Axenic Culture of Caenorhabditis elegans Alters Lysosomal/Proteasomal Balance and Increases Neuropeptide Expression
Abstract
1. Introduction
2. Results
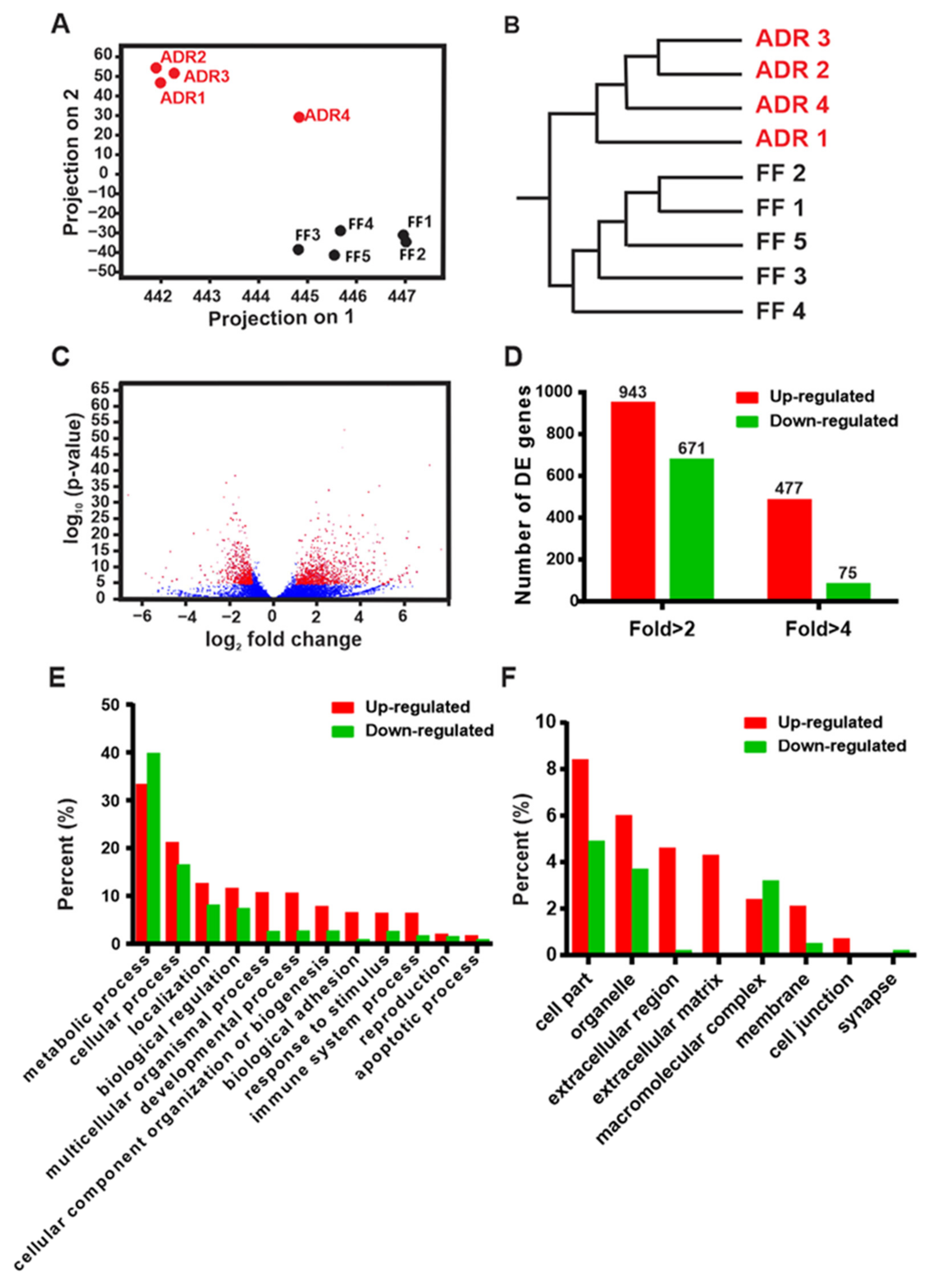
2.1. Transcriptomic Profiling of Fully Fed (FF) and Axenic Dietary Restricted (ADR) Worms
2.2. Multiple Biological Processes and Pathways Are Modulated by ADR
2.3. Increased Lysosomal Activity May Link to the Breakdown of Nutrients from Axenic Medium
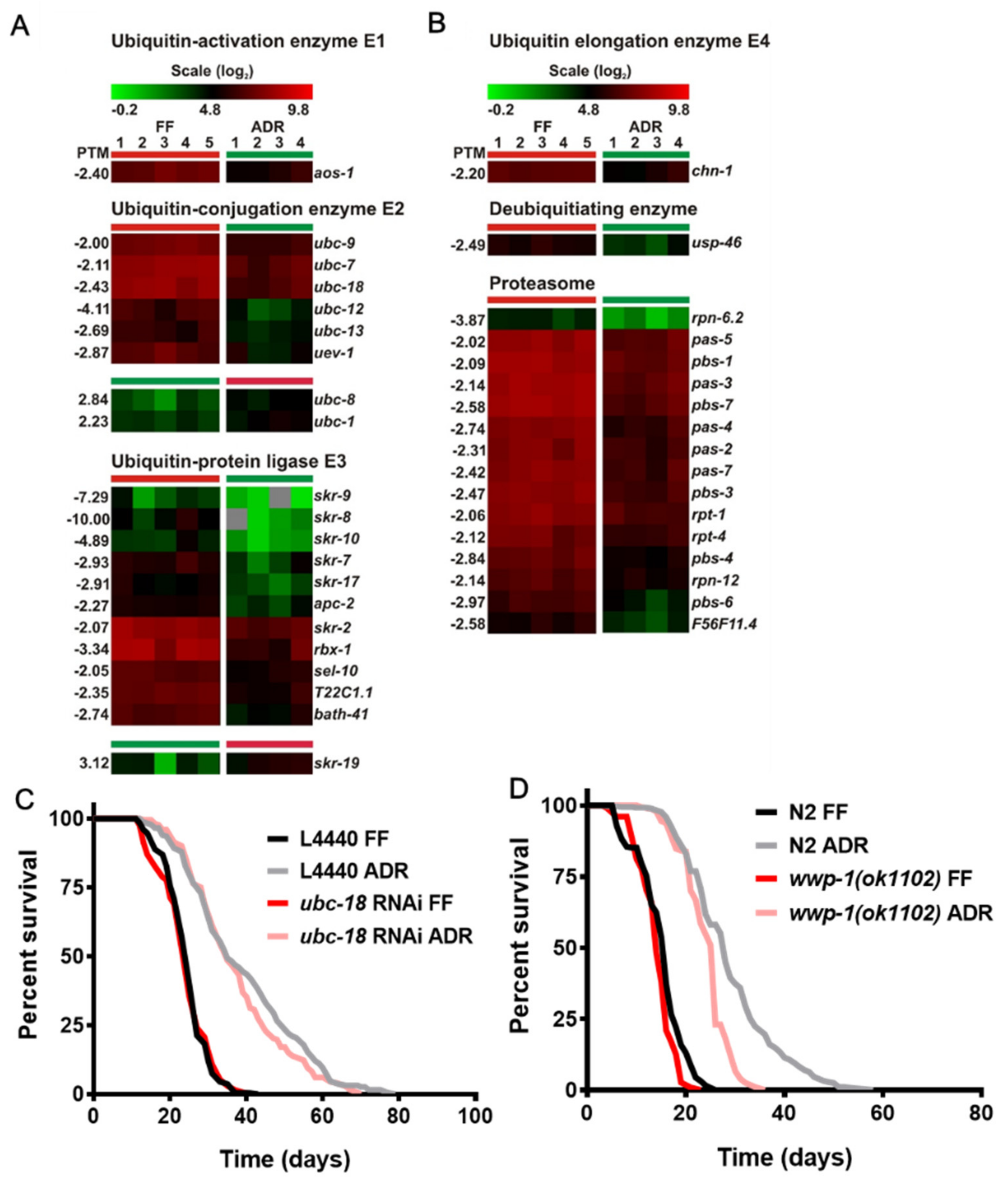
2.4. Decreased Expression of Genes Coding for the Ubiquitin–Proteasome System
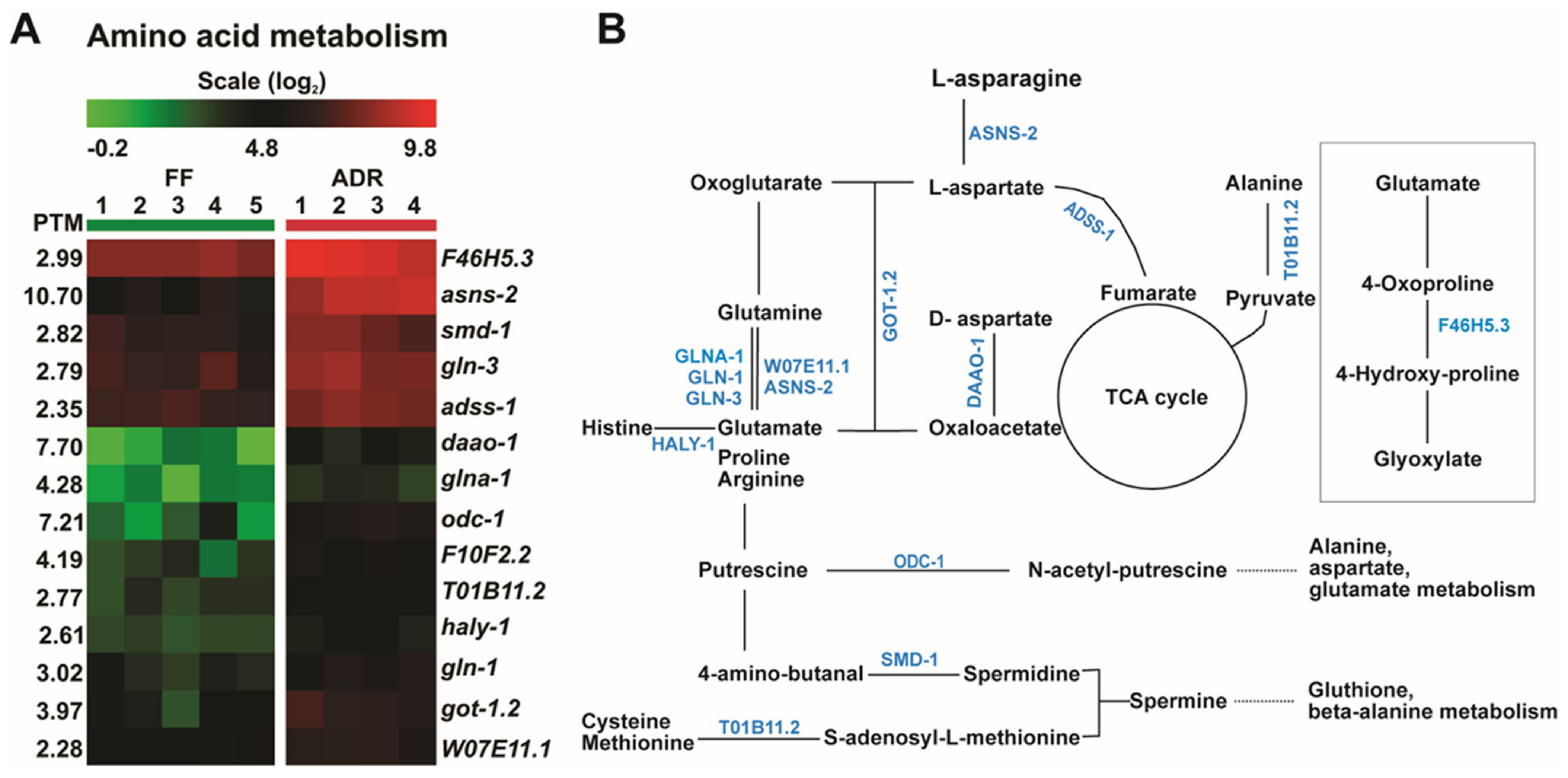
2.5. ADR Causes an Overall Increase in Amino Acid Metabolism
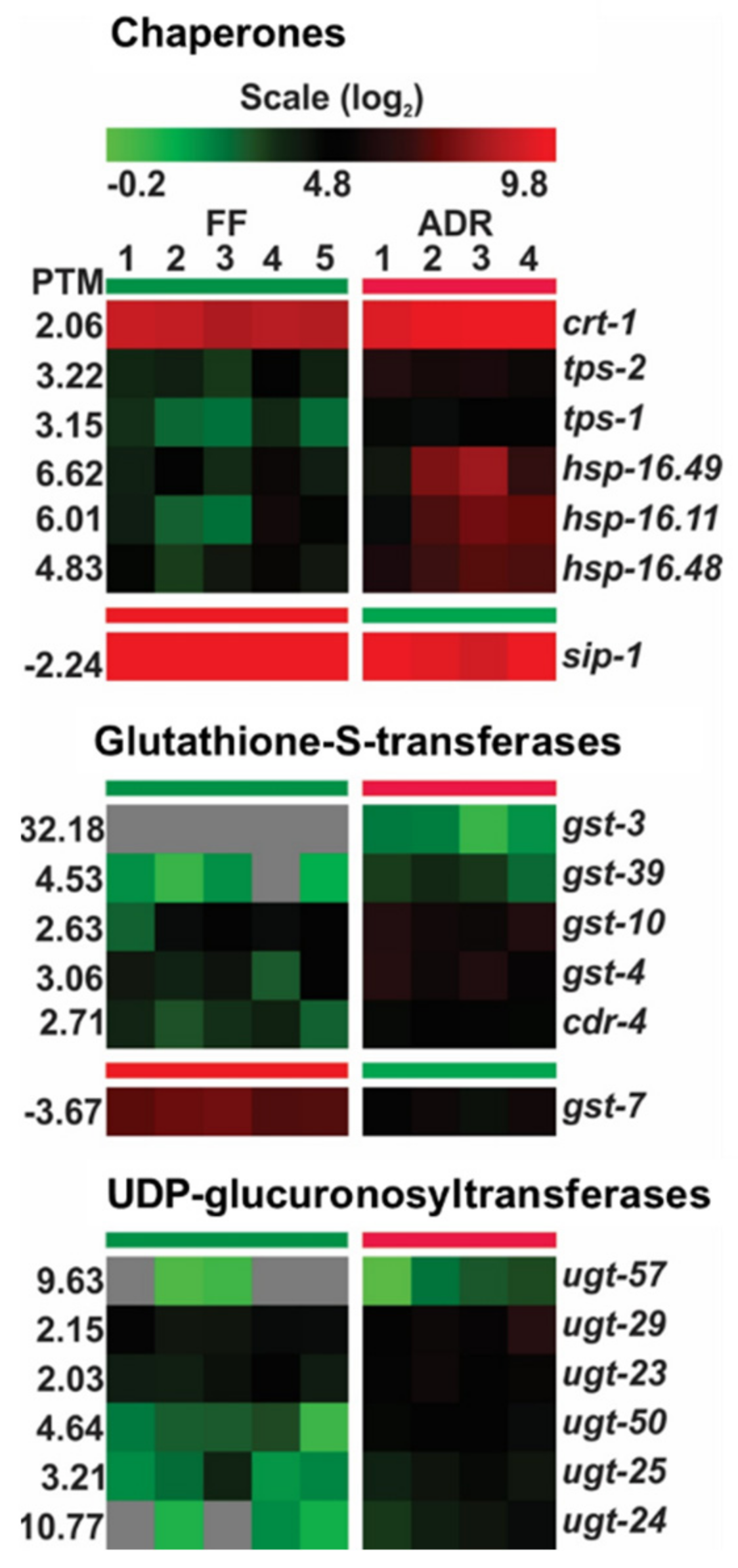
2.6. Chaperones and Detoxifying Enzymes Are Up-Regulated in ADR Worms
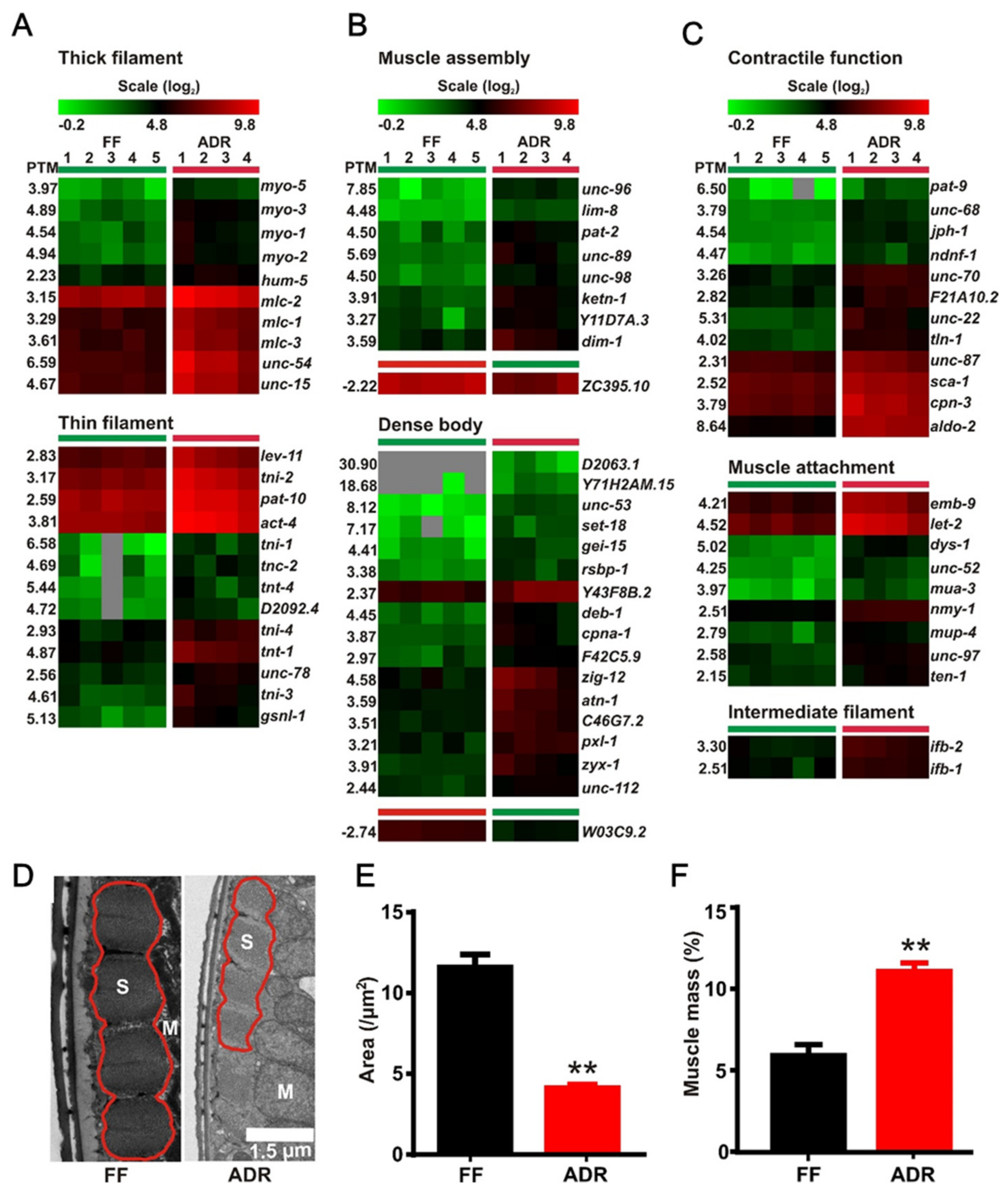
2.7. Axenic Culture Enhances Expression of Genes Involved in Muscle Function
2.8. Increased Expression of Extracellular Matrix Factors and Collagens in ADR Worms
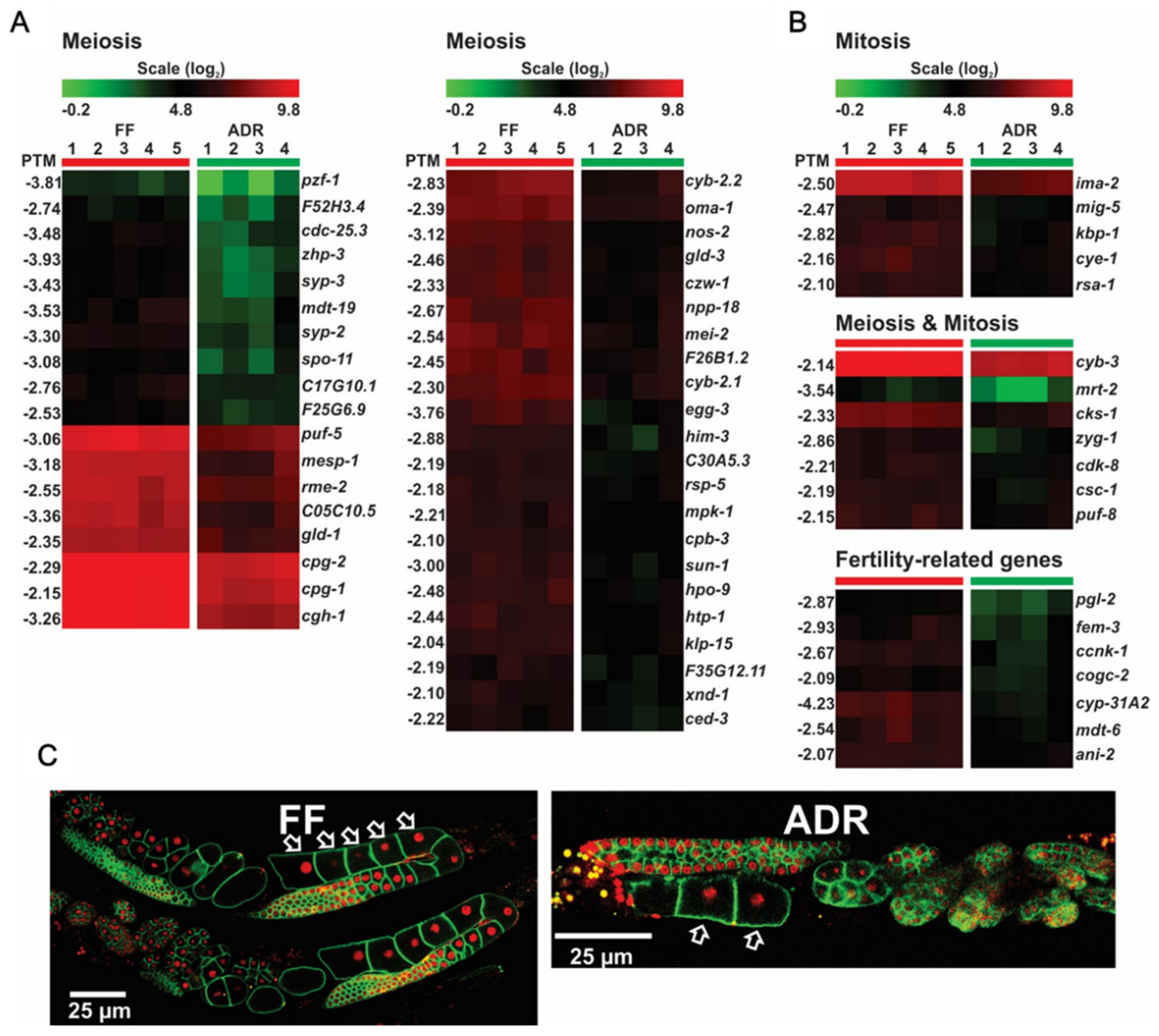
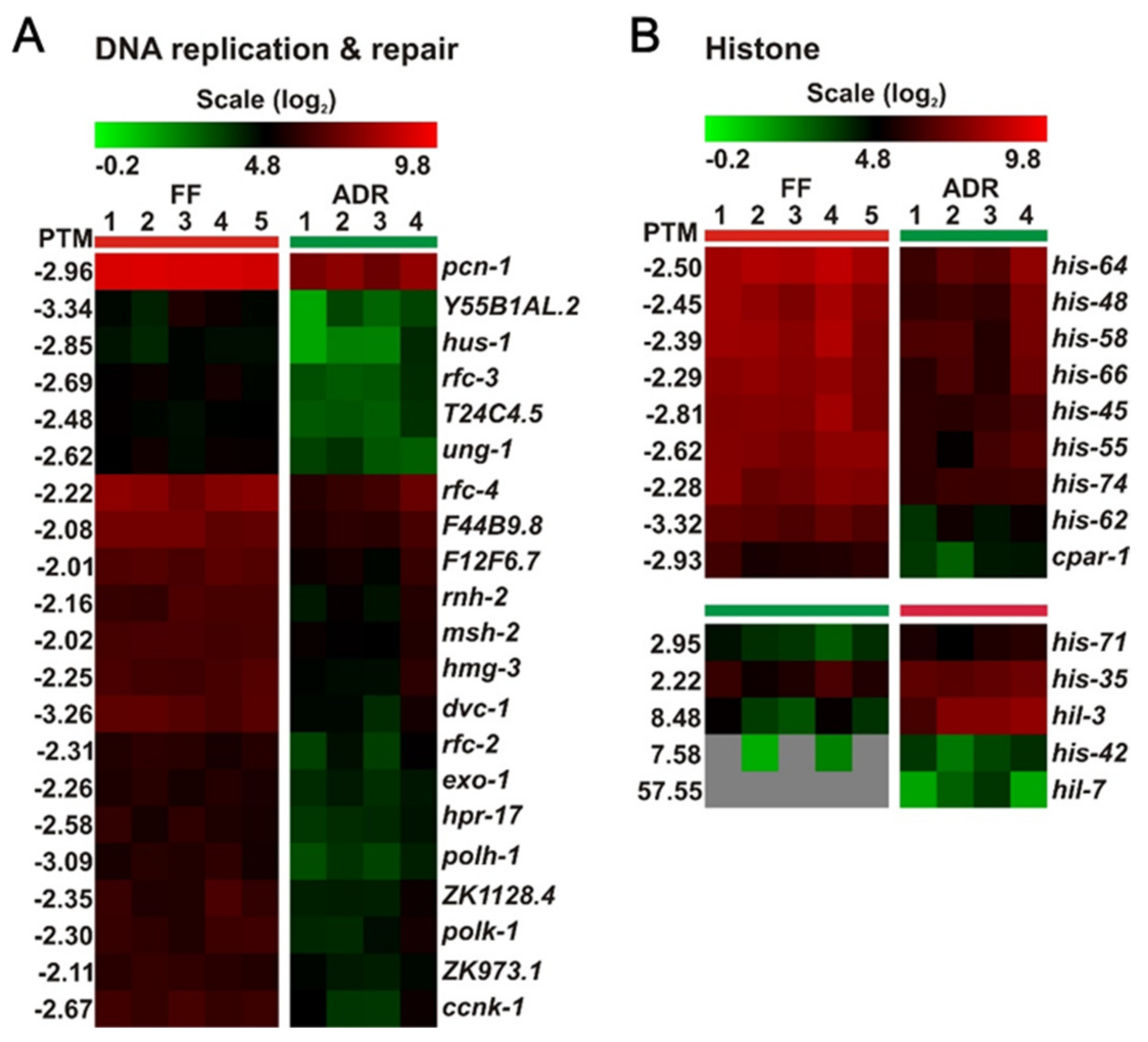
2.9. Down-Regulation of DNA Replication, Repair, and Cell Cycle Genes May Link to Reduced Fertility
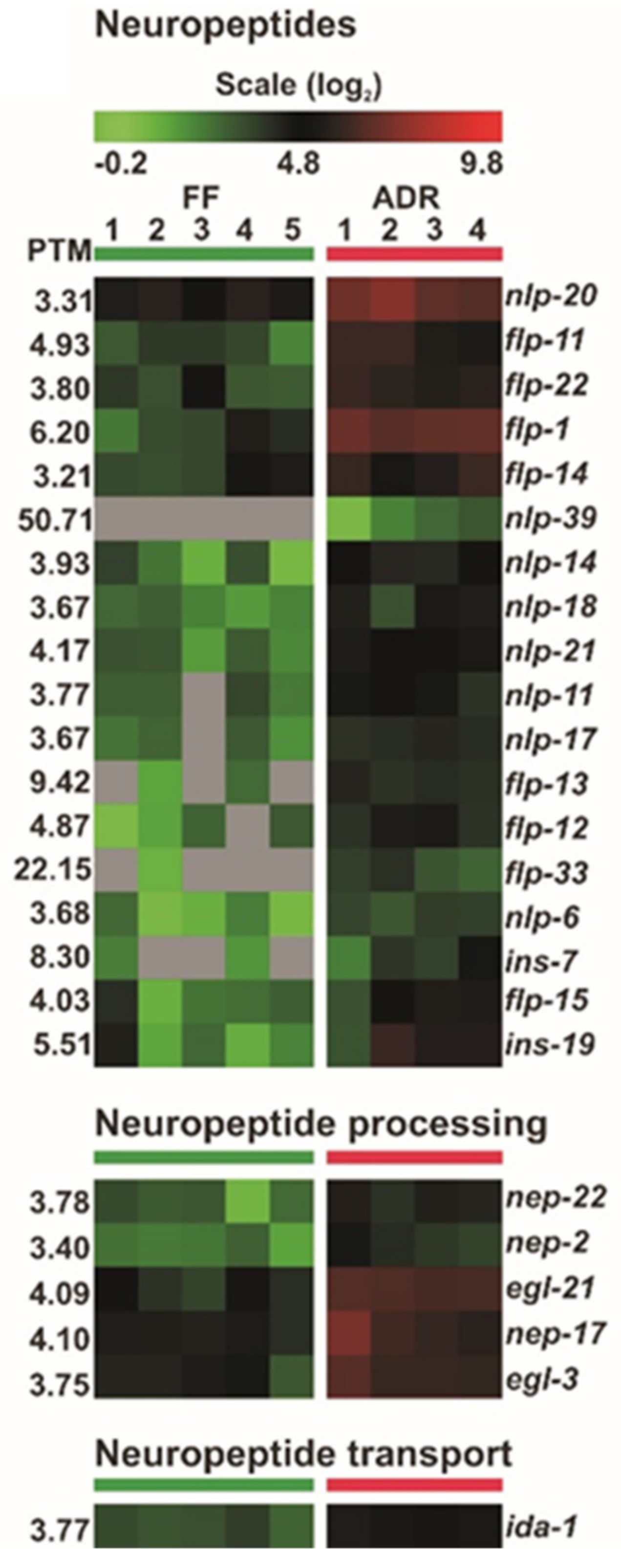
2.10. Neuroendocrine Signaling May Orchestrate ADR-Mediated Longevity
2.11. Differentially Expressed Transcription Factors (TF) and Cofactors in ADR Worms
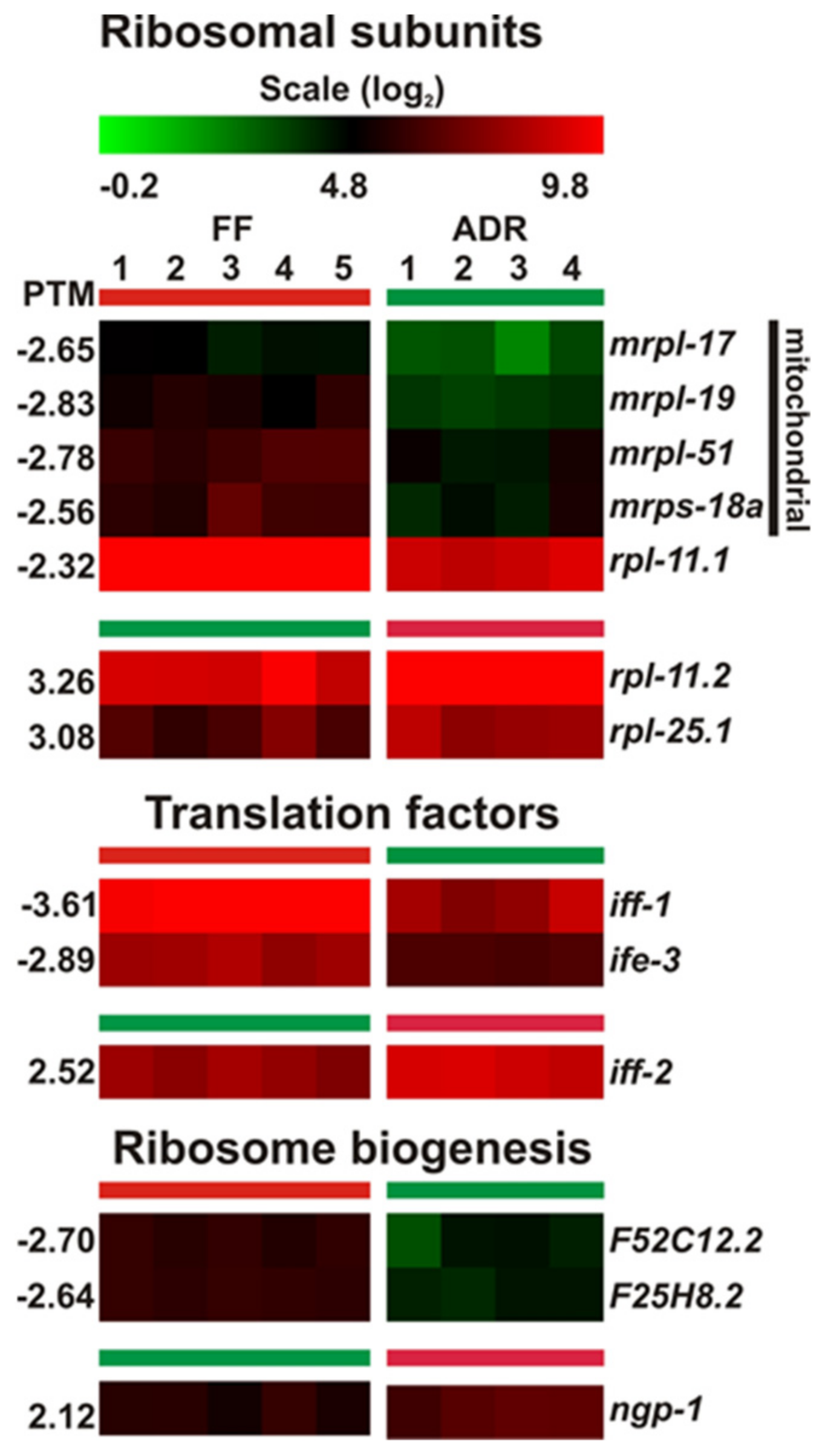
2.12. No Clear Patterns in DEGs Involved in Protein Translation
3. Discussion
4. Materials and Methods
4.1. C. elegans Strains and RNAi Bacteria
4.2. Worm Maintenance
4.3. Worm Sampling and Library Preparation
4.4. Sequence Analysis
4.5. Functional Analysis and Data Visualization
4.6. Lysosomal Activity Assay
4.7. Acid Phosphatase Assay
4.8. Cathepsin Activity Measurement
4.9. Confocal Imaging
4.10. Transmission Electron Microscopy (TEM) Analysis
4.11. Survival Assays
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dougherty, E.C.; Calhoun, H.G. Possible significance of free-living nematodes in genetic research. Nature 1948, 161, 29. [Google Scholar] [CrossRef] [PubMed]
- Yen, C.A.; Curran, S.P. Gene-diet interactions and aging in C. elegans. Exp. Gerontol. 2016, 86, 106–112. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, L.S.; Walhout, A.J. Worms, bacteria, and micronutrients: An elegant model of our diet. Trends Genet. 2014, 30, 496–503. [Google Scholar] [CrossRef] [PubMed]
- Watson, E.; MacNeil, L.T.; Ritter, A.D.; Yilmaz, L.S.; Rosebrock, A.P.; Caudy, A.A.; Walhout, A.J.M. Interspecies Systems Biology Uncovers Metabolites Affecting C. elegans Gene Expression and Life History Traits. Cell 2014, 156, 1336–1337. [Google Scholar] [CrossRef] [PubMed]
- MacNeil, L.T.; Watson, E.; Arda, H.E.; Zhu, L.J.; Walhout, A.J. Diet-induced developmental acceleration independent of TOR and insulin in C. elegans. Cell 2013, 153, 240–252. [Google Scholar] [CrossRef] [PubMed]
- Vanfleteren, J.R. Large scale cultivation of a free-living nematode (Caenorhabditis elegans). Experientia 1976, 32, 1087–1088. [Google Scholar] [CrossRef] [PubMed]
