Genome-Wide Identification and Salt Stress Response Analysis of the bZIP Transcription Factor Family in Sugar Beet
Abstract
1. Introduction
2. Results
2.1. Members of BvbZIP Gene Family
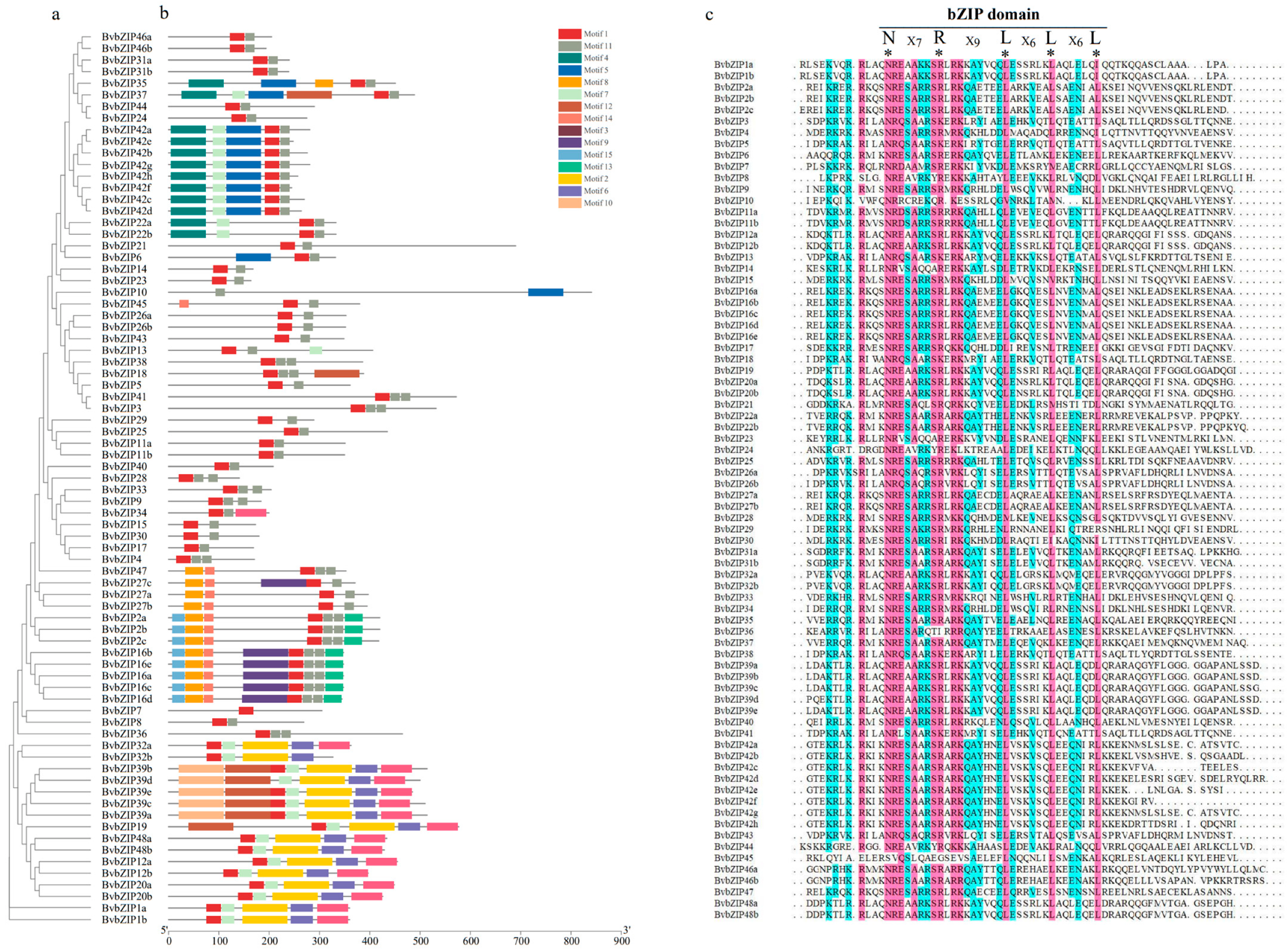
2.2. Sequence Characteristics of BvbZIP Gene Family
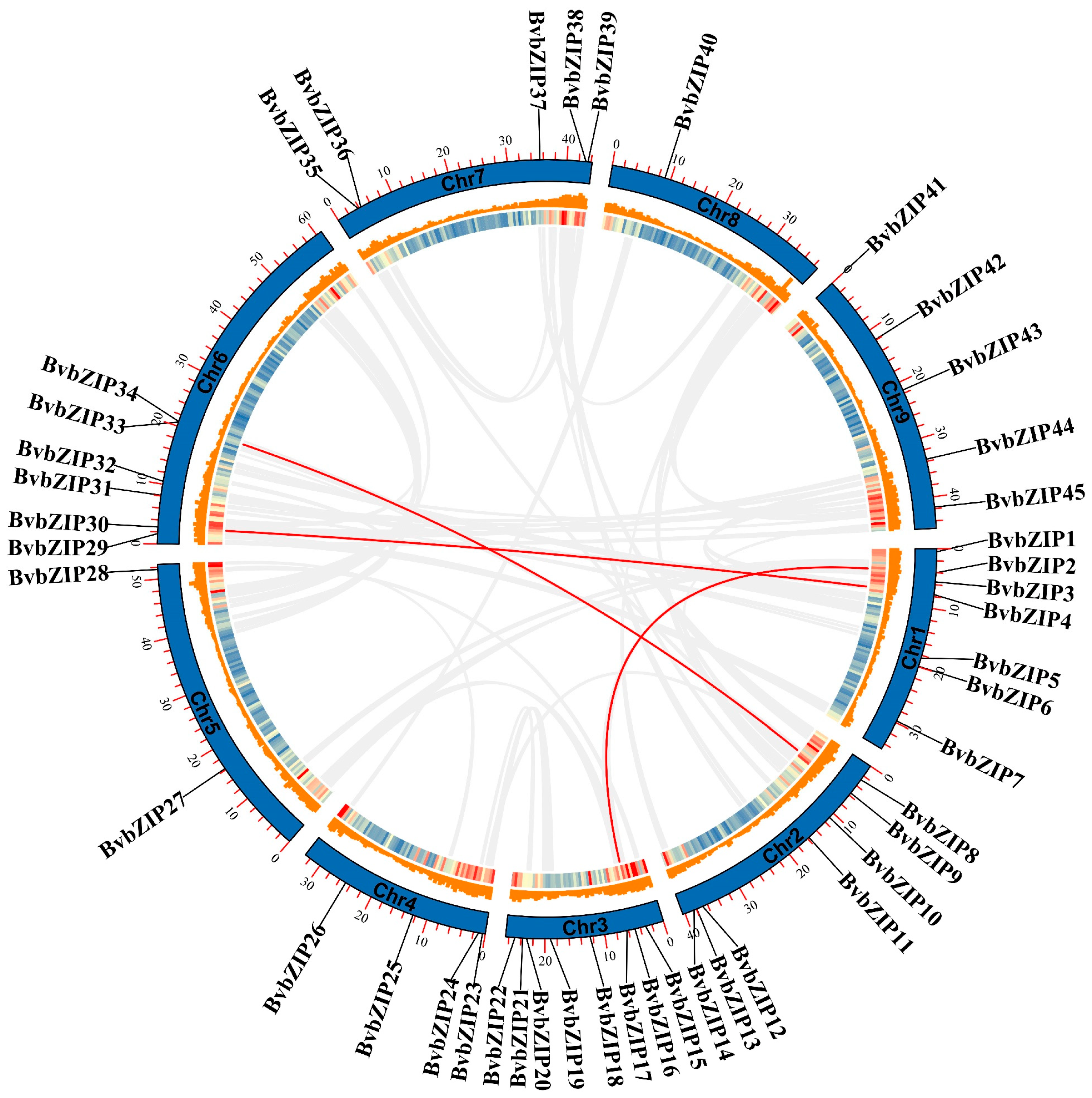
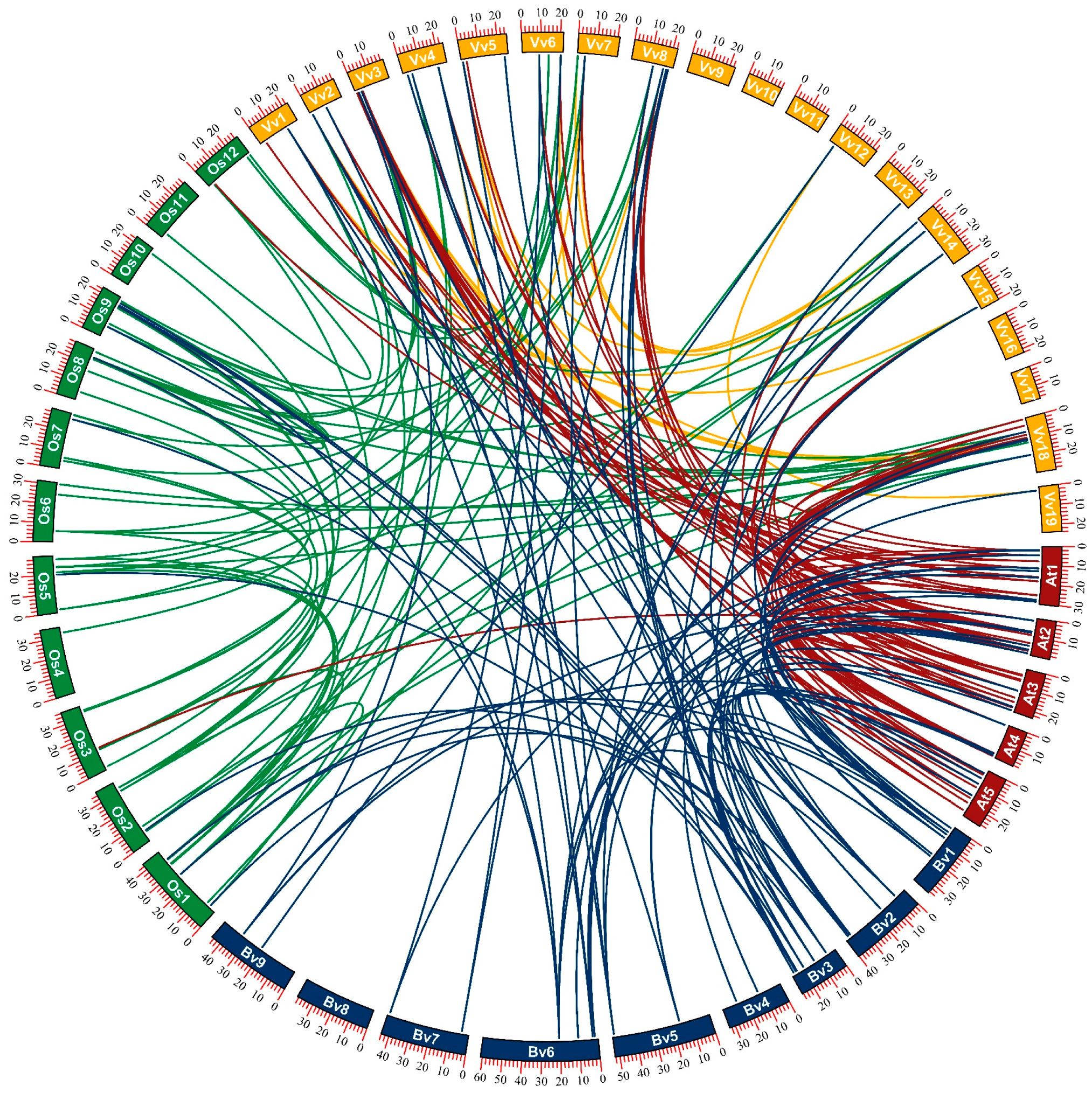
2.3. Chromosomal Localization and Collinearity Analysis of BvbZIP Genes
2.4. Phylogenetic Analysis of the bZIP Family
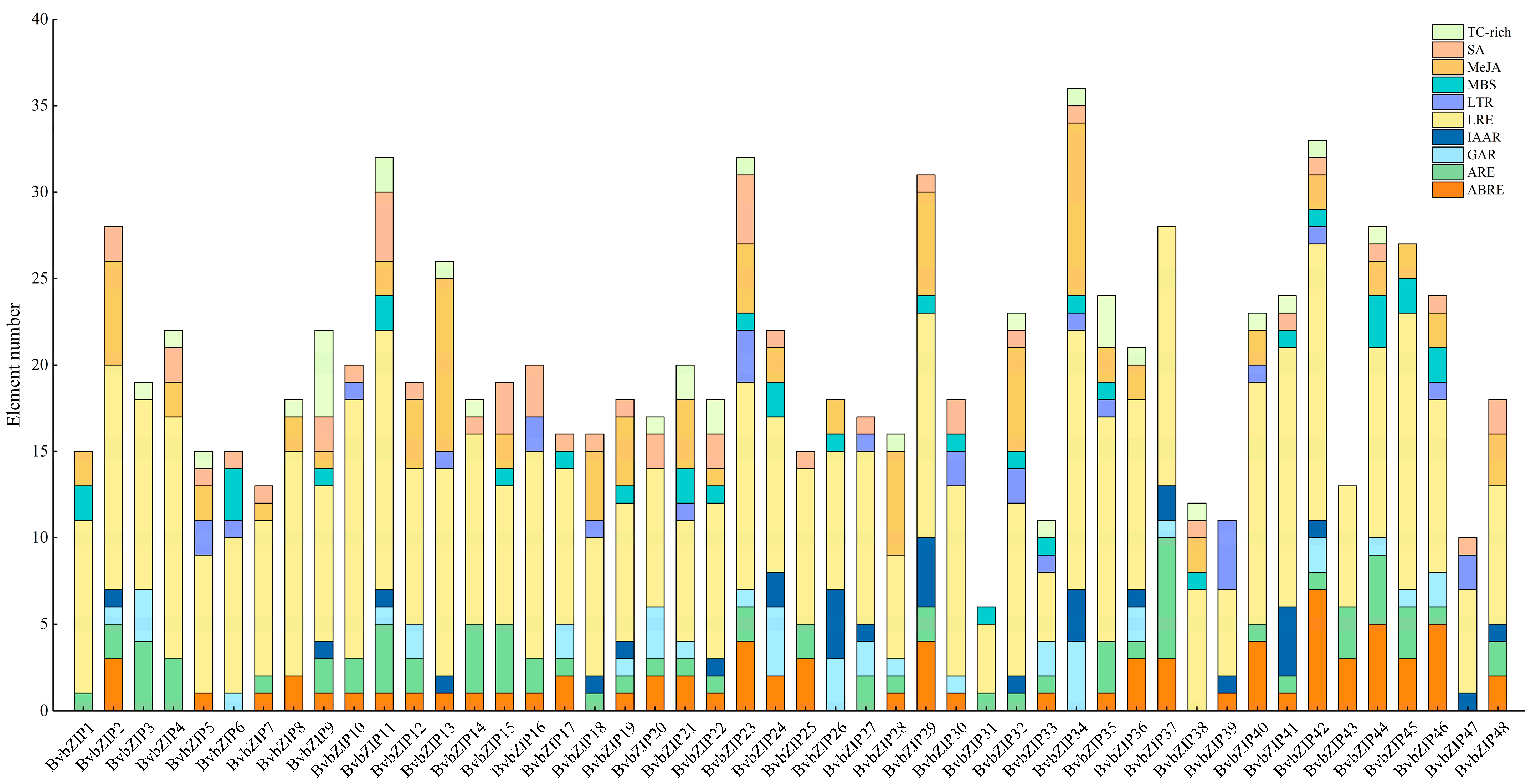
2.5. Functional Element Analysis of Promoters of BvbZIP Genes
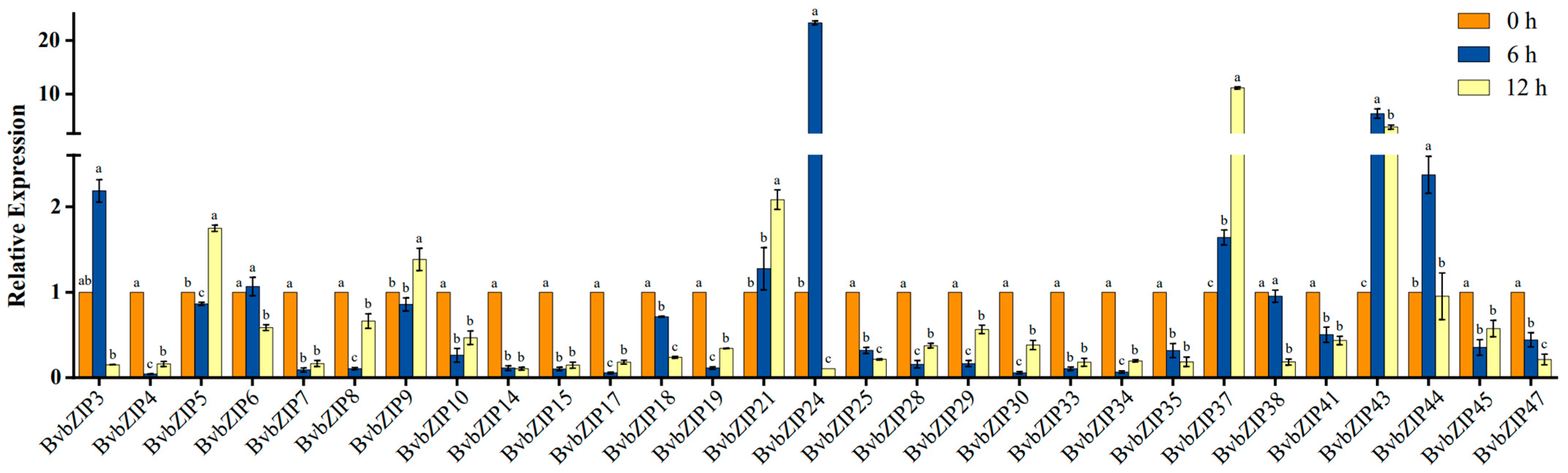
2.6. Expression Pattern Analysis of BvbZIP Genes under Salt Stress
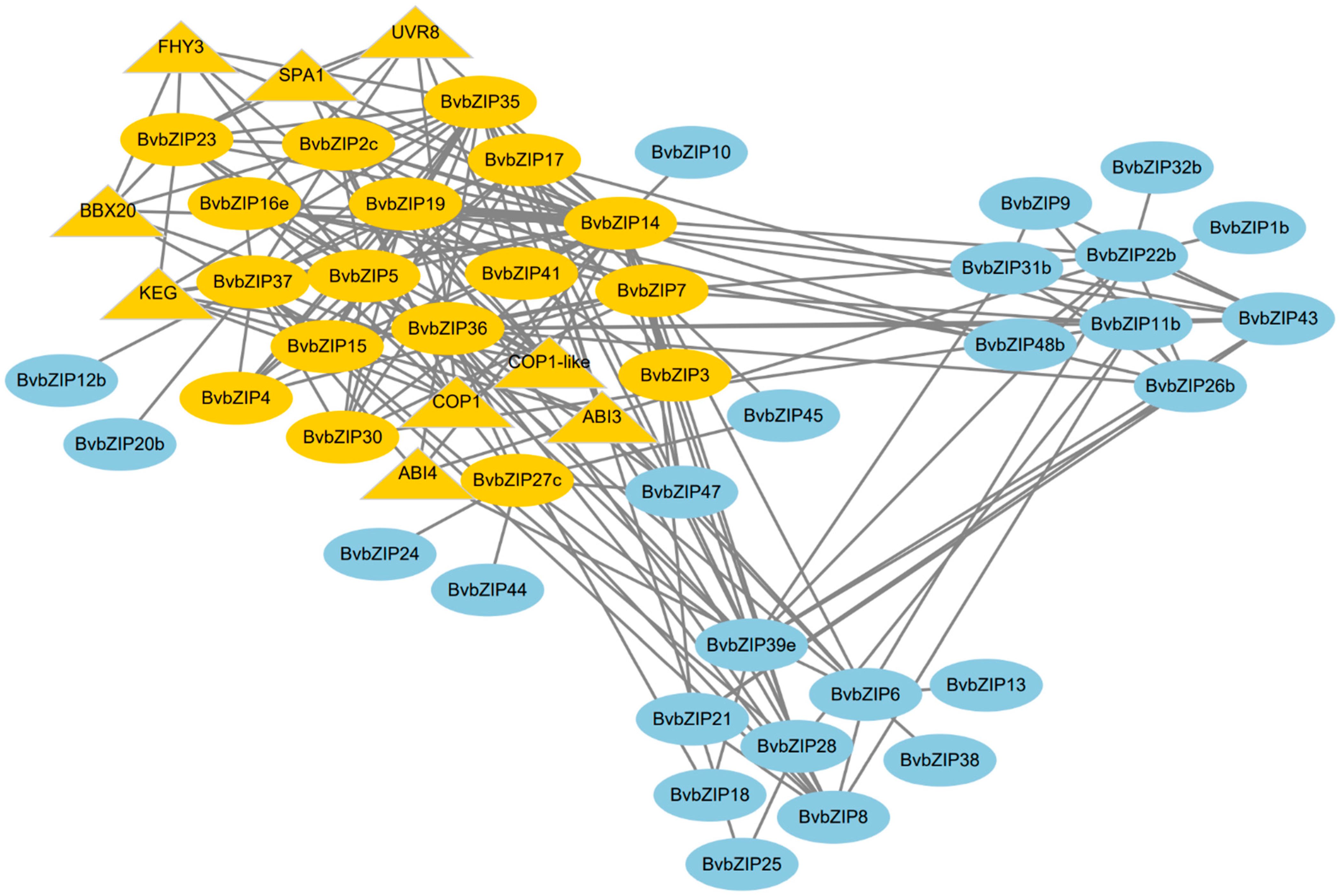
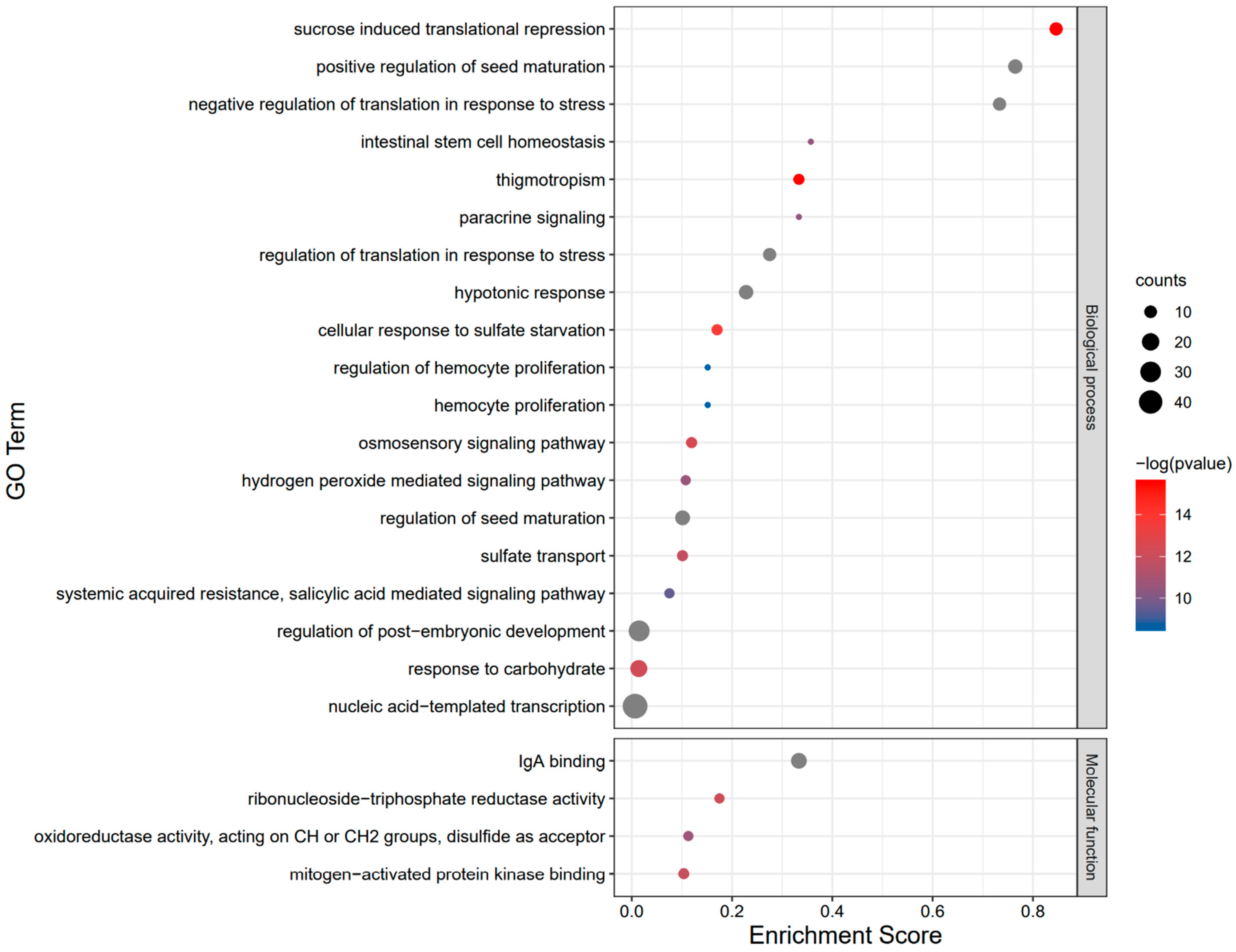
2.7. Functional Annotation and Interaction Analysis of the BvbZIP Proteins
3. Discussion
4. Materials and Methods
4.1. Plant Material Processing
4.2. Gene Family Identification
4.3. Sequence Feature Analysis
4.4. Phylogenetic Analysis
4.5. Chromosomal Localization and Collinearity Analysis
4.6. Promoter Analysis and Salt Stress Response Analysis
4.7. cDNA Acquisition and qRT-PCR Analysis
4.8. Protein Interaction Analysis and Gene Functional Annotation
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhao, J.M.; Yu, L.H.; Zhou, F.; Ma, H.X.; Yang, K.Y.; Wu, G. Synthesis and characterization of activated carbon from sugar beet residue for the adsorption of hexavalent chromium in aqueous solutions. RSC Adv. 2021, 11, 8025–8032. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.X.; Sun, L.Z.; Guo, J.R.; Liu, L.L.; Han, G.L.; Wang, B.S. Exogenous boron alleviates growth inhibition by NaCl stress by reducing Cl- uptake in sugar beet (Beta vulgaris). Plant Soil 2021, 464, 423–439. [Google Scholar] [CrossRef]
- Fishman, M.L.; Chau, H.K.; Coffin, D.R.; Cooke, P.H.; Qi, P.; Yadav, M.P.; Hotchkiss, A.T. Physico-chemical characterization of a cellulosic fraction from sugar beet pulp. Cellulose 2011, 18, 787–801. [Google Scholar] [CrossRef]
- Li, D.; Yan, J.; Liu, Z.; Liu, Z. Adsorption kinetic studies for removal of methylene blue using activated carbon prepared from sugar beet pulp. Int. J. Environ. Sci. Technol. 2016, 13, 1815–1822. [Google Scholar] [CrossRef]
- Samadi, M.T.; Rahman, A.R.; Zarrabi, M.; Shahabi, E.; Sameei, F. Adsorption of chromium (VI) from aqueous solution by sugar beet bagasse-based activated charcoal. Environ. Technol. 2009, 30, 1023–1029. [Google Scholar] [CrossRef]
- Demiral, H.; Gunduzoglu, G. Removal of nitrate from aqueous solutions by activated carbon prepared from sugar beet bagasse. Bioresour. Technol. 2010, 101, 1675–1680. [Google Scholar] [CrossRef]
