Physiological and Comparative Transcriptome Analyses of the High-Tillering Mutant mtn1 Reveal Regulatory Mechanisms in the Tillering of Centipedegrass (Eremochloa ophiuroides (Munro) Hack.)
Abstract
1. Introduction
2. Results
2.1. Morphological Analysis of the mtn1 Mutant
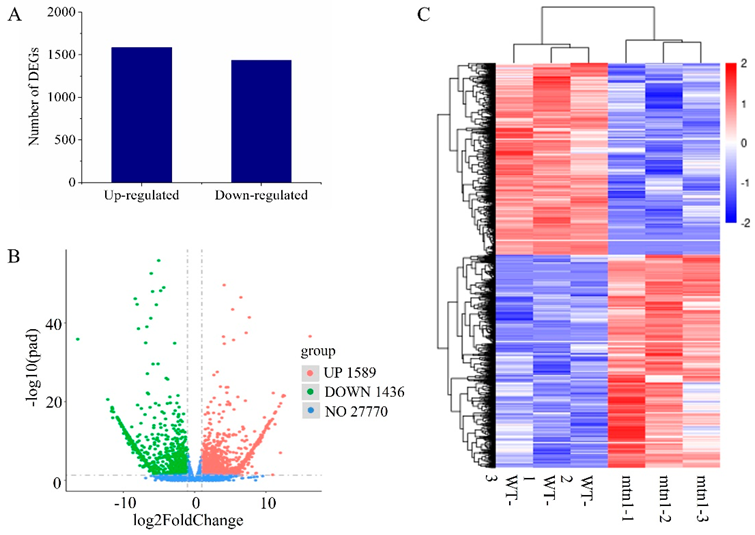
2.2. RNA-Seq Data Analysis
2.3. Identification of DEGs between the mtn1 Mutant and WT
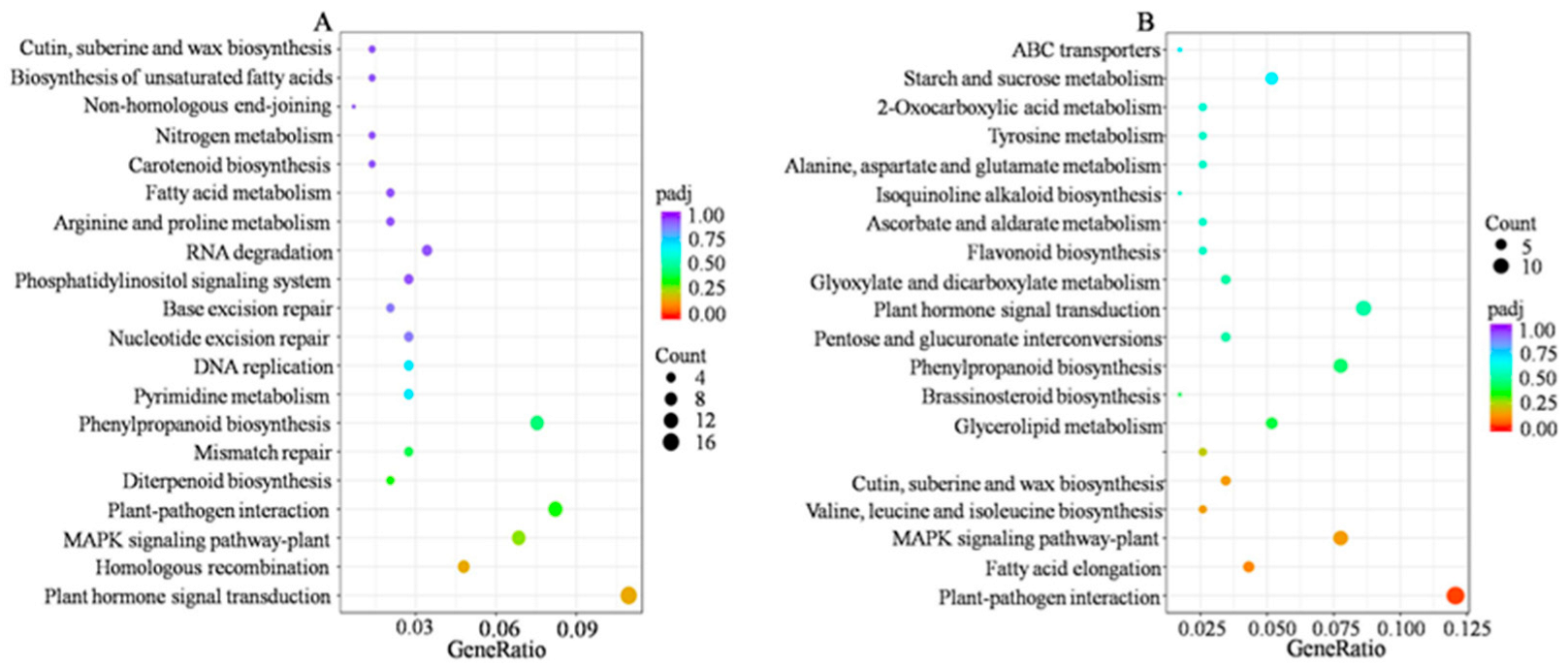
2.4. Functional Analysis of DEGs
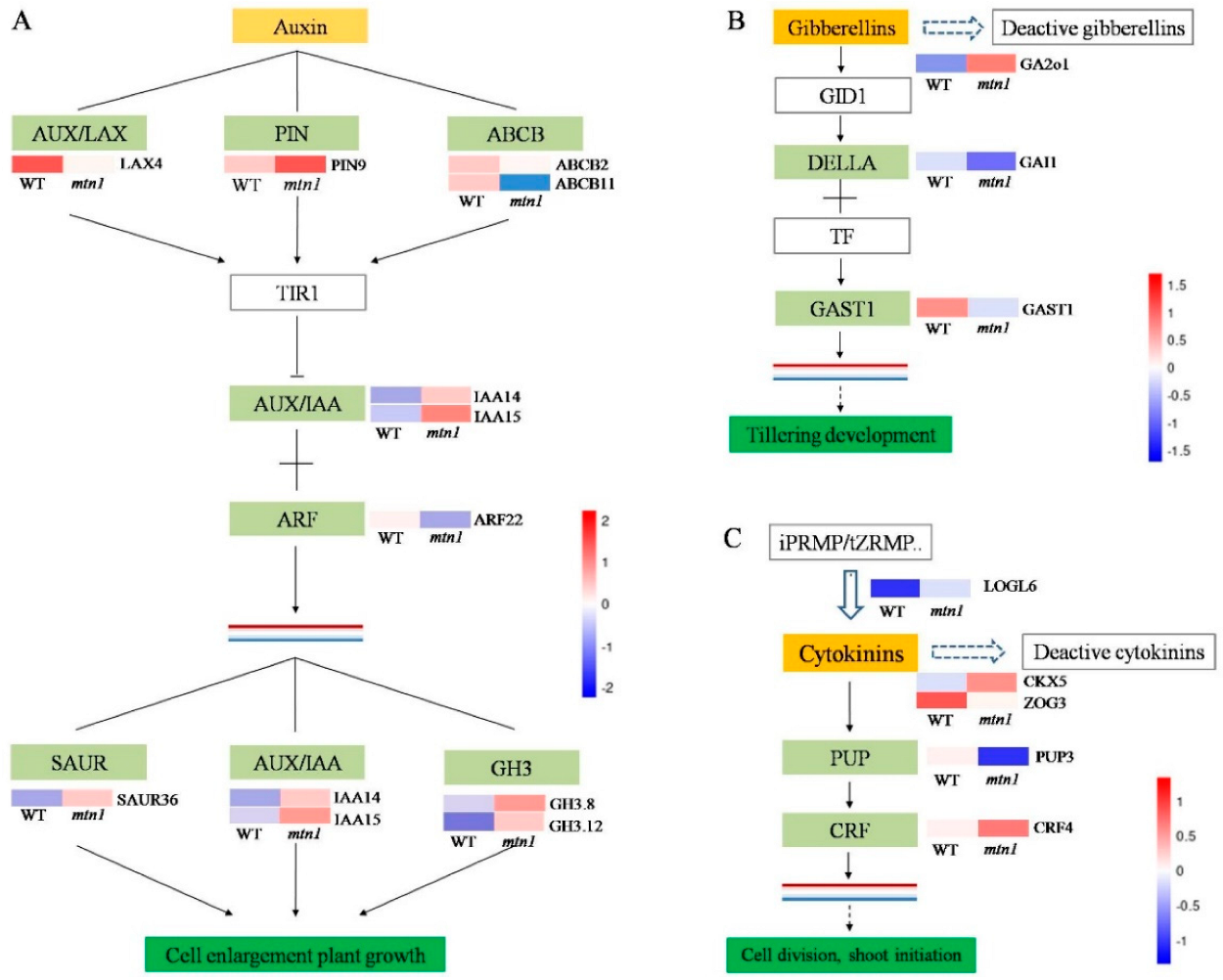
2.5. Regulatory Pathways of DEGs Related to Plant Hormones
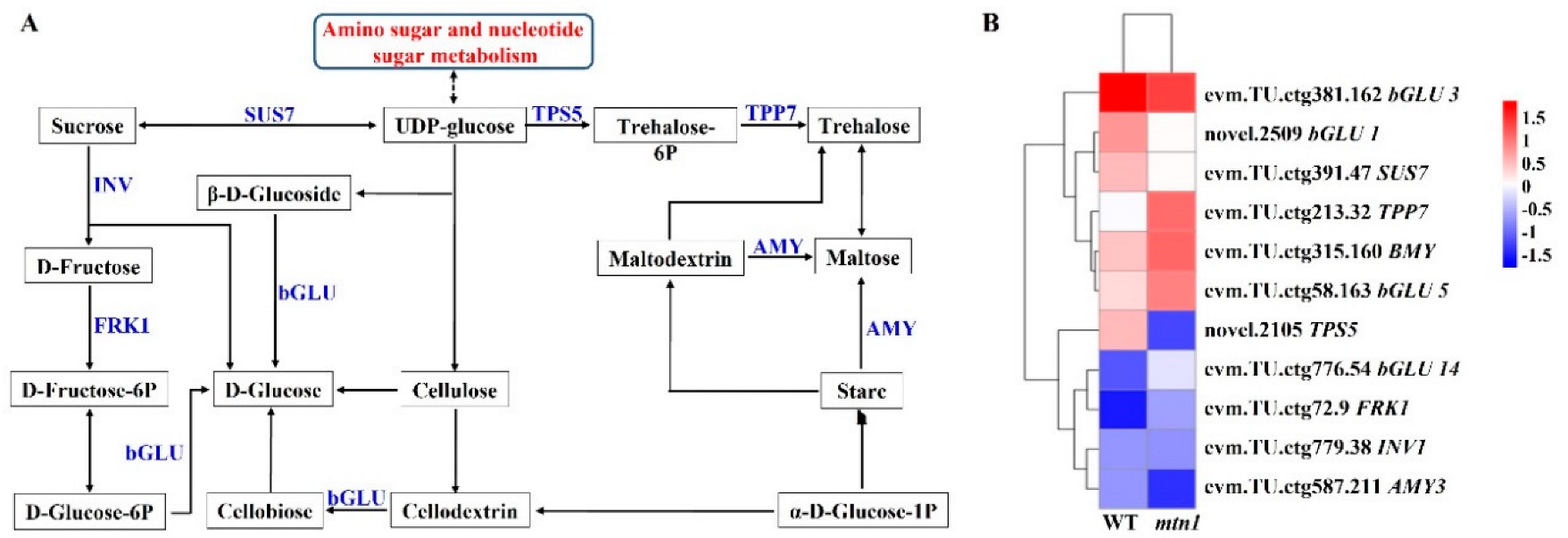
2.6. Regulatory Pathways of DEGs Involved in Starch and Sucrose Metabolism
2.7. Confirmation of RNA-Seq Data
2.8. IAA, GA, CTK and Soluble Sugar Contents and Their Influence on the Tillering Number
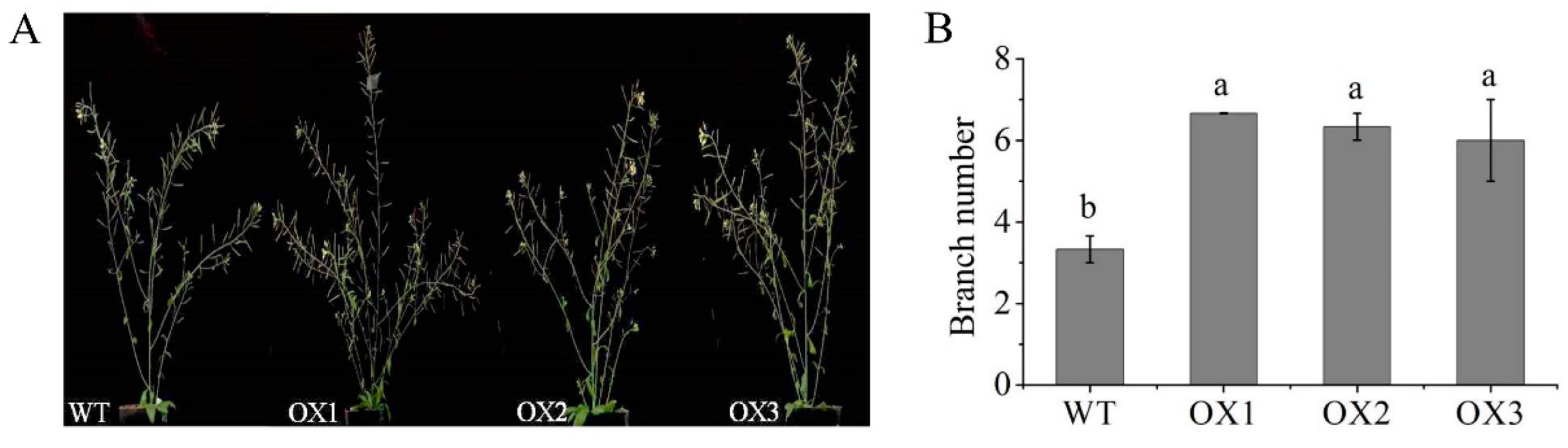
2.9. Overexpression of EoABCB11 in Arabidopsis Affected Branch Number
2.10. Identification of SNPs
3. Discussion
3.1. Regulatory Role of Plant Hormones in Centipedegrass Tillering
3.2. Regulatory Role of Sugars in Centipedegrass Tillering
3.3. EoABCB11 Was Involved in the Tillering of Centipedegrass
3.4. SNPs Identification from mtn1 Mutant and WT
4. Materials and Methods
4.1. Plant Materials
4.2. Experimental Design
4.3. Phenotypic Characteristics and Yields
4.4. RNA-Seq and Data Analyses
4.5. Quantitative Real-Time PCR Analysis of Expression of Candidate Genes
4.6. Analysis of Plant Hormones and Total Soluble Sugar Contents
4.7. Influence of IAA, GA, CTK and Soluble Sugar on Tillering Number
4.8. EoABCB11 Genetic Transformation
4.9. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Liu, J.; Hanna, W.; Elsner, E. Morphological and seed set characteristics of centipedegrass accessions collected in China. Econ. Bot. 2003, 57, 380–388. [Google Scholar] [CrossRef]
- Li, J.J.; Guo, H.L.; Zong, J.Q.; Chen, J.B.; Li, D.D.; Liu, J.X. Genetic diversity in centipedegrass [Eremochloa ophiuroides (Munro) Hack.]. Hortic. Res. 2020, 7, 4. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.; Hirata, M. Centipedegrass (Eremochloa ophiuroides (Munro) Hack.): Growth behavior and multipurpose usages. Grassl. Sci. 2005, 51, 183–190. [Google Scholar] [CrossRef]
