Down-Regulation of Lipid Metabolism in the Hepatopancreas of Shrimp Litopenaeus vannamei upon Light and Heavy Infection of Enterocytozoon hepatopenaei: A Comparative Proteomic Study
Abstract
1. Introduction
2. Results
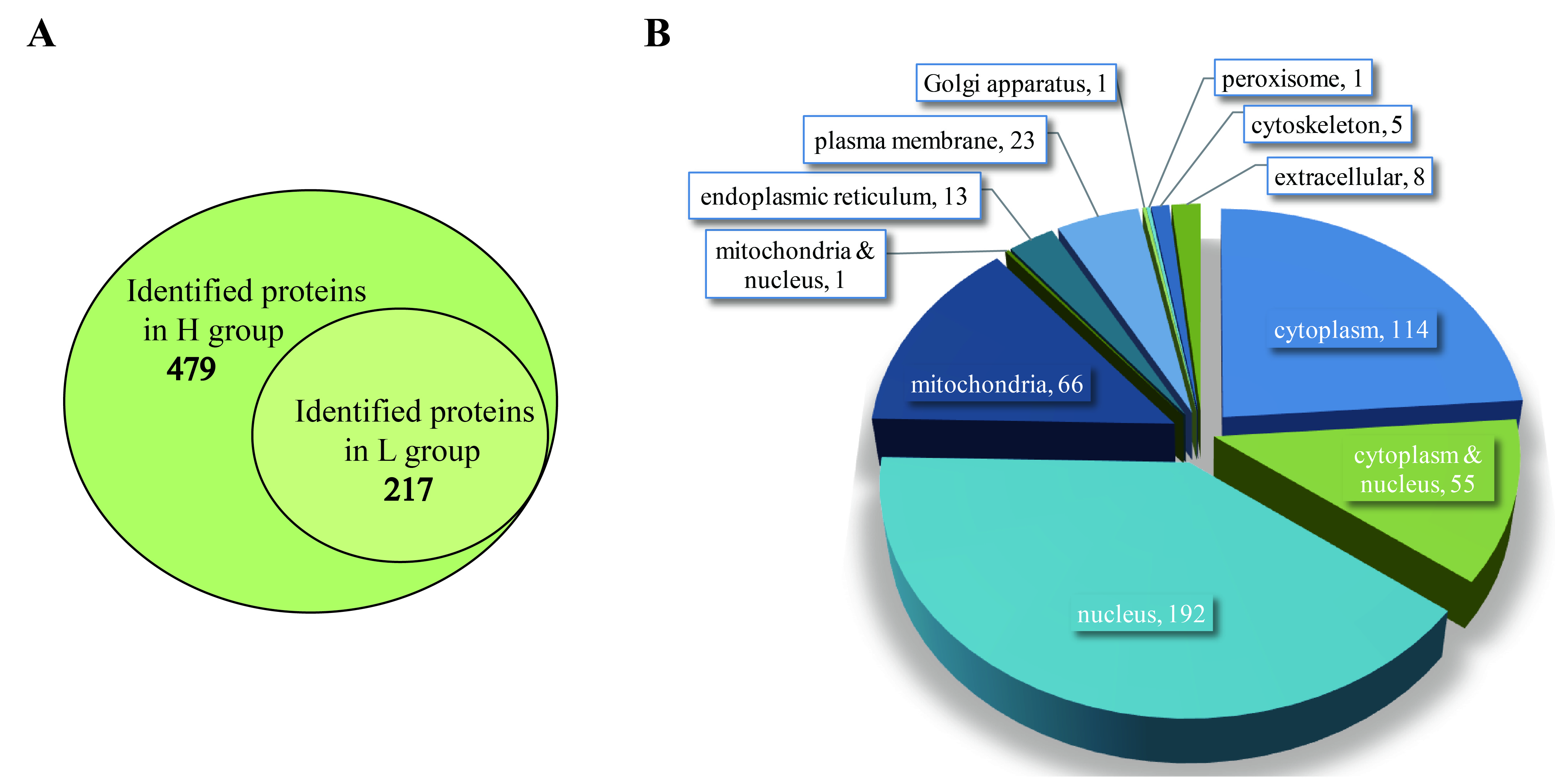
2.1. Basic Information of Samples
2.2. EHP-Infected Individuals Exhibited a Significant Down-Regulation in Lipid and Amino Acid Metabolism
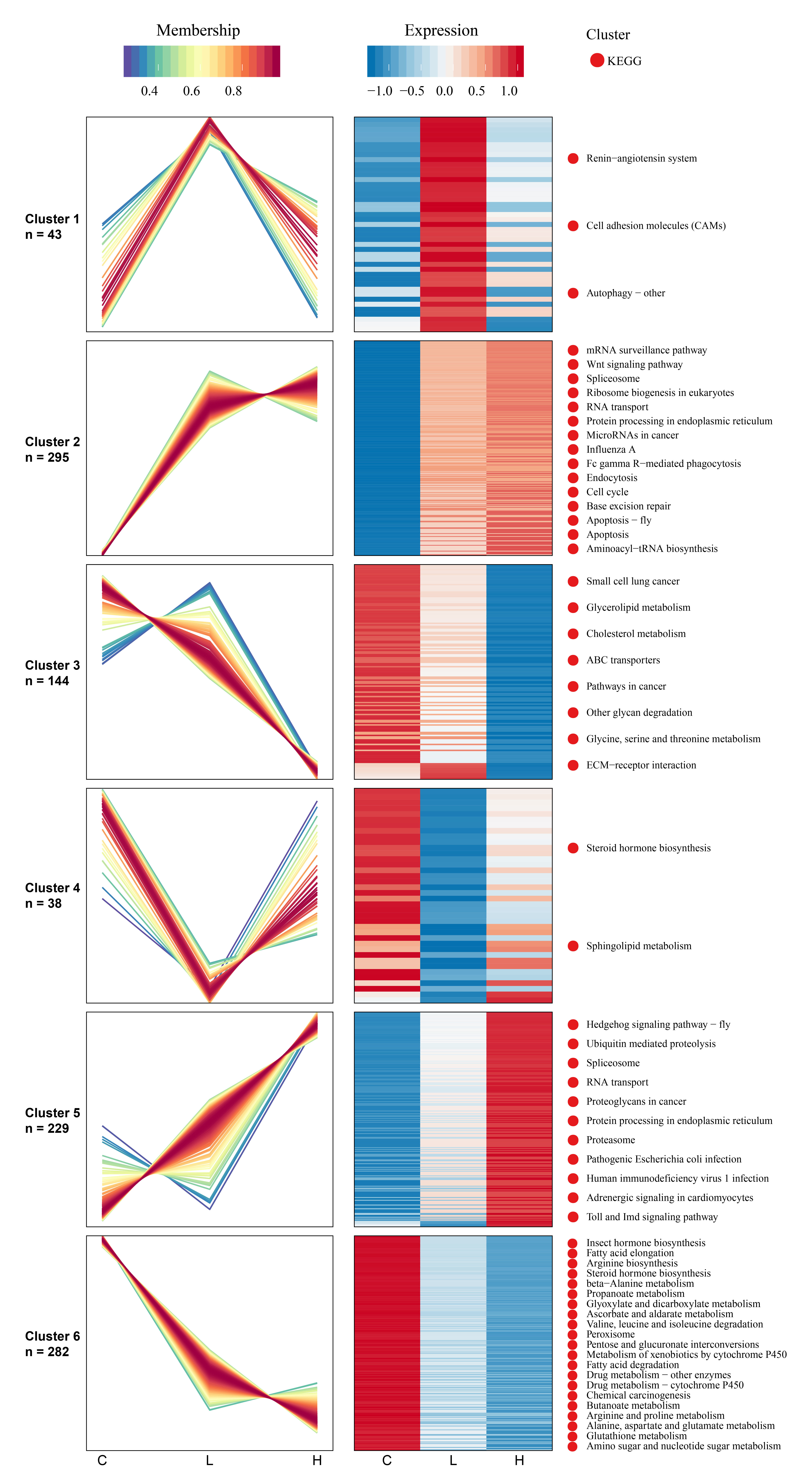
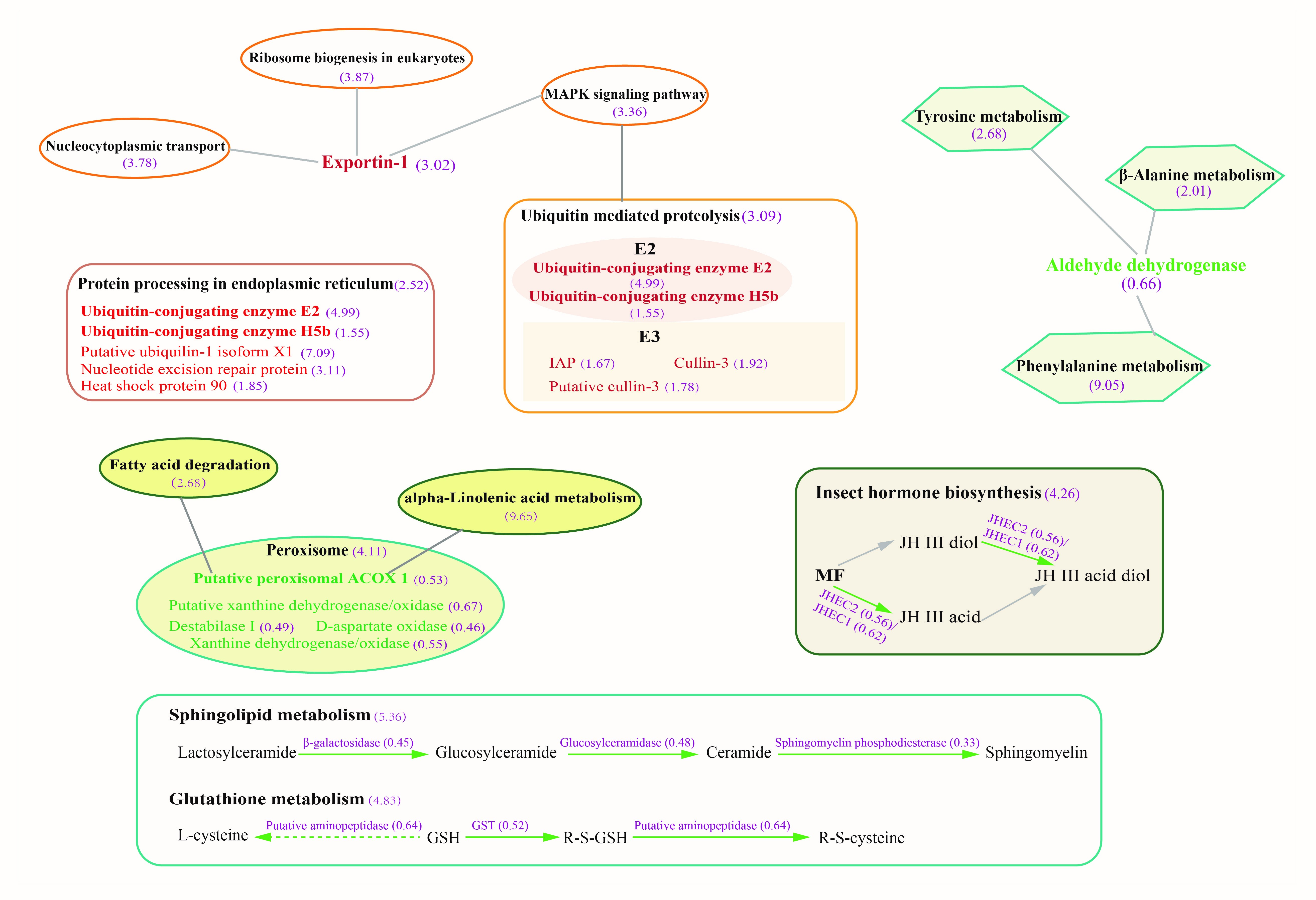
2.3. The Host Response Varies with the Severity of EHP Infection
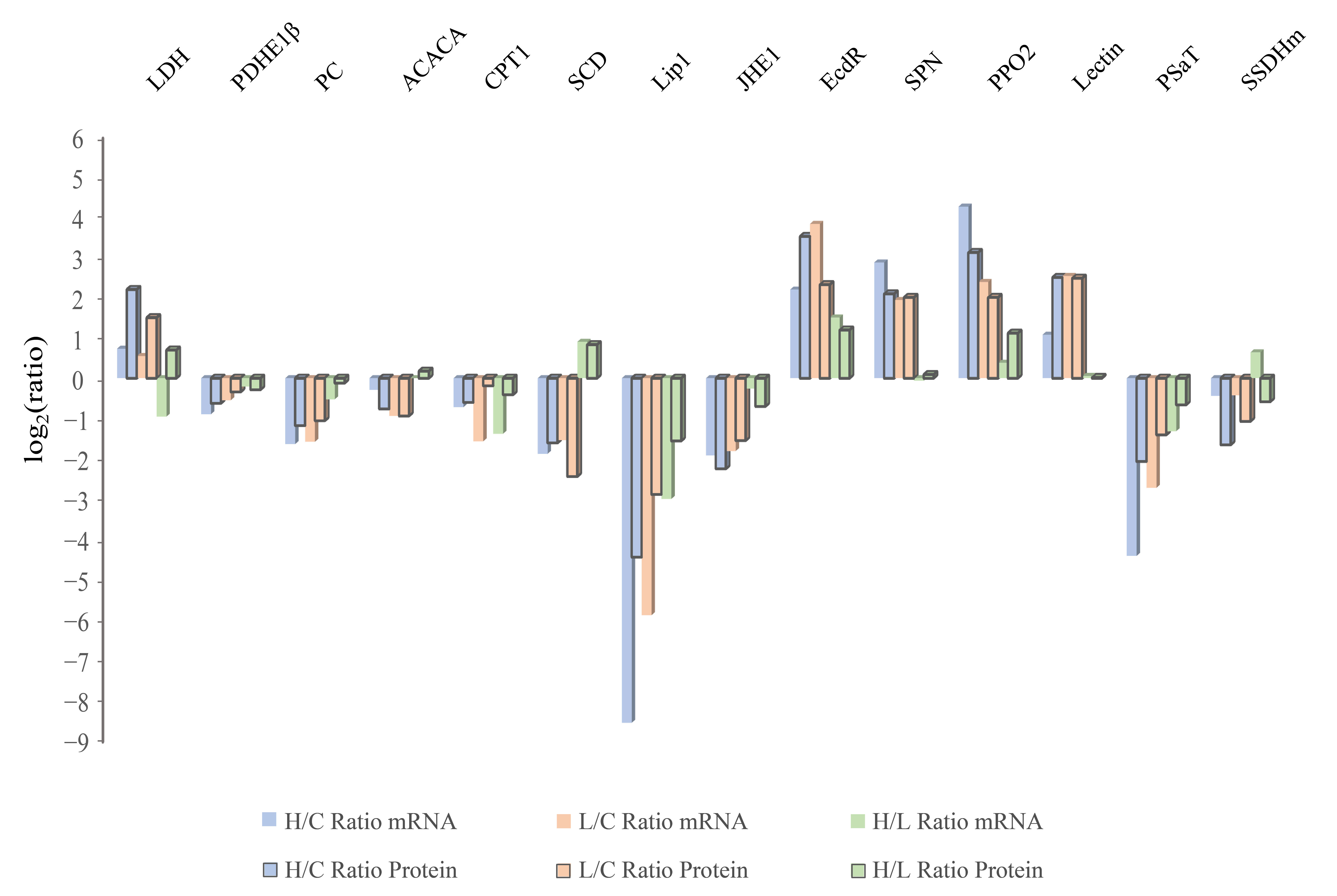
2.4. Infected Groups Showed a Coincidental Transcription Level Change Compared with those in the Proteome
2.5. Pathogen Proteins That Participate in Pathogenesis were Identified in EHP-Infected Hepatopancreas
3. Discussion
3.1. EHP Infection Resulted in Unbalance of Lipid Absorption, Metabolism, and Mediated Cell Signaling Process
3.2. Down-Regulation of Energy Metabolism May Result from Long-Term Parasitism of EHP
3.3. The Relevance of EHP Infection to Shrimp Growth
4. Materials and Methods
4.1. Sampling and EHP Detection
4.2. Protein Extraction and Enzymolysis
4.3. Liquid Chromatography-Mass Spectrometry (LC-MS/MS) Analyses
4.4. Spectrum Data Analysis
4.5. Protein Annotation Methods
4.6. Function Enrichment and the Cluster of Differentially Expressed Proteins (DEPs)
4.7. Mfuzz Analysis
4.8. RT-qPCR Verifying the mRNA Level of DEPs
4.9. Transporter and Secreted Protein Prediction
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chaijarasphong, T.; Munkongwongsiri, N.; Stentiford, G.D.; Aldama-Cano, D.J.; Thansa, K.; Flegel, T.W.; Sritunyalucksana, K.; Itsathitphaisarn, O. The shrimp microsporidian Enterocytozoon hepatopenaei (EHP): Biology, pathology, diagnostics and control. J. Invertebr. Pathol. 2020, 186, 107458. [Google Scholar] [CrossRef]
