miR-140-5p and miR-140-3p: Key Actors in Aging-Related Diseases?
Abstract
1. Introduction
2. miR-140-5p and miR-140-3p Biogenesis
3. Role of miR-140-5p/-3p in Neurodegenerative Diseases/Cognitive Impairment
4. Role of miR-140-5p/-3p in Atherosclerosis
5. miR-140-5p/-3p and Visual Acuity Loss
6. Role of miR-140-5p/-3p in Osteoporosis
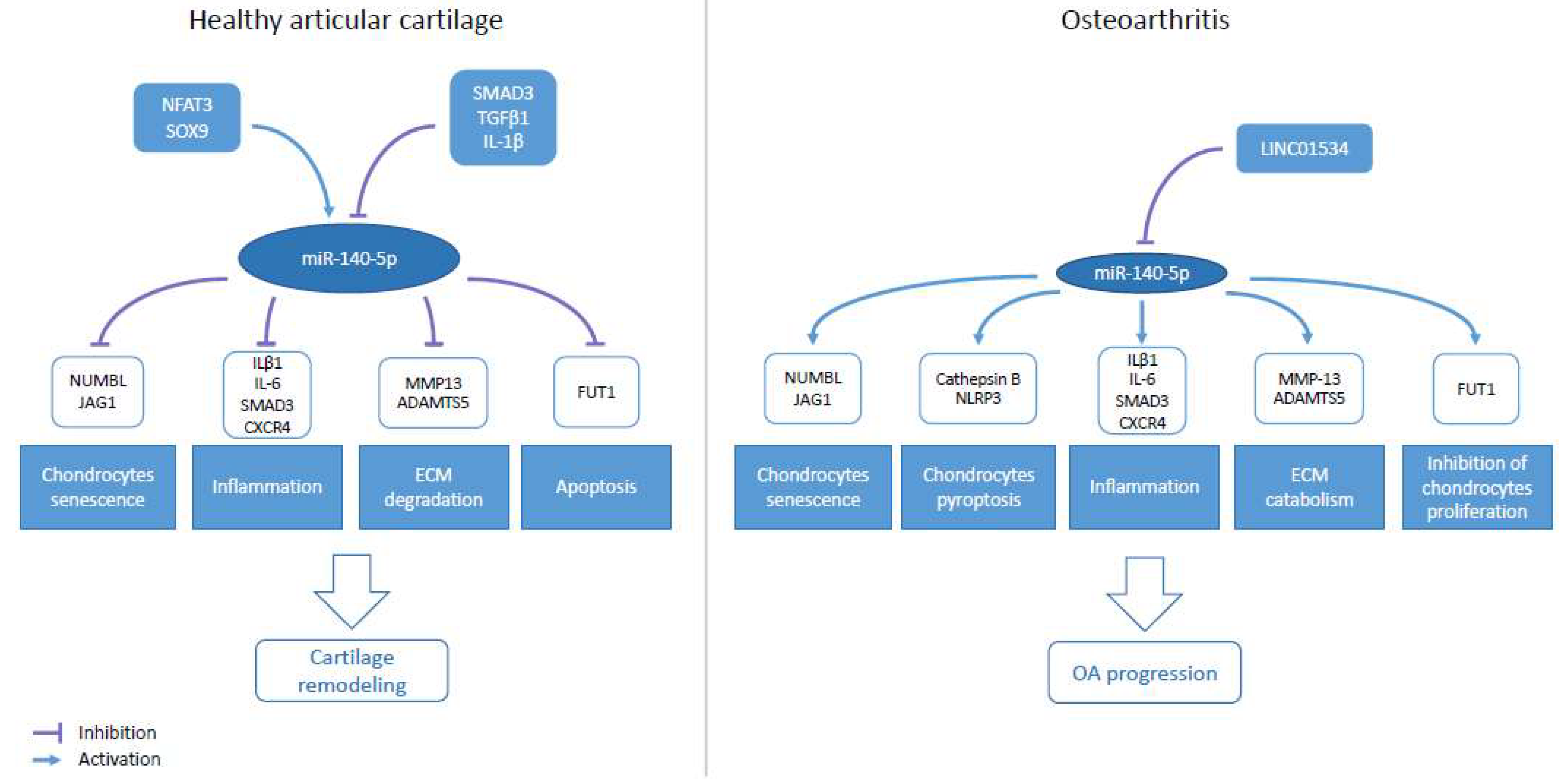
7. Role of miR-140-5p/-3p in Osteoarthritis
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. Elegans Heterochronic Gene Lin-4 Encodes Small RNAs with Antisense Complementarity to Lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Wightman, B.; Ha, I.; Ruvkun, G. Posttranscriptional Regulation of the Heterochronic Gene Lin-14 by Lin-4 Mediates Temporal Pattern Formation in C. Elegans. Cell 1993, 75, 855–862. [Google Scholar] [CrossRef]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. MiRBase: From MicroRNA Sequences to Function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef] [PubMed]
- Plotnikova, O.; Baranova, A.; Skoblov, M. Comprehensive Analysis of Human MicroRNA–MRNA Interactome. Front. Genet. 2019, 10, 933. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef]
- Hsu, P.W.C. MiRNAMap: Genomic Maps of MicroRNA Genes and Their Target Genes in Mammalian Genomes. Nucleic Acids Res. 2006, 34, D135–D139. [Google Scholar] [CrossRef]
- Rodriguez, A.; Griffiths-Jones, S.; Ashurst, J.L.; Bradley, A. Identification of Mammalian MicroRNA Host Genes and Transcription Units. Genome Res. 2004, 14, 1902–1910. [Google Scholar] [CrossRef]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Rådmark, O.; Kim, S.; et al. The Nuclear RNase III Drosha Initiates MicroRNA Processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef]
- Ha, M.; Kim, V.N. Regulation of MicroRNA Biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef]
- Denli, A.M.; Tops, B.B.J.; Plasterk, R.H.A.; Ketting, R.F.; Hannon, G.J. Processing of Primary MicroRNAs by the Microprocessor Complex. Nature 2004, 432, 231–235. [Google Scholar] [CrossRef]