- Loescher, W.; Chan, Z.L.; Grumet, R. Options for developing salt-tolerant crops. Hortscience 2011, 46, 1085–1092. [Google Scholar] [CrossRef]
- Li, X.; Wang, A.C.; Wan, W.J.; Luo, X.S.; Zheng, L.X.; He, G.W.; Huang, D.Q.; Chen, W.L.; Huang, Q.Y. High salinity inhibits soil bacterial community mediating nitrogen cycling. Appl. Environ. Microbiol. 2021, 87, e0136621. [Google Scholar] [CrossRef]
- Van Zelm, E.; Zhang, Y.; Testerink, C. Salt tolerance mechanisms of plants. Annu. Rev. Plant. Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef]
- Lv, X.Y.; Chen, S.X.; Wang, Y.G. Advances in understanding the physiological and molecular responses of sugar beet to salt stress. Front. Plant Sci. 2019, 10, 1431. [Google Scholar] [CrossRef]
- Ben Hsouna, A.; Ghneim-Herrera, T.; Ben Romdhane, W.; Dabbous, A.; Ben Saad, R.; Brini, F.; Abdelly, C.; Ben Hamed, K. Early effects of salt stress on the physiological and oxidative status of the halophyte Lobularia maritima. Funct. Plant Biol. 2020, 47, 912–924. [Google Scholar] [CrossRef]
- Alotaibi, F.; Bamagoos, A.A.; Ismaeil, F.M.; Zhang, W.; Abou-Elwafa, S.F. Application of beet sugar byproducts improves sugar beet biofortification in saline soils and reduces sugar losses in beet sugar processing. Environ. Sci. Pollut. Res. Int. 2021, 28, 30303–30311. [Google Scholar] [CrossRef]
- Aljabri, M.; Alharbi, S.; Al-Qthanin, R.N.; Ismaeil, F.M.; Chen, J.; Abou-Elwafa, S.F. Recycling of beet sugar byproducts and wastes enhances sugar beet productivity and salt redistribution in saline soils. Environ. Sci. Pollut. Res. Int. 2021, 28, 45745–45755. [Google Scholar] [CrossRef]
- Liu, C.; Wu, Y.; Wang, X. bZIP transcription factor OsbZIP52/RISBZ5: A potential negative regulator of cold and drought stress response in rice. Planta 2012, 235, 1157–1169. [Google Scholar] [CrossRef]
- Nakano, T.; Suzuki, K.; Fujimura, T.; Shinshi, H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 2006, 140, 411–432. [Google Scholar] [CrossRef]
- Hsieh, W.P.; Hsieh, H.L.; Wu, S.H. Arabidopsis bZIP16 transcription factor integrates light and hormone signaling pathways to regulate early seedling development. Plant Cell 2012, 24, 3997–4011. [Google Scholar] [CrossRef][Green Version]
- Cheng, Z.; Zhao, X.Y.; Shao, X.; Wang, F.; Zhou, C.; Liu, Y.-G.; Zhang, Y.; Zhang, X. Abscisic acid regulates early seed development in arabidopsis by ABI5-Mediated transcription of SHORT HYPOCOTYL Under BLUE1. Plant Cell 2014, 26, 1053–1068. [Google Scholar] [CrossRef]
- Alves, M.; Dadalto, S.; Gonçalves, A.; Souza, G.; Barros, V.; Fietto, L. Plant bZIP transcription factors responsive to pathogens: A review. Int. J. Mol. Sci. 2013, 14, 7815–7828. [Google Scholar] [CrossRef]