- Tourtip, S.; Wongtripop, S.; Stentiford, G.D.; Bateman, K.S.; Sriurairatana, S.; Chavadej, J.; Sritunyalucksana, K.; Withyachumnarnkul, B. Enterocytozoon hepatopenaei sp. nov. (Microsporida: Enterocytozoonidae), a parasite of the black tiger shrimp Penaeus monodon (Decapoda: Penaeidae): Fine structure and phylogenetic relationships. J. Invertebr. Pathol. 2009, 102, 21–29. [Google Scholar] [CrossRef]
- Caro, L.F.A.; Mai, H.N.; Pichardo, O.; Cruz-Flores, R.; Hanggono, B.; Dhar, A.K. Evidences supporting Enterocytozoon hepatopenaei association with white feces syndrome in farmed Penaeus vannamei in Venezuela and Indonesia. Dis. Aquat. Org. 2020, 141, 71–78. [Google Scholar] [CrossRef]
- Chayaburakul, K.; Nash, G.; Pratanpipat, P.; Sriruairatana, S.; Withyachumnarnkul., B. Multiple pathogens found in growth-retarded black tiger shrimp Penaeus monodon cultivated in Thailand. Dis. Aquat. Org. 2004, 60, 89–96. [Google Scholar] [CrossRef]
- Thamizhvanan, S.; Sivakumar, S.; Kumar, S.S.; Kumar, D.V.; Suryakodi, S.; Balaji, K.; Rajkumar, T.; Vimal, S.; Majeed, S.A.; Taju, G.; et al. Multiple infections caused by white spot syndrome virus and Enterocytozoon hepatopenaei in pond-reared Penaeus vannamei in India and multiplex PCR for their simultaneous detection. J. Fish. Dis. 2019, 42, 447–454. [Google Scholar] [CrossRef]
- Lee, S.C.; Corradi, N.; Byrnes III, E.J.; Torres-Martinez, S. Microsporidia evolved from ancestral sexual fungi. Curr. Biol. 2009, 18, 1675–1679. [Google Scholar] [CrossRef] [PubMed]
- Stentiford, G.D.; Bass, D.; Williams, B.A.P. Ultimate opportunists—The emergent Enterocytozoon group Microsporidia. PLOS Pathog. 2019, 15, e1007668. [Google Scholar] [CrossRef]
- Vavra, J.; Lukes, J. Microsporidia and ‘the art of living together’. Adv. Parasitol. 2013, 82, 253–319. [Google Scholar] [CrossRef] [PubMed]
- Stentiford, G.D.; Becnel, J.J.; Weiss, L.M.; Keeling, P.J.; Didier, E.S.; Williams, B.A.P.; Bjornson, S.; Kent, M.L.; Freeman, M.A.; Brown, M.J.F.; et al. Microsporidia: Emergent Pathogens in the Global Food Chain. Trends Parasitol. 2016, 32, 336–348. [Google Scholar] [CrossRef]