- Alarcón, C.R.; Lee, H.; Goodarzi, H.; Halberg, N.; Tavazoie, S.F. N6-Methyladenosine Marks Primary MicroRNAs for Processing. Nature 2015, 519, 482–485. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Lee, Y.; Yeom, K.-H.; Kim, Y.-K.; Jin, H.; Kim, V.N. The Drosha-DGCR8 Complex in Primary MicroRNA Processing. Genes Dev. 2004, 18, 3016–3027. [Google Scholar] [CrossRef]
- Okada, C.; Yamashita, E.; Lee, S.J.; Shibata, S.; Katahira, J.; Nakagawa, A.; Yoneda, Y.; Tsukihara, T. A High-Resolution Structure of the Pre-MicroRNA Nuclear Export Machinery. Science 2009, 326, 1275–1279. [Google Scholar] [CrossRef] [PubMed]
- Bohnsack, M.T.; Czaplinski, K.; Gorlich, D. Exportin 5 Is a RanGTP-Dependent DsRNA-Binding Protein That Mediates Nuclear Export of Pre-MiRNAs. RNA 2004, 10, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Yi, R.; Qin, Y.; Macara, I.G.; Cullen, B.R. Exportin-5 Mediates the Nuclear Export of Pre-MicroRNAs and Short Hairpin RNAs. Genes Dev. 2003, 17, 3011–3016. [Google Scholar] [CrossRef]
- Zhang, H.; Kolb, F.A.; Jaskiewicz, L.; Westhof, E.; Filipowicz, W. Single Processing Center Models for Human Dicer and Bacterial RNase III. Cell 2004, 118, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Hutvágner, G.; McLachlan, J.; Pasquinelli, A.E.; Bálint, É.; Tuschl, T.; Zamore, P.D. A Cellular Function for the RNA-Interference Enzyme Dicer in the Maturation of the Let-7 Small Temporal RNA. Science 2001, 293, 834–838. [Google Scholar] [CrossRef]
- Kobayashi, H.; Tomari, Y. RISC Assembly: Coordination between Small RNAs and Argonaute Proteins. Biochim. Et Biophys. Acta (BBA)—Gene Regul. Mech. 2016, 1859, 71–81. [Google Scholar] [CrossRef]
- Meijer, H.A.; Smith, E.M.; Bushell, M. Regulation of MiRNA Strand Selection: Follow the Leader? Biochem. Soc. Trans. 2014, 42, 1135–1140. [Google Scholar] [CrossRef]
- Ruby, J.G.; Jan, C.H.; Bartel, D.P. Intronic MicroRNA Precursors That Bypass Drosha Processing. Nature 2007, 448, 83–86. [Google Scholar] [CrossRef]