- Liu, M.; Yang, J.; Lu, S.Y.; Guo, Z.F.; Lin, X.P.; Wu, H. Somatic embryogenesis and plant regeneration in centipedegrass (Eremochloa ophiuroides [Munro] Hack.). Vitr. Cell. Dev. Biol. 2008, 44, 100–104. [Google Scholar] [CrossRef]
- Li, L.; Chen, J.B.; Wang, H.R.; Guo, H.L.; Li, D.D.; Li, J.J.; Liu, J.X.; Shao, H.L.; Zong, J.Q. Cold plasma treatment improves seed germination and accelerates the establishment of centipedegrass. Crop Sci. 2021, 61, 2827–2836. [Google Scholar] [CrossRef]
- Jewiss, O.R. Tillering in grasses-its significance and control. Grass Forage Sci. 1972, 27, 65–82. [Google Scholar] [CrossRef]
- Wang, B.; Smith, S.M.; Li, J. Genetic regulation of shoot architecture. Ann. Rev. Plant Biol. 2018, 69, 437–468. [Google Scholar] [CrossRef]
- Hou, M.M.; Luo, F.F.; Wu, D.X.; Zhang, X.H.; Lou, M.M.; Shen, D.F.; Mao, C.Z.; Mao, C.Z.; Fan, X.R.; Xu, G.H. OsPIN9, an auxin efflux carrier, is required for the regulation of rice tiller bud outgrowth by ammonium. New Phytol. 2021, 229, 935–949. [Google Scholar] [CrossRef]
- Wang, Q.; Kohlen, W.; Rossmann, S.; Vernoux, T.; Theres, K. Auxin depletion from the leaf axil conditions competence for axillary meristem formation in Arabidopsis and tomato. Plant Cell 2014, 26, 2068–2079. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.X.; Zha, M.R.; Li, Y.; Ding, Y.F.; Ding, C.Q.; Wang, S.H. The interaction between nitrogen availability and auxin, cytokinin, and strigolactone in the control of shoot branching in rice (Oryza sativa L.). Plant Cell Rep. 2015, 34, 1647–1662. [Google Scholar] [CrossRef]
- Chen, Y.; Fan, X.; Song, W.; Zhang, Y.; Xu, G. Over-expression of OsPIN2 leads to increased tiller numbers, angle and shorter plant height through suppression of OsLAZY1. Plant Biotechnol. J. 2012, 10, 139–149. [Google Scholar] [CrossRef] [PubMed]
- Overvoorde, P.J.; Yoko Okushima, Y.; Alonso, J.M.; Chan, A.; Chang, C.; Ecker, J.R.; Hughes, B.; Liu, A.; Onodera, C.; Quach, H.; et al. Functional genomic analysis of the AUXIN/INDOLE-3-ACID gene family members in Arabidopsis thaliana. Plant Cell 2005, 17, 3282–3300. [Google Scholar] [CrossRef] [PubMed]