- Lenaerts, I.; Walker, G.A.; Van Hoorebeke, L.; Gems, D.; Vanfleteren, J.R. Dietary restriction of Caenorhabditis elegans by axenic culture reflects nutritional requirement for constituents provided by metabolically active microbes. J. Gerontol. A Biol. Sci. Med. Sci. 2008, 63, 242–252. [Google Scholar] [CrossRef]
- Vanfleteren, J.R.; Braeckman, B.P. Mechanisms of life span determination in Caenorhabditis elegans. Neurobiol. Aging 1999, 20, 487–502. [Google Scholar] [CrossRef]
- Castelein, N.; Cai, H.; Rasulova, M.; Braeckman, B.P. Lifespan regulation under axenic dietary restriction: A close look at the usual suspects. Exp. Gerontol. 2014, 58, 96–103. [Google Scholar] [CrossRef]
- Cai, H.; Rasulova, M.; Vandemeulebroucke, L.; Meagher, L.; Vlaeminck, C.; Dhondt, I.; Braeckman, B.P. Life-Span Extension by Axenic Dietary Restriction Is Independent of the Mitochondrial Unfolded Protein Response and Mitohormesis in Caenorhabditis elegans. J. Gerontol. A Biol. Sci. Med. Sci. 2017, 72, 1311–1318. [Google Scholar] [CrossRef]
- Castelein, N.; Hoogewijs, D.; De Vreese, A.; Braeckman, B.P.; Vanfleteren, J.R. Dietary restriction by growth in axenic medium induces discrete changes in the transcriptional output of genes involved in energy metabolism in Caenorhabditis elegans. Biotechnol. J. 2008, 3, 803–812. [Google Scholar] [CrossRef]
- Houthoofd, K.; Braeckman, B.P.; Lenaerts, I.; Brys, K.; De Vreese, A.; Van Eygen, S.; Vanfleteren, J.R. Axenic growth up-regulates mass-specific metabolic rate, stress resistance, and extends life span in Caenorhabditis elegans. Exp. Gerontol. 2002, 37, 1371–1378. [Google Scholar] [CrossRef]
- Mi, H.; Muruganujan, A.; Huang, X.; Ebert, D.; Mills, C.; Guo, X.; Thomas, P.D. Protocol Update for large-scale genome and gene function analysis with the PANTHER classification system (v.14.0). Nat. Protoc. 2019, 14, 703–721. [Google Scholar] [CrossRef]
- Mi, H.; Muruganujan, A.; Casagrande, J.T.; Thomas, P.D. Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 2013, 8, 1551–1566. [Google Scholar] [CrossRef]
- Thomas, D.M.; Kannabiran, C.; Balasubramanian, D. Identification of Key Genes and Pathways in Persistent Hyperplastic Primary Vitreous of the Eye Using Bioinformatic Analysis. Front. Med. 2021, 8, 690594. [Google Scholar]
- Huang, D.W.; Sherman, B.T.; Tan, Q.; Kir, J.; Liu, D.; Bryant, D.; Guo, Y.; Stephens, R.; Baseler, M.W.; Lane, H.C.; et al. DAVID Bioinformatics Resources: Expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007, 35, W169–W175. [Google Scholar] [CrossRef]
- Finkbeiner, S. The Autophagy Lysosomal Pathway and Neurodegeneration. Cold Spring Harb. Perspect. Biol. 2020, 12, a033993. [Google Scholar] [CrossRef]
- Taylor, R.C.; Dillin, A. Aging as an event of proteostasis collapse. Cold Spring Harb. Perspect. Biol. 2011, 3, a004440. [Google Scholar] [CrossRef]