- Wang, J.; Li, Q.; Mao, X.; Li, A.; Jing, R. Wheat transcription factor TaAREB3 participates in drought and freezing tolerances in Arabidopsis. Int. J. Biol. Sci. 2016, 12, 257–269. [Google Scholar] [CrossRef]
- Jakoby, M.; Weisshaar, B.; Dröge-Laser, W.; Vicente-Carbajosa, J.; Tiedemann, J.; Kroj, T.; Parcy, F. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002, 7, 106–111. [Google Scholar] [CrossRef]
- Zhang, Y.P.; Zhou, J.H.; Wang, L. Mini review roles of the bZIP gene family in rice. Genet. Mol. Res. 2014, 13, 3025–3036. [Google Scholar] [CrossRef]
- Riechmann, J.; Heard, J.; Martin, G.; Reuber, T.; Jiang, C.-Z.; Keddie, J.; Adam, L.; Pineda, O.J.; Ratcliffe, O.; Samaha, R.R.; et al. Arabidopsis transcription factors: Genome-wide comparative analysis among Eukaryotes. Science 2001, 290, 2105–2110. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Liu, Y.; Shi, H.; Guo, M.; Chai, M.; He, Q.; Yan, M.; Cao, D.; Zhao, L.; Cai, H.; et al. Evolutionary and expression analyses of soybean basic Leucine zipper transcription factor family. BMC Genom. 2018, 19, 159. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-H.; Jeong, J.S.; Lee, K.H.; Kim, Y.S.; Do Choi, Y.; Kim, J.-K. OsbZIP23 and OsbZIP45, members of the rice basic leucine zipper transcription factor family, are involved in drought tolerance. Plant Biotechnol. Rep. 2015, 9, 89–96. [Google Scholar] [CrossRef]
- Wei, K.F.; Chen, J.; Wang, Y.M.; Chen, Y.H.; Chen, S.X.; Lin, Y.N.; Pan, S.; Zhong, X.J.; Xie, D.X. Genome-wide analysis of bZIP-encoding genes in maize. DNA Res. 2012, 19, 463–476. [Google Scholar] [CrossRef]
- Wang, J.Z.; Zhou, J.X.; Zhang, B.L.; Vanitha, J.; Ramachandran, S.; Jiang, S.Y. Genome-wide expansion and expression divergence of the basic leucine zipper transcription factors in higher plants with an emphasis on sorghum. J. Integr. Plant Biol. 2011, 53, 212–231. [Google Scholar] [CrossRef]
- Liu, J.Y.; Chen, N.N.; Chen, F.; Cai, B.; Dal Santo, S.; Tornielli, G.B.; Pezzotti, M.; Cheng, Z.M.M. Genome-wide analysis and expression profile of the bZIP transcription factor gene family in grapevine (Vitis vinifera). BMC Genom. 2014, 15, 281. [Google Scholar] [CrossRef]
- Perez-Rodriguez, P.; Riano-Pachon, D.M.; Correa, L.G.G.; Rensing, S.A.; Kersten, B.; Mueller-Roeber, B. PInTFDB: Updated content and new features of the plant transcription factor database. Nucleic Acids Res. 2010, 38, D822–D827. [Google Scholar] [CrossRef]
- Droge-Laser, W.; Snoek, B.L.; Snel, B.; Weiste, C. The Arabidopsis bZIP transcription factor family—An update. Curr. Opin. Plant Biol. 2018, 45, 36–49. [Google Scholar] [CrossRef]