- Elfving, D.C.; Visser, D.B.; Henry, J.L. Gibberellins stimulate lateral branch development in young sweet cherry trees in the orchard. Int. J. Fruit Sci. 2011, 11, 41–54. [Google Scholar] [CrossRef]
- Ni, J.; Zhao, M.L.; Chen, M.S.; Pan, B.Z.; Tao, Y.B.; Xu, Z.F. Comparative transcriptome analysis of axillary buds in response to the shoot branching regulators gibberellins A3 and 6-benzyladenine in Jatropha Curcas. Sci. Rep. 2017, 7, 1141. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, L.l.; Ge, Y.; Wang, J.; Yu, J.J.; Yang, Z.M.; Huang, B.R. Gibberellic acid inhibition of tillering in tall fescue involving crosstalks with cytokinins and transcriptional regulation of genes controlling axillary bud outgrowth. Plant Sci. 2019, 287, 110168. [Google Scholar] [CrossRef]
- Xu, X.H.; Feng, G.Y.; Liang, Y.Y.; Shuang, Y.; Liu, Q.X.; Nie, G.; Yang, Z.F.; Hang, L.K.; Zhang, X.Q. Comparative transcriptome analyses reveal different mechanism of high- and low-tillering genotypes controlling tiller growth in orchardgrass (Dactylis glomerata L.). BMC Plant Biol. 2020, 20, 369. [Google Scholar] [CrossRef]
- Martinezbello, L.; Moritz, T.; Lopezdiaz, I. Silencing C19-GA 2-oxidases induces parthenocarpic development and inhibits lateral branching in tomato plants. J. Exp. Bot. 2015, 66, 5897–5910. [Google Scholar] [CrossRef]
- Shang, X.L.; Xie, R.R.; Tian, H.; Wang, Q.L.; Guo, F.Q. Putative zeatin O-glucosyltransferase OscZOG1 regulates root and shoot development and formation of agronomic traits in rice. J. Integr. Plant Biol. 2016, 58, 627–641. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Fang, J.; Fan, X.; Wei, W.; Sun, X. CYTOKININ OXIDASE/DEHYDROGENASE4 integrates cytokinin and auxin signaling to control rice crown root formation. Plant Physiol. 2014, 165, 1035–1046. [Google Scholar] [CrossRef] [PubMed]
- Zhu, K.M.; Xu, S.; Li, K.X.; Chen, S.; Zafar, S.; Cao, W.; Wang, Z.; Ding, L.N.; Yang, Y.H.; Li, Y.M.; et al. Transcriptome analysis of the irregular shape of shoot apical meristem in dt (dou tou) mutant of Brassica napus L. Mol. Breed. 2019, 39, 39. [Google Scholar] [CrossRef]
- Mason, M.G.; Ross, J.J.; Babst, B.A.; Wienclaw, B.N.; Beveridge, C.A. Sugar demand, not auxin, is the initial regulator of apical dominance. Proc. Natl. Acad. Sci. USA 2014, 111, 6092–6097. [Google Scholar] [CrossRef] [PubMed]
