossRef]
- Sainio, A.; Järveläinen, H. Extracellular Matrix-Cell Interactions: Focus on Therapeutic Applications. Cell. Signal. 2020, 66, 109487. [Google Scholar] [CrossRef]
- Scadden, D.T. The Stem-Cell Niche as an Entity of Action. Nature 2006, 441, 1075–1079. [Google Scholar] [CrossRef]
- Pineda, E.T.; Nerem, R.M.; Ahsan, T. Differentiation Patterns of Embryonic Stem Cells in Two- versus Three-Dimensional Culture. Cells. Tissues. Organs 2013, 197, 399–410. [Google Scholar] [CrossRef]
- Hatani, T.; Miki, K.; Yoshida, Y. Induction of Human Induced Pluripotent Stem Cells to Cardiomyocytes Using Embryoid Bodies. Methods Mol. Biol. 2018, 1816, 79–92. [Google Scholar] [CrossRef]
- Gaetani, R.; Yin, C.; Srikumar, N.; Braden, R.; Doevendans, P.A.; Sluijter, J.P.G.; Christman, K.L. Cardiac-Derived Extracellular Matrix Enhances Cardiogenic Properties of Human Cardiac Progenitor Cells. Cell Transplant. 2016, 25, 1653–1663. [Google Scholar] [CrossRef]
- Gaetani, R.; Feyen, D.A.M.; Verhage, V.; Slaats, R.; Messina, E.; Christman, K.L.; Giacomello, A.; Doevendans, P.A.F.M.; Sluijter, J.P.G. Epicardial Application of Cardiac Progenitor Cells in a 3D-Printed Gelatin/Hyaluronic Acid Patch Preserves Cardiac Function after Myocardial Infarction. Biomaterials 2015, 61, 339–348. [Google Scholar] [CrossRef]
- Pesce, M.; Messina, E.; Chimenti, I.; Beltrami, A.P. Cardiac Mechanoperception: A Life-Long Story from Early Beats to Aging and Failure. Stem Cells Dev. 2017, 26, 77–90. [Google Scholar] [CrossRef] [PubMed]
- Engler, A.J.; Sen, S.; Sweeney, H.L.; Discher, D.E. Matrix Elasticity Directs Stem Cell Lineage Specification. Cell 2006, 126, 677–689. [Google Scholar] [CrossRef] [PubMed]
- Choi, M.Y.; Kim, J.T.; Lee, W.J.; Lee, Y.; Park, K.M.; Yang, Y.I.; Park, K.D. Engineered Extracellular Microenvironment with a Tunable Mechanical Property for Controlling Cell Behavior and Cardiomyogenic Fate of Cardiac Stem Cells. Acta Biomater. 2017, 50, 234–248. [Google Scholar] [CrossRef]
- Stoppel, W.L.; Kaplan, D.L.; Black, L.D. Electrical and Mechanical Stimulation of Cardiac Cells and Tissue Constructs. Adv. Drug Deliv. Rev. 2016, 96, 135–155. [Google Scholar] [CrossRef] [PubMed]
- Ruan, J.L.; Tulloch, N.L.; Razumova, M.V.; Saiget, M.; Muskheli, V.; Pabon, L.; Reinecke, H.; Regnier, M.; Murry, C.E. Mechanical Stress Conditioning and Electrical Stimulation Promote Contractility and Force Maturation of Induced Pluripotent Stem Cell-Derived Human Cardiac Tissue. Circulation 2016, 134, 1557–1567. [Google Scholar] [CrossRef]
- Nunes, S.S.; Miklas, J.W.; Liu, J.; Aschar-Sobbi, R.; Xiao, Y.; Zhang, B.; Jiang, J.; Massé, S.; Gagliardi, M.; Hsieh, A.; et al. Biowire: A Platform for Maturation of Human Pluripotent Stem Cell-Derived Cardiomyocytes. Nat. Methods 2013, 10, 781–787. [Google Scholar] [CrossRef]
- Ronaldson-Bouchard, K.; Yeager, K.; Teles, D.; Chen, T.; Ma, S.; Song, L.J.; Morikawa, K.; Wobma, H.M.; Vasciaveo, A.; Ruiz, E.C.; et al. Engineering of Human Cardiac Muscle Electromechanically Matured to an Adult-like Phenotype. Nat. Protoc. 2019, 14, 2781–2817. [Google Scholar] [CrossRef]