- Dean, P.; Hirt, R.P.; Embley, T.M. Microsporidia: Why Make Nucleotides if You can steal them. PLOS Pathog. 2016, 12, e1005870. [Google Scholar] [CrossRef] [PubMed]
- Peyretaillade, E.; Alaoui, H.E.; Diogon, M.; Polonais, V. Extreme reduction and compaction of microsporidian genomes. Res. Microbiol. 2011, 162, 598–606. [Google Scholar] [CrossRef] [PubMed]
- Nakjang, S.; Williams, T.A.; Heinz, E.; Watson, A.K. Reduction and expansion in microsporidian genome evolution: New insights from comparative genomics. Genome. Biol. Evol. 2013, 5, 2285–2303. [Google Scholar] [CrossRef]
- Wiredu Boakye, D.; Jaroenlak, P.; Prachumwat, A.; Williams, T.A.; Bateman, K.S.; Itsathitphaisarn, O.; Sritunyalucksana, K.; Paszkiewicz, K.H.; Moore, K.A.; Stentiford, G.D.; et al. Decay of the glycolytic pathway and adaptation to intranuclear parasitism within Enterocytozoonidae microsporidia. Environ. Microbiol. 2017, 19, 2077–2089. [Google Scholar] [CrossRef]
- Yuan, J.; Zhang, X.; Li, F.; Xiang, J. Genome Sequencing and Assembly Strategies and a Comparative Analysis of the Genomic Characteristics in Penaeid Shrimp Species. Front. Genet. 2021, 12, 658619. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Yuan, J.; Sun, Y.; Li, S. Penaeid shrimp genome provides insights into benthic adaptation and frequent molting. Nat. Commun. 2019, 10, 356. [Google Scholar] [CrossRef] [PubMed]
- Thitamadee, S.; Prachumwat, A.; Srisala, J.; Jaroenlak, P. Review of current disease threats for cultivated penaeid shrimp in Asia. Aquaculture 2016, 452, 69–87. [Google Scholar] [CrossRef]
- Salachan, P.V.; Jaroenlak, P.; Thitamadee, S.; Itsathitphaisarn, O.; Sritunyalucksana, K. Laboratory cohabitation challenge model for shrimp hepatopancreatic microsporidiosis (HPM) caused by Enterocytozoon hepatopenaei (EHP). BMC Vet. Res. 2017, 13, 9. [Google Scholar] [CrossRef]