- Barbosa, E.G.G.; Leite, J.P.; Marin, S.R.R.; Marinho, J.P.; Carvalho, J.D.C.; Fuganti-Pagliarini, R.; Farias, J.R.B.; Neumaier, N.; Marcelino-Guimaraes, F.C.; de Oliveira, M.C.N.; et al. Overexpression of the ABA-dependent AREB1 transcription factor from Arabidopsis thaliana improves soybean tolerance to water deficit. Plant Mol. Biol. Rep. 2013, 31, 719–730. [Google Scholar] [CrossRef]
- Ji, C.; Mao, X.; Hao, J.; Wang, X.; Xue, J.; Cui, H.; Li, R. Analysis of bZIP transcription factor family and their expressions under salt stress in Chlamydomonas reinhardtii. Int. J. Mol. Sci. 2018, 19, 2800. [Google Scholar] [CrossRef]
- Liu, J.-X.; Srivastava, R.; Che, P.; Howell, S. Salt stress responses in Arabidopsis utilize a signal transduction pathway related to ER stress signaling. Plant J. 2007, 51, 897–909. [Google Scholar] [CrossRef]
- Tang, W.; Page, M.; Fei, Y.J.; Liu, L.C.; Xu, F.; Cai, X.D.; Yuan, L.Y.; Wu, Q.S.; Zhou, M.Q. Overexpression of AtbZIP60deltaC gene alleviates salt-induced oxidative damage in transgenic cell cultures. Plant Mol. Biol. Rep. 2012, 30, 1183–1195. [Google Scholar] [CrossRef]
- Liu, C.T.; Mao, B.G.; Ou, S.J.; Wang, W.; Liu, L.C.; Wu, Y.B.; Chu, C.C.; Wang, X.P. OsbZIP71, a bZIP transcription factor, confers salinity and drought tolerance in rice. Plant Mol. Biol. 2014, 84, 19–36. [Google Scholar] [CrossRef]
- Zhang, Z.S.; Liu, W.X.; Qi, X.; Liu, Z.P.; Xie, W.G.; Wang, Y.R. Genome-wide identification, expression profiling, and SSR marker development of the bZIP transcription factor family in Medicago truncatula. Biochem. Syst. Ecol. 2015, 61, 218–228. [Google Scholar] [CrossRef]
- Wang, Z.H.; Cheng, K.; Wan, L.Y.; Yan, L.Y.; Jiang, H.F.; Liu, S.Y.; Lei, Y.; Liao, B.S. Genome-wide analysis of the basic leucine zipper (bZIP) transcription factor gene family in six legume genomes. BMC Genomics 2015, 16, 1053. [Google Scholar] [CrossRef]
- Kumar, J.; Singh, S.; Singh, M.; Srivastava, P.; Mishra, R.; Singh, D.V.; Prasad, S. Transcriptional regulation of salinity stress in plants: A short review. Plant Gene 2017, 11, 160–169. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, L.; Meng, H.; Wen, H.T.; Fan, Y.L.; Zhao, J. Maize ABP9 enhances tolerance to multiple stresses in transgenic Arabidopsis by modulating ABA signaling and cellular levels of reactive oxygen species. Plant Mol. Biol. 2011, 75, 365–378. [Google Scholar] [CrossRef]
- García, M.; Giammaria, V.; Grandellis, C.; Tellez, M.; Ulloa, R.; Capiati, D. Characterization of StABF1, a stress-responsive bZIP transcription factor from Solanum tuberosum L. that is phosphorylated by StCDPK2 in vitro. Planta 2011, 235, 761–778. [Google Scholar] [CrossRef]