- Kong, M.; Lee, J.; Yazdi, I.K.; Miri, A.K.; Lin, Y.D.; Seo, J.; Zhang, Y.S.; Khademhosseini, A.; Shin, S.R. Cardiac Fibrotic Remodeling on a Chip with Dynamic Mechanical Stimulation. Adv. Healthc. Mater. 2019, 8, 1801146. [Google Scholar] [CrossRef]
- Garoffolo, G.; Casaburo, M.; Amadeo, F.; Salvi, M.; Bernava, G.; Piacentini, L.; Chimenti, I.; Zaccagnini, G.; Milcovich, G.; Zuccolo, E.; et al. Reduction of Cardiac Fibrosis by Interference with YAP-Dependent Transactivation. Circ. Res. 2022, 131, 239–257. [Google Scholar] [CrossRef]
- Pinto, A.R.; Ilinykh, A.; Ivey, M.J.; Kuwabara, J.T.; D’antoni, M.L.; Debuque, R.; Chandran, A.; Wang, L.; Arora, K.; Rosenthal, N.A.; et al. Revisiting Cardiac Cellular Composition. Circ. Res. 2016, 118, 400–409. [Google Scholar] [CrossRef]
- Litviňuková, M.; Talavera-López, C.; Maatz, H.; Reichart, D.; Worth, C.L.; Lindberg, E.L.; Kanda, M.; Polanski, K.; Heinig, M.; Lee, M.; et al. Cells of the Adult Human Heart. Nature 2020, 588, 466–472. [Google Scholar] [CrossRef] [PubMed]
- Saini, H.; Navaei, A.; Van Putten, A.; Nikkhah, M. 3D Cardiac Microtissues Encapsulated with the Co-Culture of Cardiomyocytes and Cardiac Fibroblasts. Adv. Healthc. Mater. 2015, 4, 1961–1971. [Google Scholar] [CrossRef] [PubMed]
- Navaei, A.; Truong, D.; Heffernan, J.; Cutts, J.; Brafman, D.; Sirianni, R.W.; Vernon, B.; Nikkhah, M. PNIPAAm-Based Biohybrid Injectable Hydrogel for Cardiac Tissue Engineering. Acta Biomater. 2016, 32, 10–23. [Google Scholar] [CrossRef] [PubMed]
- de Lange, W.J.; Farrell, E.T.; Kreitzer, C.R.; Jacobs, D.R.; Lang, D.; Glukhov, A.V.; Carter Ralphe, J. Human IPSC-Engineered Cardiac Tissue Platform Faithfully Models Important Cardiac Physiology. Am. J. Physiol.-Heart Circ. Physiol. 2021, 320, H1670–H1686. [Google Scholar] [CrossRef] [PubMed]
- Plikus, M.V.; Wang, X.; Sinha, S.; Forte, E.; Thompson, S.M.; Herzog, E.L.; Driskell, R.R.; Rosenthal, N.; Biernaskie, J.; Horsley, V. Fibroblasts: Origins, Definitions, and Functions in Health and Disease. Cell 2021, 184, 3852–3872. [Google Scholar] [CrossRef] [PubMed]
- Pagano, F.; Picchio, V.; Angelini, F.; Iaccarino, A.; Peruzzi, M.; Cavarretta, E.; Biondi-Zoccai, G.; Sciarretta, S.; De Falco, E.; Chimenti, I.; et al. The Biological Mechanisms of Action of Cardiac Progenitor Cell Therapy. Curr. Cardiol. Rep. 2018, 20, 84. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro da Silva, A.; Neri, E.A.; Turaça, L.T.; Dariolli, R.; Fonseca-Alaniz, M.H.; Santos-Miranda, A.; Roman-Campos, D.; Venturini, G.; Krieger, J.E. NOTCH1 Is Critical for Fibroblast-Mediated Induction of Cardiomyocyte Specialization into Ventricular Conduction System-like Cells in Vitro. Sci. Rep. 2020, 10, 16163. [Google Scholar] [CrossRef]
- Giacomelli, E.; Meraviglia, V.; Campostrini, G.; Cochrane, A.; Cao, X.; van Helden, R.W.J.; Krotenberg Garcia, A.; Mircea, M.; Kostidis, S.; Davis, R.P.; et al. Human-IPSC-Derived Cardiac Stromal Cells Enhance Maturation in 3D Cardiac Microtissues and Reveal Non-Cardiomyocyte Contributions to Heart Disease. Cell Stem Cell 2020, 26, 862–879.e11. [Google Scholar] [CrossRef]