- Barbier, F.; Péron, T.; Lecerf, M.; Perez-Garcia, M.D.; Barrière, Q.; Rolčík, J.; Boutet-Mercey, S.; Citerne, S.; Lemoine, R.; Porcheron, B.; et al. Sucrose is an early modulator of the key hormonal mechanisms controlling bud outgrowth in Rosa hybrida. J. Exp. Bot. 2015, 66, 2569–2582. [Google Scholar] [CrossRef] [PubMed]
- Fichtner, F.; Barbier, F.F.; Feil, R.; Watanabe, M.; Annunziata, M.G.; Chabikwa, T.G.; Höfgen, R.; Stitt, M.; Beveridge, C.A.; Lunn, J.E. Trehalose 6-phosphate is involved in triggering axillary bud outgrowth in garden pea (Pisum sativum L.). Plant J. 2017, 92, 611–623. [Google Scholar] [CrossRef] [PubMed]
- Franziska, F.; Barbier, F.; Annunziata, M.G.; Feil, R.; Olas, J.J.; Bernd Mueller-Roeber, B.M.; Stitt, M.; Beveridge, C.A.; Lunn, J.E. Regulation of shoot branching in arabidopsis by trehalose 6-phosphate. New Phytol. 2021, 229, 2135–2151. [Google Scholar]
- Claeys, H.; Vi, S.; Xu, X.; Satoh-Nagasawa, N.; Eveland, A.; Goldshmidt, A.; Feil, R.; Beggs, G.; Sakai, H.; Brennan, R.; et al. Control of meristem determinacy by trehalose-6-phosphate phosphatases is uncoupled from enzymatic activity. Nat. Plants 2019, 5, 352. [Google Scholar] [CrossRef] [PubMed]
- Bertheloot, J.; Barbier, F.; Boudon, F.; Perez-Garcia, M.D.; Peron, T.; Citerne, S.; Dun, E.; Beveridge, C.; Godin, C.; Sakr, S. Sugar availability suppresses the auxin-induced strigolactone pathway to promote bud outgrowth. New Phytol. 2020, 225, 866–879. [Google Scholar] [CrossRef]
- Geisler, M.; Aryal, B.; Donato, M.D.; Hao, P.C. A critical view on ABC transporters and their interacting partners in auxin transport. Plant Cell Physiol. 2017, 58, 1601–1614. [Google Scholar] [CrossRef] [PubMed]
- Rongen, M.; Bennett, T.; Ticchiarelli, F.; Leyser, O. Connective auxin transport contributes to strigolactone-mediated shoot branching control independent of the transcription factor BRC1. PLoS Genet. 2019, 15, e1008023. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Wang, D.; Zhang, C.; Kong, N.; Ma, H.; Chen, Q. Comparative analysis of the PIN auxin transporter gene family in different plant species: A focus on structural and expression profiling of PINs in Solanum tuberosum. Int. J. Mol. Sci. 2019, 20, 3270. [Google Scholar] [CrossRef] [PubMed]