- Orellana, S.; Yañez, M.; Espinoza, A.; Verdugo, I.; González, E.; Ruiz-Lara, S.; Casaretto, J. The transcription factor SlAREB1 confers drought, salt stress tolerance and regulates biotic and abiotic stress-related genes in tomato. Plant Cell Environ. 2010, 33, 2191–2208. [Google Scholar] [CrossRef]
- Diao, W.; Snyder, J.C.; Wang, S.; Liu, J.; Pan, B.; Guo, G.; Ge, W.; Dawood, M. Genome-wide analyses of the NAC transcription factor gene family in pepper (Capsicum annuum L.): Chromosome location, phylogeny, structure, expression patterns, cis-elements in the promoter, and interaction network. Int. J. Mol. Sci. 2018, 19, 1028. [Google Scholar] [CrossRef]
- Sun, Q.; Gao, F.; Zhao, L.; Li, K.; Zhang, J. Identification of a new 130 bp cis-acting element in the TsVP1 promoter involved in the salt stress response from Thellungiella halophila. BMC Plant Biol. 2010, 10, 90. [Google Scholar] [CrossRef]
- Wang, Z.; Zhu, J.; Yuan, W.; Wang, Y.; Hu, P.; Jiao, C.; Xia, H.; Wang, D.; Cai, Q.; Li, J.; et al. Genome-wide characterization of bZIP transcription factors and their expression patterns in response to drought and salinity stress in Jatropha curcas. Int. J. Biol. Macromol. 2021, 181, 1207–1223. [Google Scholar] [CrossRef]
- Lv, X.; Jin, Y.; Wang, Y. De Novo transcriptome assembly and identification of salt-responsive genes in sugar beet M14. Comput. Biol. Chem. 2018, 75, 1–10. [Google Scholar] [CrossRef]
- Bowers, J.E.; Chapman, B.A.; Rong, J.; Paterson, A.H. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 2003, 422, 433–438. [Google Scholar] [CrossRef]
- Zhao, P.; Wang, D.D.; Wang, R.Q.; Kong, N.N.; Zhang, C.; Yang, C.H.; Wu, W.T.; Ma, H.L.; Chen, Q. Genome-wide analysis of the potato Hsp20 gene family: Identification, genomic organization and expression profiles in response to heat stress. BMC Genom. 2018, 19, 1–13. [Google Scholar] [CrossRef]
- Yang, L.X.; Su, D.Y.; Chang, X.; Foster, C.S.P.; Sun, L.H.; Huang, C.H.; Zhou, X.F.; Zeng, L.P.; Ma, H.; Zhong, B.J. Phylogenomic insights into deep phylogeny of angiosperms based on broad nuclear gene sampling. Plant Commun. 2020, 1, 100027. [Google Scholar] [CrossRef]
- He, S.; Shan, W.; Kuang, J.F.; Xie, H.; Xiao, Y.Y.; Lu, W.J.; Chen, J.Y. Molecular characterization of a stress-response bZIP transcription factor in banana. Plant Cell Tissue Organ Cult. 2013, 113, 173–187. [Google Scholar] [CrossRef]
- Onodera, Y.; Suzuki, A.; Wu, C.-Y.; Washida, H.; Takaiwa, F. A rice functional transcriptional activator, RISBZ1, responsible for endosperm-specific expression of storage protein genes through GCN4 motif. J. Biol. Chem. 2001, 276, 14139–14152. [Google Scholar] [CrossRef] [PubMed]
- Stotz, H.U.; Mueller, S.; Zoeller, M.; Mueller, M.J.; Berger, S. TGA transcription factors and jasmonate-independent COI1 signalling regulate specific plant responses to reactive oxylipins. J. Exp. Bot. 2013, 64, 963–975. [Google Scholar] [CrossRef] [PubMed]