- Gelino, S.; Chang, J.T.; Kumsta, C.; She, X.; Davis, A.; Nguyen, C.; Panowski, S.; Hansen, M. Intestinal Autophagy Improves Healthspan and Longevity in C. elegans During Dietary Restriction. PLoS Genet. 2016, 12, e1006271. [Google Scholar]
- Gelino, S.; Hansen, M. Autophagy—An Emerging Anti-Aging Mechanism. J. Clin. Exp. Pathol. 2012, (Suppl. 4), 6. [Google Scholar] [CrossRef]
- Aman, Y.; Schmauck-Medina, T.; Hansen, M.; Morimoto, R.I.; Simon, A.K.; Bjedov, I.; Palikaras, K.; Simonsen, A.; Johansen, T.; Tavernarakis, N.; et al. Autophagy in healthy aging and disease. Nat. Aging 2021, 1, 634–650. [Google Scholar] [CrossRef]
- Lapierre, L.R.; De Magalhaes Filho, C.D.; McQuary, P.R.; Chu, C.C.; Visvikis, O.; Chang, J.T.; Gelino, S.; Ong, B.; Davis, A.E.; Irazoqui, J.E.; et al. The TFEB orthologue HLH-30 regulates autophagy and modulates longevity in Caenorhabditis elegans. Nat. Commun. 2013, 4, 2267. [Google Scholar] [CrossRef]
- Bedford, L.; Lowe, J.; Dick, L.R.; Mayer, R.J.; Brownell, J.E. Ubiquitin-like protein conjugation and the ubiquitin-proteasome system as drug targets. Nat. Rev. Drug. Discov. 2011, 10, 29–46. [Google Scholar] [CrossRef]
- Yu, Z.; Li, H.; Zhu, J.; Wang, H.; Jin, X. The roles of E3 ligases in Hepatocellular carcinoma. Am. J. Cancer Res. 2022, 12, 1179–1214. [Google Scholar]
- Tomaru, U.; Takahashi, S.; Ishizu, A.; Miyatake, Y.; Gohda, A.; Suzuki, S.; Ono, A.; Ohara, J.; Baba, T.; Murata, S.; et al. Decreased proteasomal activity causes age-related phenotypes and promotes the development of metabolic abnormalities. Am. J. Pathol. 2012, 180, 963–972. [Google Scholar] [CrossRef]
- Koyuncu, S.; Loureiro, R.; Lee, H.J.; Wagle, P.; Krueger, M.; Vilchez, D. Rewiring of the ubiquitinated proteome determines ageing in C. elegans. Nature 2021, 596, 285–290. [Google Scholar] [CrossRef]
- Carrano, A.C.; Liu, Z.; Dillin, A.; Hunter, T. A conserved ubiquitination pathway determines longevity in response to diet restriction. Nature 2009, 460, 396–399. [Google Scholar] [CrossRef]
- Vilchez, D.; Morantte, I.; Liu, Z.; Douglas, P.M.; Merkwirth, C.; Rodrigues, A.P.; Manning, G.; Dillin, A. RPN-6 determines C.elegans longevity under proteotoxic stress conditions. Nature 2012, 489, 263–268. [Google Scholar] [CrossRef]
- Li, X.; Matilainen, O.; Jin, C.; Glover-Cutter, K.M.; Holmberg, C.I.; Blackwell, T.K. Specific SKN-1/Nrf stress responses to perturbations in translation elongation and proteasome activity. PLoS Genet. 2011, 7, e1002119. [Google Scholar] [CrossRef]
- Ijomone, O.M.; Miah, M.R.; Akingbade, G.T.; Bucinca, H.; Aschner, M. Nickel-Induced Developmental Neurotoxicity in C. elegans Includes Cholinergic, Dopaminergic and GABAergic Degeneration, Altered Behaviour, and Increased SKN-1 Activity. Neurotox. Res. 2020, 37, 1018–1028. [Google Scholar] [CrossRef]
- Blackwell, T.K.; Steinbaugh, M.J.; Hourihan, J.M.; Ewald, C.Y.; Isik, M. SKN-1/Nrf, stress responses, and aging in Caenorhabditis elegans. Free Radic. Biol. Med. 2015, 88, 290–301. [Google Scholar] [CrossRef] [PubMed]