- Robert, H.S.; Friml, J. Auxin and other signals on the move in plants. Nat. Chem. Biol. 2009, 5, 325–332. [Google Scholar] [CrossRef]
- Noh, B.; Murphy, A.; Spalding, E.P. Multidrug resistance-like genes of Arabidopsis required for auxin transport and auxin-mediated development. Plant Cell 2001, 13, 2441–2454. [Google Scholar]
- Dai, Z.Y.; Wang, J.; Yang, X.F.; Lu, H.; Miao, X.; Shi, Z.Y. Modulation of plant architecture by the miR156f-OsSPL7-OsGH3.8 pathway in rice. J. Exp. Bot. 2018, 69, 5117–5130. [Google Scholar] [CrossRef]
- Liu, S.D.; Hu, Q.N.; Luo, S.; Li, Q.Q.; Yang, X.Y.; Wang, X.L.; Wang, S.C. Expression of wild-type PtrIAA14.1, a poplar Aux/IAA gene causes morphological changes in Arabidopsis. Front. Plant Sci. 2015, 6, 388. [Google Scholar] [CrossRef]
- Staswick, P.E.; Serban, B.; Rowe, M.; Tiryaki, I.; Maldonado, M.T.; Maldonado, M.C.; Suza, W. Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 2005, 17, 616–627. [Google Scholar] [CrossRef]
- Ni, J.; Gao, C.; Chen, M.S.; Pan, B.Z.; Ye, K.; Xu, Z.F. Gibberellin promotes shoot branching in the perennial wood plant Jatropha curcas. Plant Cell Physiol. 2015, 56, 1655–1666. [Google Scholar] [CrossRef]
- Cheng, J.; Ma, N.; Zheng, X.; Feng, J.S. Functional analysis of the gibberellin 2-oxidase gene family in peach. Front. Plant Sci. 2021, 12, 619158. [Google Scholar] [CrossRef]
- Rameau, C.; Bertheloot, J.; Leduc, N.; Andrieu, B.; Foucher, F.; Sakr, S. Multiple pathways regulate shoot branching. Front. Plant Sci. 2015, 5, 741. [Google Scholar] [CrossRef]
- Mauriat, M.; Sandberg, L.G.; Moritz, T. Proper gibberellin localization in vascular tissue is required to control auxin-dependent leaf development and bud outgrowth in hybrid aspen. Plant J. 2011, 67, 805–816. [Google Scholar] [CrossRef]
- Agharkar, M.; Lomba, P.; Altpeter, F.; Zhang, H.; Kenworthy, K.; Lange, T. Stable expression of AtGA2ox1 in a low-input turfgrass (Paspalum notatum Flugge) reduces bioactive gibberellin levels and improves turf quality under field conditions. Plant Biotechnol. J. 2007, 5, 791–801. [Google Scholar] [CrossRef]