- Zander, M.; Chen, S.; Imkampe, J.; Thurow, C.; Gatz, C. Repression of the Arabidopsis thaliana jasmonic acid/ethylene-induced defense pathway by TGA-interacting glutaredoxins depends on their C-terminal ALWL motif. Mol. Plant 2012, 5, 831–840. [Google Scholar] [CrossRef] [PubMed]
- Wellmer, F.; Schäfer, E.; Harter, K. The DNA binding properties of the parsley bZIP transcription factor CPRF4a are regulated by light. J. Biol. Chem. 2001, 276, 6274–6279. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, A.K.; Senapati, D.; Srivastava, A.; Chakraborty, M.; Gangappa, S.N.; Chattopadhyay, S. Short hypocotyl in white light1 interacts with elongated hypocotyl5 (HY5) and constitutive photomorphogenic1 (COP1) and Promotes COP1-mediated degradation of HY5 during Arabidopsis seedling development. Plant Physiol. 2015, 169, 2922–2934. [Google Scholar] [CrossRef] [PubMed]
- Rook, F.; Gerrits, N.; Kortstee, A.; van Kampen, M.; Borrias, M.; Weisbeek, P.; Smeekens, S. Sucrose-specifc signalling represses translation of the Arabidopsis ATB2 bZIP transcription factor gene. Plant J. 1998, 15, 253–263. [Google Scholar] [CrossRef]
- Ma, J.; Hanssen, M.; Lundgren, K.; Hernandez, L.; Delatte, T.; Ehlert, A.; Liu, C.M.; Schluepmann, H.; Droge-Laser, W.; Moritz, T.; et al. The sucrose-regulated Arabidopsis transcription factor bZIP11 reprograms metabolism and regulates trehalose metabolism. New Phytol. 2011, 191, 733–745. [Google Scholar] [CrossRef]
- Chang, Q.Y.; Lu, X.; Liu, Z.; Zheng, Z.M.; Yu, S. Identification and characterization of the bZIP transcription factor family in yellowhorn. J. For. Res. 2021, 32, 273–284. [Google Scholar] [CrossRef]
- Yang, Y.; Yu, T.F.; Ma, J.; Chen, J.; Zhou, Y.B.; Chen, M.; Ma, Y.Z.; Wei, W.L.; Xu, Z.S. The soybean bZIP transcription factor gene GmbZIP2 confers drought and salt resistances in transgenic plants. Int. J. Mol. Sci. 2020, 21, 670. [Google Scholar] [CrossRef]
- Rolly, N.K.; Imran, Q.M.; Lee, I.J.; Yun, B.W. Salinity stress-mediated suppression of expression of salt overly sensitive signaling pathway genes suggests negative regulation by AtbZIP62 transcription factor in Arabidopsis thaliana. Int. J. Mol. Sci. 2020, 21, 1726. [Google Scholar] [CrossRef]
- Lv, S.L.; Wang, D.L.; Jiang, P.; Jia, W.T.; Li, Y.X. Variation of PHT families adapts salt cress to phosphate limitation under salinity. Plant Cell Environ. 2021, 44, 1549–1564. [Google Scholar] [CrossRef]
- Zhang, M.; Liu, Y.H.; Cai, H.Y.; Guo, M.L.; Chai, M.N.; She, Z.Y.; Ye, L.; Cheng, Y.; Wang, B.R.; Qin, Y. The bZIP transcription factor GmbZIP15 negatively regulates salt- and drought-stress responses in soybean. Int. J. Mol. Sci. 2020, 21, 7778. [Google Scholar] [CrossRef]