- Carsten, L.D.; Watts, T.; Markow, T.A. Gene expression patterns accompanying a dietary shift in Drosophila melanogaster. Mol. Ecol. 2005, 14, 3203–3208. [Google Scholar] [CrossRef] [PubMed]
- Ganguly, A.; Dey, M.; Scott, C.; Duong, V.K.; Arun Dahanukar, A. Dietary Macronutrient Imbalances Lead to Compensatory Changes in Peripheral Taste via Independent Signaling Pathways. J. Neurosci. 2021, 41, 10222–10246. [Google Scholar] [CrossRef] [PubMed]
- Elashry, M.I.; Eldaey, A.; Glenske, K.; Matsakas, A.; Wenisch, S.; Arnhold, S.; Patel, K. The effect of high-fat diet on the morphological properties of the forelimb musculature in hypertrophic myostatin null mice. J. Anat. 2019, 235, 825–835. [Google Scholar] [CrossRef]
- Tsverov, J.; Yegorov, K.; Powers, T. Identification of defined structural elements within TOR2 kinase required for TOR complex 2 assembly and function in Saccharomyces cerevisiae. Mol. Biol. Cell 2022, 33, ar44. [Google Scholar] [CrossRef]
- Dato, S.; Hoxha, E.; Crocco, P.; Iannone, F.; Passarino, G.; Rose, G. Amino acids and amino acid sensing: Implication for aging and diseases. Biogerontology 2019, 20, 17–31. [Google Scholar] [CrossRef]
- Xu, S.; Cai, Y.; Wei, Y. mTOR Signaling from Cellular Senescence to Organismal Aging. Aging Dis. 2014, 5, 263–273. [Google Scholar] [CrossRef]
- Depuydt, G.; Xie, F.; Petyuk, V.A.; Smolders, A.; Brewer, H.M.; Camp, D.G., 2nd; Smith, R.D.; Braeckman, B.P. LC-MS proteomics analysis of the insulin/IGF-1-deficient Caenorhabditis elegans daf-2(e1370) mutant reveals extensive restructuring of intermediary metabolism. J. Proteome Res. 2014, 13, 1938–1956. [Google Scholar] [CrossRef]
- Katane, M.; Saitoh, Y.; Seida, Y.; Sekine, M.; Furuchi, T.; Homma, H. Comparative characterization of three D-aspartate oxidases and one D-amino acid oxidase from Caenorhabditis elegans. Chem. Biodivers. 2010, 7, 1424–1434. [Google Scholar] [CrossRef]
- Katane, M.; Seida, Y.; Sekine, M.; Furuchi, T.; Homma, H. Caenorhabditis elegans has two genes encoding functional d-aspartate oxidases. FEBS J. 2007, 274, 137–149. [Google Scholar] [CrossRef]
- Gems, D.; McElwee, J.J. Broad spectrum detoxification: The major longevity assurance process regulated by insulin/IGF-1 signaling? Mech. Ageing Dev. 2005, 126, 381–387. [Google Scholar] [CrossRef]
- Oakley, A. Glutathione transferases: A structural perspective. Drug Metab. Rev. 2011, 43, 138–151. [Google Scholar] [CrossRef]
- Walther, D.M.; Kasturi, P.; Zheng, M.; Pinkert, S.; Vecchi, G.; Ciryam, P.; Morimoto, R.I.; Dobson, C.M.; Vendruscolo, M.; Mann, M.; et al. Widespread Proteome Remodeling and Aggregation in Aging C. elegans. Cell 2017, 168, 944. [Google Scholar] [CrossRef]
- David, D.C.; Ollikainen, N.; Trinidad, J.C.; Cary, M.P.; Burlingame, A.L.; Kenyon, C. Widespread protein aggregation as an inherent part of aging in C. elegans. PLoS Biol. 2010, 8, e1000450. [Google Scholar] [CrossRef]