- Stevens, K.R.; Kreutziger, K.L.; Dupras, S.K.; Korte, F.S.; Regnier, M.; Muskheli, V.; Nourse, M.B.; Bendixen, K.; Reinecke, H.; Murry, C.E. Physiological Function and Transplantation of Scaffold-Free and Vascularized Human Cardiac Muscle Tissue. Proc. Natl. Acad. Sci. USA 2009, 106, 16568–16573. [Google Scholar] [CrossRef]
- Forte, E.; Furtado, M.B.; Rosenthal, N. The Interstitium in Cardiac Repair: Role of the Immune-Stromal Cell Interplay. Nat. Rev. Cardiol. 2018, 15, 601–616. [Google Scholar] [CrossRef]
- Forte, E.; Skelly, D.A.; Chen, M.; Daigle, S.; Morelli, K.A.; Hon, O.; Philip, V.M.; Costa, M.W.; Rosenthal, N.A.; Furtado, M.B. Dynamic Interstitial Cell Response during Myocardial Infarction Predicts Resilience to Rupture in Genetically Diverse Mice. Cell Rep. 2020, 30, 3149–3163.e6. [Google Scholar] [CrossRef] [PubMed]
- Fiaschi, T.; Magherini, F.; Gamberi, T.; Lucchese, G.; Faggian, G.; Modesti, A.; Modesti, P.A. Hyperglycemia and Angiotensin II Cooperate to Enhance Collagen I Deposition by Cardiac Fibroblasts through a ROS-STAT3-Dependent Mechanism. Biochim. Biophys. Acta 2014, 1843, 2603–2610. [Google Scholar] [CrossRef] [PubMed]
- Wang, E.Y.; Rafatian, N.; Zhao, Y.; Lee, A.; Lai, B.F.L.; Lu, R.X.; Jekic, D.; Davenport Huyer, L.; Knee-Walden, E.J.; Bhattacharya, S.; et al. Biowire Model of Interstitial and Focal Cardiac Fibrosis. ACS Cent. Sci. 2019, 5, 1146–1158. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Méndez, J.D.; Méndez-Valenzuela, V.; Aguilar-Hernández, M.M. Cellular Signalling of the Receptor for Advanced Glycation End Products (RAGE). Cell. Signal. 2013, 25, 2185–2197. [Google Scholar] [CrossRef]
- Edmondson, R.; Broglie, J.J.; Adcock, A.F.; Yang, L. Three-Dimensional Cell Culture Systems and Their Applications in Drug Discovery and Cell-Based Biosensors. Assay Drug Dev. Technol. 2014, 12, 207–218. [Google Scholar] [CrossRef] [PubMed]
- Polonchuk, L.; Chabria, M.; Badi, L.; Hoflack, J.C.; Figtree, G.; Davies, M.J.; Gentile, C. Cardiac Spheroids as Promising in Vitro Models to Study the Human Heart Microenvironment. Sci. Rep. 2017, 7, 7005. [Google Scholar] [CrossRef]
- Sacchi, M.; Bansal, R.; Rouwkema, J. Bioengineered 3D Models to Recapitulate Tissue Fibrosis. Trends Biotechnol. 2020, 38, 623–636. [Google Scholar] [CrossRef]
- Hofer, M.; Lutolf, M.P. Engineering Organoids. Nat. Rev. Mater. 2021, 6, 402–420. [Google Scholar] [CrossRef]
- Nugraha, B.; Buono, M.F.; von Boehmer, L.; Hoerstrup, S.P.; Emmert, M.Y. Human Cardiac Organoids for Disease Modeling. Clin. Pharmacol. Ther. 2019, 105, 79–85. [Google Scholar] [CrossRef]
- Figtree, G.A.; Bubb, K.J.; Tang, O.; Kizana, E.; Gentile, C. Vascularized Cardiac Spheroids as Novel 3D in Vitro Models to Study Cardiac Fibrosis. Cells Tissues Organs 2017, 204, 191–198. [Google Scholar] [CrossRef]