- Weiss, L.M.; Becnel, J.J. Microsporidia: Pathogens of Opportunity; WILEY Blackwell: Oxford, UK, 2014. [Google Scholar]
- Al-Mohanna, S.Y.; Nott, J.A.; Lane, D.J.W. Mitotic E- and Secretory F-Cells in the Hepatopancreas of the Shrimp Penaeus Semisulcatus (Crustacea: Decapoda). J. Mar. Biol. Ass. UK 1985, 65, 901–910. [Google Scholar] [CrossRef]
- Vogt, G. Functional cytology of the hepatopancreas of decapod crustaceans. J. Morphol. 2019, 280, 1405–1444. [Google Scholar] [CrossRef]
- Rőszer, T. The invertebrate midintestinal gland (“hepatopancreas”) is an evolutionary forerunner in the integration of immunity and metabolism. Cell Tissue Res. 2014, 358, 685–695. [Google Scholar] [CrossRef]
- Yepiz-Plascencia, G.; Galvan, T.G.; Vargas-Albores, F.; Garcia-Banuelos, M. Synthesis of Hemolymph High-Density Lipoprotein beta-Glucan Binding Protein by Penaeus vannamei Shrimp Hepatopancreas. Mar. Biotechnol. 2000, 2, 485–492. [Google Scholar] [CrossRef]
- Ding, Z. Lipid metabolism disorders contribute to the pathogenesis of Hepatospora eriocheir in the crab Eriocheir sinensis. J. Fish Dis. 2021, 44, 305–313. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Paz, A.; García-Carreño, F.; Hernández-López, J.; Muhlia-Almazán, A.; Yepiz-Plascencia, G. Effect of short-term starvation on hepatopancreas and plasma energy reserves of the Pacific white shrimp (Litopenaeus vannamei). J. Exp. Mar. Biol. Ecol. 2007, 340, 184–193. [Google Scholar] [CrossRef]
- Hacker, C.; Howell, M.; Bhella, D.; Lucocq, J. Strategies for maximizing ATP supply in the microsporidian Encephalitozoon cuniculi: Direct binding of mitochondria to the parasitophorous vacuole and clustering of the mitochondrial porin VDAC. Cell Microbiol. 2014, 16, 565–579. [Google Scholar] [CrossRef]
- Tsaousis, A.D.; Kunji, E.R.S.; Goldberg, A.V.; Lucocq, J.M. A novel route for ATP acquisition by the remnant mitochondria of Encephalitozoon cuniculi. Nature 2008, 453, 553–556. [Google Scholar] [CrossRef] [PubMed]