- Sinnige, T.; Yu, A.; Morimoto, R.I. Challenging Proteostasis: Role of the Chaperone Network to Control Aggregation-Prone Proteins in Human Disease. Adv. Exp. Med. Biol. 2020, 1243, 53–68. [Google Scholar]
- McElwee, J.J.; Schuster, E.; Blanc, E.; Thomas, J.H.; Gems, D. Shared transcriptional signature in Caenorhabditis elegans Dauer larvae and long-lived daf-2 mutants implicates detoxification system in longevity assurance. J. Biol. Chem. 2004, 279, 44533–44543. [Google Scholar] [CrossRef]
- Halaschek-Wiener, J.; Khattra, J.S.; McKay, S.; Pouzyrev, A.; Stott, J.M.; Yang, G.S.; Holt, R.A.; Jones, S.J.; Marra, M.A.; Brooks-Wilson, A.R.; et al. Analysis of long-lived C. elegans daf-2 mutants using serial analysis of gene expression. Genome Res. 2005, 15, 603–615. [Google Scholar] [CrossRef]
- Dhondt, I.; Petyuk, V.A.; Cai, H.; Vandemeulebroucke, L.; Vierstraete, A.; Smith, R.D.; Depuydt, G.; Braeckman, B.P. FOXO/DAF-16 Activation Slows Down Turnover of the Majority of Proteins in C. elegans. Cell. Rep. 2016, 16, 3028–3040. [Google Scholar] [CrossRef]
- Dong, M.Q.; Venable, J.D.; Au, N.; Xu, T.; Park, S.K.; Cociorva, D.; Johnson, J.R.; Dillin, A.; Yates, J.R., 3rd. Quantitative mass spectrometry identifies insulin signaling targets in C. elegans. Science 2007, 317, 660–663. [Google Scholar] [CrossRef]
- Depuydt, G.; Xie, F.; Petyuk, V.A.; Shanmugam, N.; Smolders, A.; Dhondt, I.; Brewer, H.M.; Camp, D.G., 2nd; Smith, R.D.; Braeckman, B.P. Reduced insulin/insulin-like growth factor-1 signaling and dietary restriction inhibit translation but preserve muscle mass in Caenorhabditis elegans. Mol. Cell. Proteomics 2013, 12, 3624–3639. [Google Scholar] [CrossRef]
- Simmer, F.; Moorman, C.; van der Linden, A.M.; Kuijk, E.; van den Berghe, P.V.; Kamath, R.S.; Fraser, A.G.; Ahringer, J.; Plasterk, R.H. Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLoS Biol. 2003, 1, E12. [Google Scholar] [CrossRef]
- Ewald, C.Y.; Landis, J.N.; Porter Abate, J.; Murphy, C.T.; Blackwell, T.K. Dauer-independent insulin/IGF-1-signalling implicates collagen remodelling in longevity. Nature 2015, 519, 97–101. [Google Scholar] [CrossRef]
- Ewald, C.Y. The Matrisome during Aging and Longevity: A Systems-Level Approach toward Defining Matreotypes Promoting Healthy Aging. Gerontology 2020, 66, 266–274. [Google Scholar] [CrossRef]
- Herndon, L.A.; Schmeissner, P.J.; Dudaronek, J.M.; Brown, P.A.; Listner, K.M.; Sakano, Y.; Paupard, M.C.; Hall, D.H.; Driscoll, M. Stochastic and genetic factors influence tissue-specific decline in ageing C. elegans. Nature 2002, 419, 808–814. [Google Scholar] [CrossRef]
- Gems, D.; de la Guardia, Y. Alternative Perspectives on Aging in Caenorhabditis elegans: Reactive Oxygen Species or Hyperfunction? Antioxid. Redox Signal. 2013, 19, 321–329. [Google Scholar] [CrossRef]
- Herbert, M.; Kalleas, D.; Cooney, D.; Lamb, M.; Lister, L. Meiosis and maternal aging: Insights from aneuploid oocytes and trisomy births. Cold Spring Harb. Perspect. Biol. 2015, 7, a017970. [Google Scholar] [CrossRef]