- van Spreeuwel, A.C.C.; Bax, N.A.M.; van Nierop, B.J.; Aartsma-Rus, A.; Goumans, M.J.T.H.; Bouten, C.V.C. Mimicking Cardiac Fibrosis in a Dish: Fibroblast Density Rather than Collagen Density Weakens Cardiomyocyte Function. J. Cardiovasc. Transl. Res. 2017, 10, 116–127. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mainardi, A.; Carminati, F.; Ugolini, G.S.; Occhetta, P.; Isu, G.; Robles Diaz, D.; Reid, G.; Visone, R.; Rasponi, M.; Marsano, A. A Dynamic Microscale Mid-Throughput Fibrosis Model to Investigate the Effects of Different Ratios of Cardiomyocytes and Fibroblasts. Lab Chip 2021, 21, 4177–4195. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.O.; Jung, K.B.; Jo, S.J.; Hyun, S.A.; Moon, K.S.; Seo, J.W.; Kim, S.H.; Son, M.Y. Modelling Cardiac Fibrosis Using Three-Dimensional Cardiac Microtissues Derived from Human Embryonic Stem Cells. J. Biol. Eng. 2019, 13, 15. [Google Scholar] [CrossRef] [PubMed]
- Richards, D.J.; Li, Y.; Kerr, C.M.; Yao, J.; Beeson, G.C.; Coyle, R.C.; Chen, X.; Jia, J.; Damon, B.; Wilson, R.; et al. Human Cardiac Organoids for the Modelling of Myocardial Infarction and Drug Cardiotoxicity. Nat. Biomed. Eng. 2020, 4, 446–462. [Google Scholar] [CrossRef] [PubMed]
- Murphy, S.V.; Atala, A. 3D Bioprinting of Tissues and Organs. Nat. Biotechnol. 2014, 32, 773–785. [Google Scholar] [CrossRef]
- Matai, I.; Kaur, G.; Seyedsalehi, A.; McClinton, A.; Laurencin, C.T. Progress in 3D Bioprinting Technology for Tissue/Organ Regenerative Engineering. Biomaterials 2020, 226, 119536. [Google Scholar] [CrossRef]
- Lee, A.; Hudson, A.R.; Shiwarski, D.J.; Tashman, J.W.; Hinton, T.J.; Yerneni, S.; Bliley, J.M.; Campbell, P.G.; Feinberg, A.W. 3D Bioprinting of Collagen to Rebuild Components of the Human Heart. Science 2019, 365, 482–487. [Google Scholar] [CrossRef]
- Murphy, S.V.; De Coppi, P.; Atala, A. Opportunities and Challenges of Translational 3D Bioprinting. Nat. Biomed. Eng. 2020, 4, 370–380. [Google Scholar] [CrossRef]
- Gungor-Ozkerim, P.S.; Inci, I.; Zhang, Y.S.; Khademhosseini, A.; Dokmeci, M.R. Bioinks for 3D Bioprinting: An Overview. Biomater. Sci. 2018, 6, 915–946. [Google Scholar] [CrossRef]
- Decante, G.; Costa, J.B.; Silva-Correia, J.; Collins, M.N.; Reis, R.L.; Oliveira, J.M. Engineering Bioinks for 3D Bioprinting. Biofabrication 2021, 13, 032001. [Google Scholar] [CrossRef]
- Ong, C.S.; Fukunishi, T.; Zhang, H.; Huang, C.Y.; Nashed, A.; Blazeski, A.; Disilvestre, D.; Vricella, L.; Conte, J.; Tung, L.; et al. Biomaterial-Free Three-Dimensional Bioprinting of Cardiac Tissue Using Human Induced Pluripotent Stem Cell Derived Cardiomyocytes. Sci. Rep. 2017, 7, 4566. [Google Scholar] [CrossRef] [PubMed]
- Daly, A.C.; Davidson, M.D.; Burdick, J.A. 3D Bioprinting of High Cell-Density Heterogeneous Tissue Models through Spheroid Fusion within Self-Healing Hydrogels. Nat. Commun. 2021, 12, 753. [Google Scholar] [CrossRef] [PubMed]
- Shin, Y.J.; Shafranek, R.T.; Tsui, J.H.; Walcott, J.; Nelson, A.; Kim, D.H. 3D Bioprinting of Mechanically Tuned Bioinks Derived from Cardiac Decellularized Extracellular Matrix. Acta Biomater. 2021, 119, 75–88. [Google Scholar] [CrossRef] [PubMed]