- Ning, M.; Bi, J.; Sun, W.; Xie, X.; Huang, Y.; Gu, W.; Wang, W.; Qiao, Y.; Jiang, G.; Shen, H.; et al. Linolenic acid improves growth performance and immune status of Penaeus vannamei infected by Enterocytozoon hepatopenaei. Aquaculture 2021, 535, 736397. [Google Scholar] [CrossRef]
- Zuo, R.; Ai, Q.; Mai, K.; Xu, W. Effects of conjugated linoleic acid on growth, non-specific immunity, antioxidant capacity, lipid deposition and related gene expression in juvenile large yellow croaker (Larmichthys crocea) fed soyabean oil-based diets. Br. J. Nutr. 2013, 110, 1220–1232. [Google Scholar] [CrossRef]
- Ramakrishnan, S.; Serricchio, M.; Striepen, B.; Butikofer, P. Lipid synthesis in protozoan parasites: A comparison between kinetoplastids and apicomplexans. Prog. Lipid. Res. 2013, 52, 488–512. [Google Scholar] [CrossRef] [PubMed]
- Coppens, I. Targeting lipid biosynthesis and salvage in apicomplexan parasites for improved chemotherapies. Nat. Rev. Microbiol. 2013, 11, 823–835. [Google Scholar] [CrossRef] [PubMed]
- Coppens, I. Exploitation of auxotrophies and metabolic defects in Toxoplasma as therapeutic approaches. Int. J. Parasitol. 2014, 44, 109–120. [Google Scholar] [CrossRef]
- Robibaro, B.; Hoppe, H.C.; Yang, M.; Coppens, I. Endocytosis in different lifestyles of protozoan parasitism: Role in nutrient uptake with special reference to Toxoplasma gondii. Int. J. Parasitol. 2001, 31, 1343–1353. [Google Scholar] [CrossRef]
- Miao, Q.; Ndao, M. Trypanosoma cruzi infection and host lipid metabolism. Mediat. Inflamm. 2014, 2014, 902038. [Google Scholar] [CrossRef]
- Rodriguez, N.E.; Gaur, U.; Wilson, M.E. Role of caveolae in Leishmania chagasi phagocytosis and intracellular survival in macrophages. Cell Microbiol. 2006, 8, 1106–1120. [Google Scholar] [CrossRef]
- Randall, G. Lipid Droplet Metabolism during Dengue Virus Infection. Trends Microbiol. 2018, 26, 640–642. [Google Scholar] [CrossRef]