- Melesse, M.; Bembenek, J.N. Cracking the eggshell: A novel link to intracellular signaling. Dev. Biol. 2019, 453, 107–109. [Google Scholar] [CrossRef]
- Wu, D.; Wang, Z.; Huang, J.; Huang, L.; Zhang, S.; Zhao, R.; Li, W.; Chen, D.; Ou, G. An antagonistic pleiotropic gene regulates the reproduction and longevity tradeoff. Proc. Natl. Acad. Sci. USA 2022, 119, e2120311119. [Google Scholar] [CrossRef]
- Nathoo, A.N.; Moeller, R.A.; Westlund, B.A.; Hart, A.C. Identification of neuropeptide-like protein gene families in Caenorhabditiselegans and other species. Proc. Natl. Acad. Sci. USA 2001, 98, 14000–14005. [Google Scholar] [CrossRef]
- Li, C.; Kim, K. Neuropeptide gene families in Caenorhabditis elegans. Adv. Exp. Med. Biol. 2010, 692, 98–137. [Google Scholar]
- Husson, S.J.; Clynen, E.; Baggerman, G.; Janssen, T.; Schoofs, L. Defective processing of neuropeptide precursors in Caenorhabditis elegans lacking proprotein convertase 2 (KPC-2/EGL-3): Mutant analysis by mass spectrometry. J. Neurochem. 2006, 98, 1999–2012. [Google Scholar] [CrossRef] [PubMed]
- Murphy, C.T.; McCarroll, S.A.; Bargmann, C.I.; Fraser, A.; Kamath, R.S.; Ahringer, J.; Li, H.; Kenyon, C. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 2003, 424, 277–283. [Google Scholar] [CrossRef] [PubMed]
- Bishop, N.A.; Guarente, L. Genetic links between diet and lifespan: Shared mechanisms from yeast to humans. Nat. Rev. Genet. 2007, 8, 835–844. [Google Scholar] [CrossRef] [PubMed]
- Panowski, S.H.; Wolff, S.; Aguilaniu, H.; Durieux, J.; Dillin, A. PHA-4/Foxa mediates diet-restriction-induced longevity of C. elegans. Nature 2007, 447, 550–555. [Google Scholar] [CrossRef]
- Greer, E.L.; Dowlatshahi, D.; Banko, M.R.; Villen, J.; Hoang, K.; Blanchard, D.; Gygi, S.P.; Brunet, A. An AMPK-FOXO pathway mediates longevity induced by a novel method of dietary restriction in C. elegans. Curr. Biol. 2007, 17, 1646–1656. [Google Scholar] [CrossRef]
- Steinkraus, K.A.; Smith, E.D.; Davis, C.; Carr, D.; Pendergrass, W.R.; Sutphin, G.L.; Kennedy, B.K.; Kaeberlein, M. Dietary restriction suppresses proteotoxicity and enhances longevity by an hsf-1-dependent mechanism in Caenorhabditis elegans. Aging Cell 2008, 7, 394–404. [Google Scholar] [CrossRef]
- Zhang, M.; Poplawski, M.; Yen, K.; Cheng, H.; Bloss, E.; Zhu, X.; Patel, H.; Mobbs, C.V. Role of CBP and SATB-1 in aging, dietary restriction, and insulin-like signaling. PLoS Biol. 2009, 7, e1000245. [Google Scholar] [CrossRef]
- Cai, H.; Dhondt, I.; Vandemeulebroucke, L.; Vlaeminck, C.; Rasulova, M.; Braeckman, B.P. CBP-1 Acts in GABAergic Neurons to Double Life Span in Axenically Cultured Caenorhabditis elegans. J. Gerontol. A Biol. Sci. Med. Sci. 2019, 74, 1198–1205. [Google Scholar] [CrossRef]
- Matai, L.; Sarkar, G.C.; Chamoli, M.; Malik, Y.; Kumar, S.S.; Rautela, U.; Jana, N.R.; Chakraborty, K.; Mukhopadhyay, A. Dietary restriction improves proteostasis and increases life span through endoplasmic reticulum hormesis. Proc. Natl. Acad. Sci. USA 2019, 116, 17383–17392. [Google Scholar] [CrossRef]