- Kerner, K.; Nagano, S.; Lubbe, A.; Hoecker, U. Functional comparison of the WD-repeat domains of SPA1 and COP1 in suppression of photomorphogenesis. Plant Cell Environ. 2021, 44, 3273–3282. [Google Scholar] [CrossRef]
- Wei, C.-Q.; Chien, C.-W.; Ai, L.-F.; Zhao, J.; Zhenzhen, Z.; Li, K.; Burlingame, A.; Sun, Y.; Wang, Z.-Y. The Arabidopsis B-BOX protein BZS1/BBX20 interacts with HY5 and mediates strigolactone regulation of photomorphogenesis. J. Genet. Genom. 2016, 43, 555–563. [Google Scholar] [CrossRef]
- Wang, H.; Wang, H. Multifaceted roles of FHY3 and FAR1 in light signaling and beyond. Trends Plant Sci. 2015, 20, 453–461. [Google Scholar] [CrossRef]
- Barrero, J.M.; Millar, A.A.; Griffiths, J.; Czechowski, T.; Scheible, W.R.; Udvardi, M.; Reid, J.B.; Ross, J.J.; Jacobsen, J.V.; Gubler, F. Gene expression profiling identifies two regulatory genes controlling dormancy and ABA sensitivity in Arabidopsis seeds. Plant J. 2010, 61, 611–622. [Google Scholar] [CrossRef]
- Kerchev, P.I.; Pellny, T.K.; Vivancos, P.D.; Kiddle, G.; Hedden, P.; Driscoll, S.; Vanacker, H.; Verrier, P.; Hancock, R.D.; Foyer, C.H. The transcription factor ABI4 is required for the ascorbic acid-dependent regulation of growth and regulation of jasmonate-dependent defense signaling pathways in arabidopsis. Plant Cell 2011, 23, 3319–3334. [Google Scholar] [CrossRef]
- Meng, L.S.; Li, C.; Xu, M.K.; Sun, X.D.; Wan, W.; Cao, X.Y.; Zhang, J.L.; Chen, K.M. Arabidopsis angustifolia3 (AN3) is associated with the promoter of constitutive photomorphogenic1 (COP1) to regulate light-mediated stomatal development. Plant Cell Environ. 2018, 41, 1645–1656. [Google Scholar] [CrossRef]
- Sun, Y.B.; Zhang, X.J.; Zhong, M.C.; Dong, X.; Yu, D.M.; Jiang, X.D.; Wang, D.; Cui, W.H.; Chen, J.H.; Hu, J.Y. Genome-wide identification of WD40 genes reveals a functional diversification of COP1-like genes in Rosaceae. Plant Mol. Biol. 2020, 104, 81–95. [Google Scholar] [CrossRef]
- Rizzini, L.; Favory, J.J.; Cloix, C.; Faggionato, D.; O’Hara, A.; Kaiserli, E.; Baumeister, R.; Schafer, E.; Nagy, F.; Jenkins, G.I.; et al. Perception of UV-B by the Arabidopsis UVR8 protein. Science 2011, 332, 103–106. [Google Scholar] [CrossRef]
- Liu, H.X.; Stone, S.L. Abscisic acid increases arabidopsis ABI5 transcription factor levels by promoting KEG E3 ligase self-ubiquitination and proteasomal degradation. Plant Cell 2010, 22, 2630–2641. [Google Scholar] [CrossRef]
- Wang, W.; Paik, I.; Kim, J.; Hou, X.; Sung, S.; Huq, E. Direct phosphorylation of HY5 by SPA kinases to regulate photomorphogenesis in Arabidopsis. New Phytol. 2021, 230, 2311–2326. [Google Scholar] [CrossRef]