- Schaller, G.E.; Street, I.H.; Kieber, J.J. Cytokinin and the cell cycle. Curr. Opin. Plant Biol. 2014, 21, 7–15. [Google Scholar] [CrossRef]
- Liu, J.Y.; Shi, M.J.; Wang, J.; Zhang, B.; Li, Y.S.; Wang, J.; El-Sappah, A.H.; Liang, Y. Comparative transcriptomic analysis of the development of sepal morphology in tomato (Solanum lycopersicum L.). Int. J. Mol. Sci. 2020, 21, 5914. [Google Scholar] [CrossRef] [PubMed]
- Chatfield, S.P.; Stirnberg, P.; Forde, B.G.; Leyser, O. The hormonal regulation of axillary bud growth in Arabidopsis. Plant J. 2000, 24, 159–169. [Google Scholar] [CrossRef]
- Kurakawa, T.; Ueda, N.; Maekawa, M.; Kobayashi, K.; Kojima, M.; Nagato, Y.; Hitoshi Sakakibara, H.; Kyozuka, J. Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 2007, 445, 652–655. [Google Scholar] [CrossRef]
- Rabot, A.; Henry, C.; Baaziz, K.B.; Mortreau, E.; Azri, W.; Lothier, J.; Hamama, L.; Boummaza, R.; Leduc, N.; Pelleschi-Travier, S.; et al. Insight into the role of sugars in bud burst under light in the rose. Plant Cell Physiol. 2012, 53, 1068–1082. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.B.; Wang, N.; Zhou, H.; Xu, Q.H.; Yan, G.T. Transcriptome analyses provide insights into the homeostatic regulation of axillary buds in upland cotton (G. hirsutum L.). BMC Plant Biol. 2020, 20, 228. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Pérez-Garcia, M.; Davière, J.; Barbier, F.; Ogé, L.; José Gentilhomme, J.; Voisine, L.; Péron, T.; Launay-Avon, A.; Clément, G.; et al. Axillary bud outgrowth in rose is controlled by sugar metabolic and signalling pathways. J. Exp. Bot. 2021, 72, 3044–3060. [Google Scholar] [CrossRef] [PubMed]
- Blakeslee, J.; Bandyopadhyay, A.; Lee, O.; Mravec, J.; Titapiwatanakun, B.; Sauer, M.; Makam, S.; Cheng, Y.; Bouchard, R.; Adamec, J.; et al. Interactions among PIN-FORMED and P-Glycoprotein auxin transporters in Arabidopsis. Plant Cell 2007, 19, 131–147. [Google Scholar] [CrossRef]
- Huang, S.N.; Liu, Z.Y.; Li, D.Y.; Yao, R.P.; Hou, L.; Li, X.; Feng, H. Physiological characterization and comparative transcriptome analysis of a slow-growing reduced-thylakoid mutant of Chinese cabbage (Brassica campestris ssp. pekinensis). Front. Plant Sci. 2017, 7, 3. [Google Scholar] [CrossRef]