- Hansen, M.; Taubert, S.; Crawford, D.; Libina, N.; Lee, S.J.; Kenyon, C. Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell 2007, 6, 95–110. [Google Scholar] [CrossRef]
- Pan, K.Z.; Palter, J.E.; Rogers, A.N.; Olsen, A.; Chen, D.; Lithgow, G.J.; Kapahi, P. Inhibition of mRNA translation extends lifespan in Caenorhabditis elegans. Aging Cell 2007, 6, 111–119. [Google Scholar] [CrossRef]
- Syntichaki, P.; Troulinaki, K.; Tavernarakis, N. eIF4E function in somatic cells modulates ageing in Caenorhabditis elegans. Nature 2007, 445, 922–926. [Google Scholar] [CrossRef]
- Depuydt, G.; Shanmugam, N.; Rasulova, M.; Dhondt, I.; Braeckman, B.P. Increased Protein Stability and Decreased Protein Turnover in the Caenorhabditis elegans Ins/IGF-1 daf-2 Mutant. J. Gerontol. A Biol. Sci. Med. Sci. 2016, 71, 1553–1559. [Google Scholar] [CrossRef]
- Visscher, M.; De Henau, S.; Wildschut, M.H.E.; van Es, R.M.; Dhondt, I.; Michels, H.; Kemmeren, P.; Nollen, E.A.; Braeckman, B.P.; Burgering, B.M.T.; et al. Proteome-wide Changes in Protein Turnover Rates in C. elegans Models of Longevity and Age-Related Disease. Cell Rep. 2016, 16, 3041–3051. [Google Scholar] [CrossRef]
- Houtkooper, R.H.; Mouchiroud, L.; Ryu, D.; Moullan, N.; Katsyuba, E.; Knott, G.; Williams, R.W.; Auwerx, J. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature 2013, 497, 451–457. [Google Scholar] [CrossRef]
- Croll, N.A.; Smith, J.M.; Zuckerman, B.M. The aging process of the nematode Caenorhabditis elegans in bacterial and axenic culture. Exp. Aging Res. 1977, 3, 175–189. [Google Scholar] [CrossRef]
- Greer, E.L.; Brunet, A. Different dietary restriction regimens extend lifespan by both independent and overlapping genetic pathways in C. elegans. Aging Cell 2009, 8, 113–127. [Google Scholar] [CrossRef]
- Liebl, M.P.; Hoppe, T. It’s all about talking: Two-way communication between proteasomal and lysosomal degradation pathways via ubiquitin. Am. J. Physiol. Cell Physiol. 2016, 311, C166–C178. [Google Scholar] [CrossRef]
- Massey, A.C.; Kaushik, S.; Cuervo, A.M. Lysosomal chat maintains the balance. Autophagy 2006, 2, 325–327. [Google Scholar] [CrossRef]
- Hansen, M.; Rubinsztein, D.C.; Walker, D.W. Autophagy as a promoter of longevity: Insights from model organisms. Nat. Rev. Mol. Cell. Biol. 2018, 19, 579–593. [Google Scholar] [CrossRef]
- Szewczyk, N.J.; Udranszky, I.A.; Kozak, E.; Sunga, J.; Kim, S.K.; Jacobson, L.A.; Conley, C.A. Delayed development and lifespan extension as features of metabolic lifestyle alteration in C. elegans under dietary restriction. J. Exp. Biol. 2006, 209, 4129–4139. [Google Scholar] [CrossRef]
- Henderson, S.T.; Johnson, T.E. daf-16 integrates developmental and environmental inputs to mediate aging in the nematode Caenorhabditis elegans. Curr. Biol. 2001, 11, 1975–1980. [Google Scholar] [CrossRef]