- Baillargeon, P.; Shumate, J.; Hou, S.; Fernandez-Vega, V.; Marques, N.; Souza, G.; Seldin, J.; Spicer, T.P.; Scampavia, L. Automating a Magnetic 3D Spheroid Model Technology for High-Throughput Screening. SLAS Technol. 2019, 24, 420–428. [Google Scholar] [CrossRef] [PubMed]
- Hogan, M.; Souza, G.; Birla, R.; Hogan, M.; Souza, G.; Birla, R. Assembly of a Functional 3D Primary Cardiac Construct Using Magnetic Levitation. AIMS Bioeng. 2016, 3, 277–288. [Google Scholar] [CrossRef]
- Eschenhagen, T.; Fink, C.; Remmers, U.; Scholz, H.; Wattchow, J.; Weil, J.; Zimmermann, W.; Dohmen, H.H.; Schäfer, H.; Bishopric, N.; et al. Three-Dimensional Reconstitution of Embryonic Cardiomyocytes in a Collagen Matrix: A New Heart Muscle Model System. FASEB J. 1997, 11, 683–694. [Google Scholar] [CrossRef]
- Tzatzalos, E.; Abilez, O.J.; Shukla, P.; Wu, J.C. Engineered Heart Tissues and Induced Pluripotent Stem Cells: Macro- and Microstructures for Disease Modeling, Drug Screening, and Translational Studies. Adv. Drug Deliv. Rev. 2016, 96, 234–244. [Google Scholar] [CrossRef]
- Szepes, M.; Melchert, A.; Dahlmann, J.; Hegermann, J.; Werlein, C.; Jonigk, D.; Haverich, A.; Martin, U.; Olmer, R.; Gruh, I. Dual Function of IPSC-Derived Pericyte-Like Cells in Vascularization and Fibrosis-Related Cardiac Tissue Remodeling In Vitro. Int. J. Mol. Sci. 2020, 21, 8947. [Google Scholar] [CrossRef]
- Leask, A. Getting to the Heart of the Matter: New Insights into Cardiac Fibrosis. Circ. Res. 2015, 116, 1269–1276. [Google Scholar] [CrossRef]
- Sager, H.B.; Hulsmans, M.; Lavine, K.J.; Moreira, M.B.; Heidt, T.; Courties, G.; Sun, Y.; Iwamoto, Y.; Tricot, B.; Khan, O.F.; et al. Proliferation and Recruitment Contribute to Myocardial Macrophage Expansion in Chronic Heart Failure. Circ. Res. 2016, 119, 853–864. [Google Scholar] [CrossRef]
- Suku, M.; Forrester, L.; Biggs, M.; Monaghan, M.G. Resident Macrophages and Their Potential in Cardiac Tissue Engineering. Tissue Eng. Part B Rev. 2022, 28, 579–591. [Google Scholar] [CrossRef] [PubMed]
- Mastrangeli, M.; Millet, S.; Mummery, C.; Loskill, P.; Braeken, D.; Eberle, W.; Cipriano, M.; Fernandez, L.; Graef, M.; Gidrol, X.; et al. Building Blocks for a European Organ-on-Chip Roadmap. ALTEX 2019, 36, 481–492. [Google Scholar] [CrossRef] [PubMed]
- Abulaiti, M.; Yalikun, Y.; Murata, K.; Sato, A.; Sami, M.M.; Sasaki, Y.; Fujiwara, Y.; Minatoya, K.; Shiba, Y.; Tanaka, Y.; et al. Establishment of a Heart-on-a-Chip Microdevice Based on Human IPS Cells for the Evaluation of Human Heart Tissue Function. Sci. Rep. 2020, 10, 19201. [Google Scholar] [CrossRef] [PubMed]
- Rogers, A.J.; Miller, J.M.; Kannappan, R.; Sethu, P. Cardiac Tissue Chips (CTCs) for Modeling Cardiovascular Disease. IEEE Trans. Biomed. Eng. 2019, 66, 3436–3443. [Google Scholar] [CrossRef] [PubMed]