- Alvarez, S.; Sanchez-Blanco, M.J. Long-term effect of salinity on plant quality, water relations, photosynthetic parameters and ion distribution in Callistemon citrinus. Plant Biol. 2014, 16, 757–764. [Google Scholar] [CrossRef]
- Waśkiewicz, A.; Beszterda, M.; Goliński, P. ABA: Role in plant signaling under salt stress. In Salt Stress in Plants; Springer: New York, NY, USA, 2013; pp. 175–196. [Google Scholar]
- Duan, L.; Dietrich, D.; Ng, C.H.; Chan, P.M.; Bhalerao, R.; Bennett, M.J.; Dinneny, J.R. Endodermal ABA signaling promotes lateral root quiescence during salt stress in Arabidopsis seedlings. Plant Cell 2013, 25, 324–341. [Google Scholar] [CrossRef] [PubMed]
- Vitali, V.; Sutka, M.; Ojeda, L.; Aroca, R.; Amodeo, G. Root hydraulics adjustment is governed by a dominant cell-to-cell pathway in Beta vulgaris seedlings exposed to salt stress. Plant Sci. 2021, 306, 110873. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.P.; Guo, A.Y.; Zhang, H.; Luo, J.C.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Wang, Y.P.; Tang, H.B.; DeBarry, J.D.; Tan, X.; Li, J.P.; Wang, X.Y.; Lee, T.H.; Jin, H.Z.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Rio, D.; Ares, M.; Hannon, G.; Nilsen, T. Purification of RNA Using TRIzol (TRI Reagent); CSHL Press: Cold Spring Harbor, NY, USA, 2010. [Google Scholar] [CrossRef]
- Sehrish, S.; Sumbal, W.; Xie, M.; Zhao, C.; Zuo, R.; Gao, F.; Liu, S. Genome-wide identification and characterization of SET domain family genes in Brassica napus L. Int. J. Mol. Sci. 2022, 23, 1936. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gong, Y.; Liu, X.; Chen, S.; Li, H.; Duanmu, H. Genome-Wide Identification and Salt Stress Response Analysis of the bZIP Transcription Factor Family in Sugar Beet. Int. J. Mol. Sci. 2022, 23, 11573. https://doi.org/10.3390/ijms231911573
Gong Y, Liu X, Chen S, Li H, Duanmu H. Genome-Wide Identification and Salt Stress Response Analysis of the bZIP Transcription Factor Family in Sugar Beet. International Journal of Molecular Sciences. 2022; 23(19):11573. https://doi.org/10.3390/ijms231911573
Chicago/Turabian StyleGong, Yongyong, Xin Liu, Sixue Chen, Hongli Li, and Huizi Duanmu. 2022. "Genome-Wide Identification and Salt Stress Response Analysis of the bZIP Transcription Factor Family in Sugar Beet" International Journal of Molecular Sciences 23, no. 19: 11573. https://doi.org/10.3390/ijms231911573
APA StyleGong, Y., Liu, X., Chen, S., Li, H., & Duanmu, H. (2022). Genome-Wide Identification and Salt Stress Response Analysis of the bZIP Transcription Factor Family in Sugar Beet. International Journal of Molecular Sciences, 23(19), 11573. https://doi.org/10.3390/ijms231911573