- Blanca, J.; Esteras, C.; Ziarsolo, P.; Pérez, D.; Fernádez-Pedrosa, V.; Collado, C.; Pablos, R.R.; Ballester, A.; Roig, C.; Cañizares, J.; et al. Transcriptome sequencing for SNP discovery across Cucumis melo. BMC Genomics 2012, 13, 280. [Google Scholar] [CrossRef]
- Wang, J.J.; Zi, H.L.; Wang, R.; Liu, J.X.; Wang, H.R.; Chen, R.R.; Li, L.; Guo, H.L.; Chen, J.B.; Li, J.J.; et al. A high-quality chromosome-scale assembly of the centipedegrass [Eremochloa ophiuroides (Munro) Hack.] genome provides insights into chromosomal structural evolution and prostrate growth habit. Hortic. Res. 2021, 8, 201. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357. [Google Scholar] [CrossRef]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2MMCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Kiyoshi, M.; Tanakaa, K.; Sakaic, T.; Sugawaraa, S.; Kawaideb, H.; Natsumeb, M.; Hanadaa, A.; Yaenoa, T.; Shirasua, K.; Yaod, H.; et al. The main auxin biosynthesis pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 18512–18517. [Google Scholar]
- Cai, B.D.; Zhu, J.X.; Gao, Q.; Luo, D.; Yuan, B.F.; Feng, Y.Q. Rapid and high-throughput determination of endogenous cytokinins in Oryza sativa by bare Fe3O4 nanoparticles-based magnetic solid-phase extraction. J. Chromatogr. A 2014, 1340, 146–150. [Google Scholar] [CrossRef]
- Zheng, Y.H.; Jia, N.; Ning, T.; Li, Z.; Jiang, G. Potassium nitrate application alleviates sodium chloride stress in winter wheat cultivars differing in salt tolerance. J. Plant Physiol. 2008, 165, 1455–1465. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
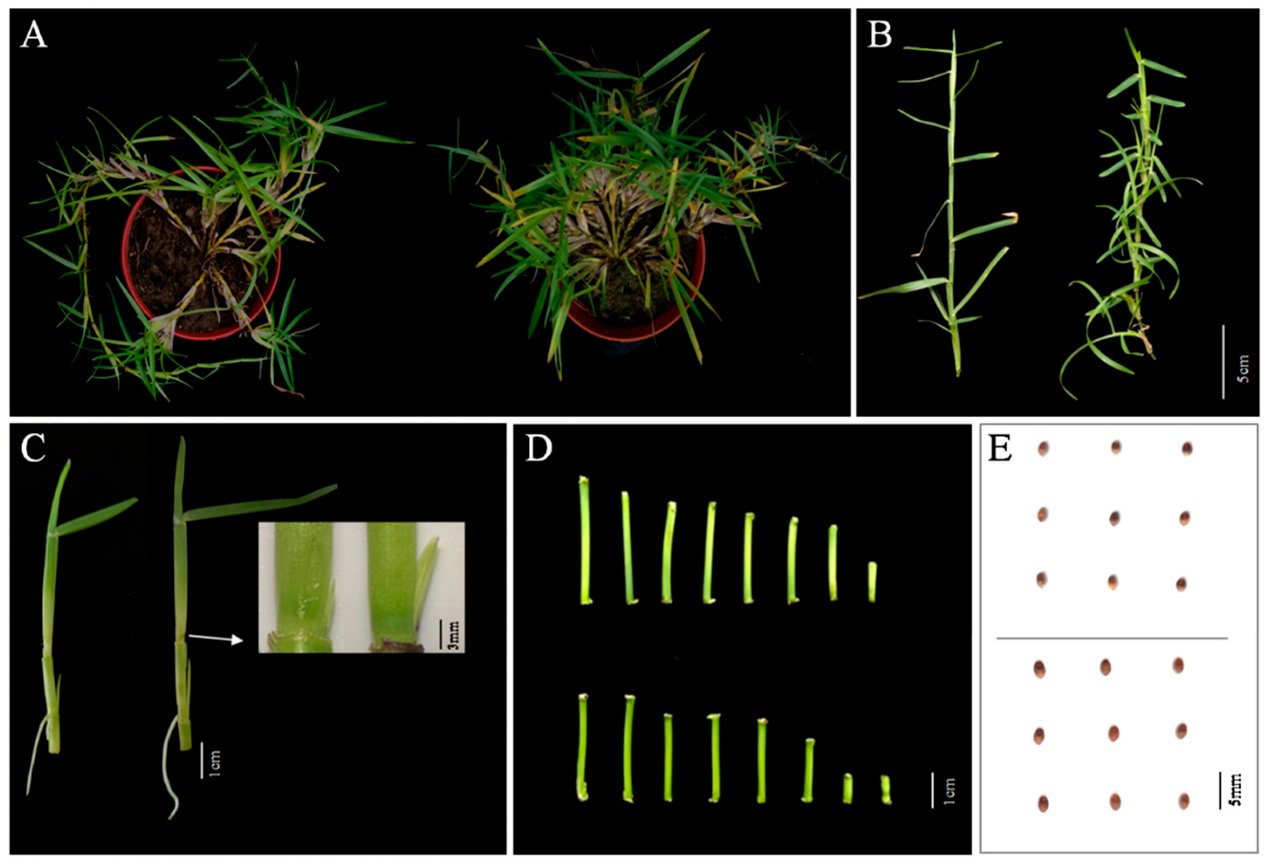

| Traits | WT | mtn1 |
|---|---|---|
| The primary tillering number | 1.33 ± 0.33 b | 2.50 ± 0.33 a |
| The second tillering number | 9.00 ± 0.58 b | 20.33 ± 1.20 a |
| Tiller bud number | 1.10 ± 0.10 a | 1.20 ± 0.13 a |
| First node of tiller bud length (mm) | 1.02 ± 0.09 b | 1.41 ± 0.13 a |
| Second node of tiller bud length (mm) | 6.30 ± 0.64 b | 11.12 ± 0.72 a |
| Third node of tiller bud length (mm) | 12.94 ± 0.68 b | 19.17 ± 0.85 a |