- Mastikhina, O.; Moon, B.U.; Williams, K.; Hatkar, R.; Gustafson, D.; Mourad, O.; Sun, X.; Koo, M.; Lam, A.Y.L.; Sun, Y.; et al. Human Cardiac Fibrosis-on-a-Chip Model Recapitulates Disease Hallmarks and Can Serve as a Platform for Drug Testing. Biomaterials 2020, 233, 119741. [Google Scholar] [CrossRef]
- Veldhuizen, J.; Chavan, R.; Moghadas, B.; Park, J.G.; Kodibagkar, V.D.; Migrino, R.Q.; Nikkhah, M. Cardiac Ischemia On-a-Chip to Investigate Cellular and Molecular Response of Myocardial Tissue under Hypoxia. Biomaterials 2022, 281, 121336. [Google Scholar] [CrossRef]
- Kuzmanov, U.; Wang, E.Y.; Vanderlaan, R.; Kim, D.H.; Lee, S.H.; Hadipour-Lakmehsari, S.; Guo, H.; Zhao, Y.; McFadden, M.; Sharma, P.; et al. Mapping Signalling Perturbations in Myocardial Fibrosis via the Integrative Phosphoproteomic Profiling of Tissue from Diverse Sources. Nat. Biomed. Eng. 2020, 4, 889–900. [Google Scholar] [CrossRef]
- Smaill, B.H.; Zhao, J.; Trew, M.L. Three-Dimensional Impulse Propagation in Myocardium: Arrhythmogenic Mechanisms at the Tissue Level. Circ. Res. 2013, 112, 834–848. [Google Scholar] [CrossRef]
- Spencer, T.M.; Blumenstein, R.F.; Pryse, K.M.; Lee, S.L.; Glaubke, D.A.; Carlson, B.E.; Elson, E.L.; Genin, G.M. Fibroblasts Slow Conduction Velocity in a Reconstituted Tissue Model of Fibrotic Cardiomyopathy. ACS Biomater. Sci. Eng. 2017, 3, 3022–3028. [Google Scholar] [CrossRef]
- Pong, T.; Adams, W.J.; Bray, M.A.; Feinberg, A.W.; Sheehy, S.P.; Werdich, A.A.; Parker, K.K. Hierarchical Architecture Influences Calcium Dynamics in Engineered Cardiac Muscle. Exp. Biol. Med. (Maywood) 2011, 236, 366–373. [Google Scholar] [CrossRef]
- Van Spreeuwel, A.C.C.; Bax, N.A.M.; Bastiaens, A.J.; Foolen, J.; Loerakker, S.; Borochin, M.; Van Der Schaft, D.W.J.; Chen, C.S.; Baaijens, F.P.T.; Bouten, C.V.C. The Influence of Matrix (an)Isotropy on Cardiomyocyte Contraction in Engineered Cardiac Microtissues. Integr. Biol. (Camb) 2014, 6, 422–429. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Picchio, V.; Floris, E.; Derevyanchuk, Y.; Cozzolino, C.; Messina, E.; Pagano, F.; Chimenti, I.; Gaetani, R. Multicellular 3D Models for the Study of Cardiac Fibrosis. Int. J. Mol. Sci. 2022, 23, 11642. https://doi.org/10.3390/ijms231911642
Picchio V, Floris E, Derevyanchuk Y, Cozzolino C, Messina E, Pagano F, Chimenti I, Gaetani R. Multicellular 3D Models for the Study of Cardiac Fibrosis. International Journal of Molecular Sciences. 2022; 23(19):11642. https://doi.org/10.3390/ijms231911642
Chicago/Turabian StylePicchio, Vittorio, Erica Floris, Yuriy Derevyanchuk, Claudia Cozzolino, Elisa Messina, Francesca Pagano, Isotta Chimenti, and Roberto Gaetani. 2022. "Multicellular 3D Models for the Study of Cardiac Fibrosis" International Journal of Molecular Sciences 23, no. 19: 11642. https://doi.org/10.3390/ijms231911642
APA StylePicchio, V., Floris, E., Derevyanchuk, Y., Cozzolino, C., Messina, E., Pagano, F., Chimenti, I., & Gaetani, R. (2022). Multicellular 3D Models for the Study of Cardiac Fibrosis. International Journal of Molecular Sciences, 23(19), 11642. https://doi.org/10.3390/ijms231911642