- Mazzon, M.; Mercer, J. Lipid interactions during virus entry and infection. Cell Microbiol. 2014, 16, 1493–1502. [Google Scholar] [CrossRef] [PubMed]
- Sviridov, D.; Bukrinsky, M. Interaction of pathogens with host cholesterol metabolism. Curr. Opin. Lipidol. 2014, 25, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Cabezas-Cruz, A.; Alberdi, P.; James, J.V.; Villar, M.; Fuente, J.D.L. Anaplasma phagocytophilum Infection Subverts Carbohydrate Metabolic Pathways in the Tick Vector, Ixodes scapularis. Front. Cell. Infect. Microbiol. 2017, 7, 23. [Google Scholar] [CrossRef]
- Voronin, D.; Schnall, E.; Grote, A.; Jawahar, S.; Ali, W.; Thomas, R.U.; Ghedin, E.; Lustigman, S. Pyruvate produced by Brugia spp. via glycolysis is essential for maintaining the mutualistic association between the parasite and its endosymbiont, Wolbachia. PLoS Pathog. 2019, 15, e1008085. [Google Scholar] [CrossRef]
- Voronin, D.; Bachu, S.; Shlossman, M.; Unnasch, T.R.; Ghedin, E.; Lustigman, S. Glucose and Glycogen Metabolism in Brugia malayi Is Associated with Wolbachia Symbiont Fitness. PLoS ONE 2016, 11, e0153812. [Google Scholar] [CrossRef]
- Krystal, A.F.; Erica, L.S.; Camarda, R.; Lagunoff, M. Dengue virus induces and requires glycolysis for Optimal Replication. J. Virol. 2015, 89, 2358–2366. [Google Scholar] [CrossRef]
- Chi, P.; Huang, W.; Chiu, H.; Li, J.; Brent, L.N.; Liu, H.-J. Avian reovirus σA-modulated suppression of lactat dehydrogenase and upregulation of glutaminolysis and the mTOC1/eIF4E/HIF-1α pathway to enhance glycolysis and the TCA cycle for virus replication. Cell Microbiol. 2018, 20, e12946. [Google Scholar] [CrossRef] [PubMed]
- Christina, A.C.; Christopher, A.D.; Malina, A.B.; Goldberg, J.; Amy, T.M.; James, J.B.; Elizabeth, S.D.; Fan, L.; David, I.H.; Joshua, Z.L.; et al. Microsporidian genome analysis reveals evolutionary strategies for obligate intracellular growth. Genome Res. 2012, 22, 2478–2488. [Google Scholar] [CrossRef]