- Houthoofd, K.; Braeckman, B.P.; Johnson, T.E.; Vanfleteren, J.R. Life extension via dietary restriction is independent of the Ins/IGF-1 signalling pathway in Caenorhabditis elegans. Exp. Gerontol. 2003, 38, 947–954. [Google Scholar] [CrossRef]
- Sun, P.Q.; Cao, X.W.; Zhang, L.S. Transcriptome Analysis of the Nematodes Caenorhabditis elegans and Litoditis marina in Different Food Environments. J. Mar. Sci. Eng. 2022, 10, 580. [Google Scholar] [CrossRef]
- Celen, I.; Doh, J.H.; Sabanayagam, C.R. Effects of liquid cultivation on gene expression and phenotype of C. elegans. BMC Genom. 2018, 19, 562. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Pavlidis, P.; Noble, W.S. Analysis of strain and regional variation in gene expression in mouse brain. Genome Biol. 2001, 2, research0042. [Google Scholar] [CrossRef]
- Yang, J.S.; Nam, H.J.; Seo, M.; Han, S.K.; Choi, Y.; Nam, H.G.; Lee, S.J.; Kim, S. OASIS: Online application for the survival analysis of lifespan assays performed in aging research. PLoS ONE 2011, 6, e23525. [Google Scholar] [CrossRef]


| KEGG Pathway Term | Fold | p-Value |
|---|---|---|
| Enrichment | ||
| Up-regulated | ||
| Alanine, aspartate, and glutamate metabolism | 4.09 | 5.22 × 10−3 |
| Calcium signaling pathway | 3.2 | 9.56 × 10−3 |
| Arginine and proline metabolism | 3.54 | 1.09 × 10−2 |
| Nitrogen metabolism | 4 | 3.09 × 10−2 |
| MAPK signaling pathway | 2.32 | 3.40 × 10−2 |
| Down-regulated | ||
| Proteasome | 5.22 | 1.77 × 10−7 |
| Mismatch repair | 6.03 | 1.49 × 10−4 |
| Ubiquitin mediated proteolysis | 2.68 | 2.86 × 10−4 |
| Nucleotide excision repair | 3.39 | 3.38 × 10−3 |
| DNA replication | 3.29 | 7.97 × 10−3 |
| TGF-beta signaling pathway | 3.02 | 1.30 × 10−2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cai, H.; Wu, P.; Vandemeulebroucke, L.; Dhondt, I.; Rasulova, M.; Vierstraete, A.; Braeckman, B.P. Axenic Culture of Caenorhabditis elegans Alters Lysosomal/Proteasomal Balance and Increases Neuropeptide Expression. Int. J. Mol. Sci. 2022, 23, 11517. https://doi.org/10.3390/ijms231911517
Cai H, Wu P, Vandemeulebroucke L, Dhondt I, Rasulova M, Vierstraete A, Braeckman BP. Axenic Culture of Caenorhabditis elegans Alters Lysosomal/Proteasomal Balance and Increases Neuropeptide Expression. International Journal of Molecular Sciences. 2022; 23(19):11517. https://doi.org/10.3390/ijms231911517
Chicago/Turabian StyleCai, Huaihan, Ping Wu, Lieselot Vandemeulebroucke, Ineke Dhondt, Madina Rasulova, Andy Vierstraete, and Bart P. Braeckman. 2022. "Axenic Culture of Caenorhabditis elegans Alters Lysosomal/Proteasomal Balance and Increases Neuropeptide Expression" International Journal of Molecular Sciences 23, no. 19: 11517. https://doi.org/10.3390/ijms231911517
APA StyleCai, H., Wu, P., Vandemeulebroucke, L., Dhondt, I., Rasulova, M., Vierstraete, A., & Braeckman, B. P. (2022). Axenic Culture of Caenorhabditis elegans Alters Lysosomal/Proteasomal Balance and Increases Neuropeptide Expression. International Journal of Molecular Sciences, 23(19), 11517. https://doi.org/10.3390/ijms231911517