| Internode length (cm) | 2.68 ± 0.085 a | 1.82 ± 0.225 b |
| Internode diameter (mm) | 2.17 ± 0.055 a | 2.12 ± 0.027 a |
| Shoot dry weight (mg) | 203.70 ± 10.72 b | 362.38 ± 12.27 a |
| Root dry weight (mg) | 77.00 ± 3.68 b | 102.70 ± 6.74 a |
| Seed length (mm) | 1.93 ± 0075 b | 2.27 ± 0.023 a |
| Seed width (mm) | 1.16 ± 0.014 a | 1.19 ± 0.026 a |
| 1000-seed weight (g) | 0.99 ± 0.012 b | 1.25 ± 0.015 a |
| SNP Type | mtn1 | WT |
|---|---|---|
| Transition | ||
| C/T | 26,436 (32.89%) | 27,274 (33.16%) |
| A/G | 25,066 (31.18%) | 25,558 (31.07%) |
| Transversion | ||
| C/G | 8275 (10.30%) | 8429 (10.25%) |
| G/T | 7370 (9.17%) | 7578 (9.21%) |
| A/C | 7331 (9.12%) | 7400 (9.00%) |
| A/T | 5905 (7.35%) | 6007 (7.30%) |
| Total | 80,384 | 82,247 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, L.; Xie, C.; Zong, J.; Guo, H.; Li, D.; Liu, J. Physiological and Comparative Transcriptome Analyses of the High-Tillering Mutant mtn1 Reveal Regulatory Mechanisms in the Tillering of Centipedegrass (Eremochloa ophiuroides (Munro) Hack.). Int. J. Mol. Sci. 2022, 23, 11580. https://doi.org/10.3390/ijms231911580
Li L, Xie C, Zong J, Guo H, Li D, Liu J. Physiological and Comparative Transcriptome Analyses of the High-Tillering Mutant mtn1 Reveal Regulatory Mechanisms in the Tillering of Centipedegrass (Eremochloa ophiuroides (Munro) Hack.). International Journal of Molecular Sciences. 2022; 23(19):11580. https://doi.org/10.3390/ijms231911580
Chicago/Turabian StyleLi, Ling, Chenming Xie, Junqin Zong, Hailin Guo, Dandan Li, and Jianxiu Liu. 2022. "Physiological and Comparative Transcriptome Analyses of the High-Tillering Mutant mtn1 Reveal Regulatory Mechanisms in the Tillering of Centipedegrass (Eremochloa ophiuroides (Munro) Hack.)" International Journal of Molecular Sciences 23, no. 19: 11580. https://doi.org/10.3390/ijms231911580
APA StyleLi, L., Xie, C., Zong, J., Guo, H., Li, D., & Liu, J. (2022). Physiological and Comparative Transcriptome Analyses of the High-Tillering Mutant mtn1 Reveal Regulatory Mechanisms in the Tillering of Centipedegrass (Eremochloa ophiuroides (Munro) Hack.). International Journal of Molecular Sciences, 23(19), 11580. https://doi.org/10.3390/ijms231911580