- Méténier, G.; Vivarès, C.P. Molecular characteristics and physiology of microsporidia. Microbes Infect. 2001, 3, 407–415. [Google Scholar] [CrossRef]
- Duan, Y.; Chen, H.; Wang, J.; Zeng, S.; Wang, Y.; Mo, Z.; Dan, X.; Li, Y. Response signatures of Litopenaeus vannamei to natural Enterocytozoon hepatopenaei infection revealed by the integration of the microbiome and transcriptome. Aquaculture 2021, 542, 736885. [Google Scholar] [CrossRef]
- Ning, M.; Wei, P.; Shen, H.; Wan, X.; Jin, M.; Li, X.; Shi, H.; Qiao, Y.; Jiang, G.; Gu, W.; et al. Proteomic and metabolomic responses in hepatopancreas of whiteleg shrimp Litopenaeus vannamei infected by microsporidian Enterocytozoon hepatopenae. Fish Shellfish Immunol. 2019, 87, 534–545. [Google Scholar] [CrossRef]
- Yang, L.G.; Wang, Y.; Wang, Y.; Fang, W.H.; Feng, G.P.; Ying, N.; Zhou, J.Y.; Li, X.C. Transcriptome analysis of pacific white shrimp (Penaeus vannamei) intestines and hepatopancreas in response to Enterocytozoon hepatopenaei (EHP) infection. J. Invertebr. Pathol. 2021, 186, 107665. [Google Scholar] [CrossRef]
- Cuzon, G.; Rosas, C.; Gaxiola, G.; Taboada, G.; Van Wormhoudt, A. Utilization of carbohydrates by shrimp. Av. Nutr. Acuícola V. Mem. V. Simp. Int. Nutr. Acuícola 2000, 328–339. [Google Scholar]
- Chen, M.M.; Zhong, L.; Zhao, C.S.; Wang, F.C.; Ji, W.J.; Zhang, B.; Liu, S.Y.; Liu, Y.Q.; Li, X.S. Characterization of an Ecdysteroid-Regulated 16 kDa Protein Gene in Chinese Oak Silkworm, Antheraea pernyi (Lepidoptera: Saturniidae). J. Insect. Sci. 2020, 20, 4. [Google Scholar] [CrossRef]
- Chen, J.; Liao, G.-l.; Wu, Y.-j.; Zhang, Q.; Xiao-juan, Y.; Xiao-dong, F.; Meng-xian, L.; Guo-qing, P.; Ze-yang, Z. Three Methods for Light Microscopic Detection of Enterocytozoon hepatopanaei. J. Southwest Univ. 2021, 43, 17–23. [Google Scholar]
- Chen, T.; Chen, X.; Zhang, S.; Zhu, J.; Tang, B.; Wang, A.; Dong, L.; Zhang, Z.; Yu, C.; Sun, Y.; et al. The Genome Sequence Archive Family: Toward Explosive Data Growth and Diverse Data Types. Genom. Proteom. Bioinform. 2021, 19, 578–583. [Google Scholar] [CrossRef]
- CNCB-NGDC Members and Partners. Database Resources of the National Genomics Data Center, China National Center for Bioinformation in 2022. Nucleic Acids Res. 2022, 50, D27–D38. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, Y.; Chen, J.; Liao, G.; Hu, M.; Zhang, Q.; Meng, X.; Li, T.; Long, M.; Fan, X.; Yu, Q.; et al. Down-Regulation of Lipid Metabolism in the Hepatopancreas of Shrimp Litopenaeus vannamei upon Light and Heavy Infection of Enterocytozoon hepatopenaei: A Comparative Proteomic Study. Int. J. Mol. Sci. 2022, 23, 11574. https://doi.org/10.3390/ijms231911574
Wu Y, Chen J, Liao G, Hu M, Zhang Q, Meng X, Li T, Long M, Fan X, Yu Q, et al. Down-Regulation of Lipid Metabolism in the Hepatopancreas of Shrimp Litopenaeus vannamei upon Light and Heavy Infection of Enterocytozoon hepatopenaei: A Comparative Proteomic Study. International Journal of Molecular Sciences. 2022; 23(19):11574. https://doi.org/10.3390/ijms231911574
Chicago/Turabian StyleWu, Yujiao, Jie Chen, Guoli Liao, Mengjiao Hu, Qing Zhang, Xianzhi Meng, Tian Li, Mengxian Long, Xiaodong Fan, Qing Yu, and et al. 2022. "Down-Regulation of Lipid Metabolism in the Hepatopancreas of Shrimp Litopenaeus vannamei upon Light and Heavy Infection of Enterocytozoon hepatopenaei: A Comparative Proteomic Study" International Journal of Molecular Sciences 23, no. 19: 11574. https://doi.org/10.3390/ijms231911574
APA StyleWu, Y., Chen, J., Liao, G., Hu, M., Zhang, Q., Meng, X., Li, T., Long, M., Fan, X., Yu, Q., Zhang, L., Pan, G., & Zhou, Z. (2022). Down-Regulation of Lipid Metabolism in the Hepatopancreas of Shrimp Litopenaeus vannamei upon Light and Heavy Infection of Enterocytozoon hepatopenaei: A Comparative Proteomic Study. International Journal of Molecular Sciences, 23(19), 11574. https://doi.org/10.3390/ijms231911574