- Wen, J.; Ladewig, E.; Shenker, S.; Mohammed, J.; Lai, E.C. Analysis of Nearly One Thousand Mammalian Mirtrons Reveals Novel Features of Dicer Substrates. PLoS Comput. Biol. 2015, 11, e1004441. [Google Scholar] [CrossRef] [PubMed]
- Wong, A.C.H.; Rasko, J.E.J. Splice and Dice: Intronic MicroRNAs, Splicing and Cancer. Biomedicines 2021, 9, 1268. [Google Scholar] [CrossRef]
- Monteys, A.M.; Spengler, R.M.; Wan, J.; Tecedor, L.; Lennox, K.A.; Xing, Y.; Davidson, B.L. Structure and Activity of Putative Intronic MiRNA Promoters. RNA 2010, 16, 495–505. [Google Scholar] [CrossRef] [PubMed]
- Marsico, A.; Huska, M.R.; Lasserre, J.; Hu, H.; Vucicevic, D.; Musahl, A.; Orom, U.; Vingron, M. PROmiRNA: A New MiRNA Promoter Recognition Method Uncovers the Complex Regulation of Intronic MiRNAs. Genome Biol. 2013, 14, R84. [Google Scholar] [CrossRef] [PubMed]
- Veronese, A.; Visone, R.; Consiglio, J.; Acunzo, M.; Lupini, L.; Kim, T.; Ferracin, M.; Lovat, F.; Miotto, E.; Balatti, V.; et al. Mutated β-Catenin Evades a MicroRNA-Dependent Regulatory Loop. Proc. Natl. Acad. Sci. USA 2011, 108, 4840–4845. [Google Scholar] [CrossRef]
- Gao, X.; Qiao, Y.; Han, D.; Zhang, Y.; Ma, N. Enemy or Partner: Relationship between Intronic Micrornas and Their Host Genes. IUBMB Life 2012, 64, 835–840. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Target Recognition and Regulatory Functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef]
- Helwak, A.; Kudla, G.; Dudnakova, T.; Tollervey, D. Mapping the Human MiRNA Interactome by CLASH Reveals Frequent Noncanonical Binding. Cell 2013, 153, 654–665. [Google Scholar] [CrossRef]
- Xu, W.; San Lucas, A.; Wang, Z.; Liu, Y. Identifying MicroRNA Targets in Different Gene Regions. BMC Bioinform. 2014, 15, S4. [Google Scholar] [CrossRef]
- Ørom, U.A.; Nielsen, F.C.; Lund, A.H. MicroRNA-10a Binds the 5′UTR of Ribosomal Protein MRNAs and Enhances Their Translation. Mol. Cell 2008, 30, 460–471. [Google Scholar] [CrossRef]
- Dharap, A.; Pokrzywa, C.; Murali, S.; Pandi, G.; Vemuganti, R. MicroRNA MiR-324-3p Induces Promoter-Mediated Expression of RelA Gene. PLoS ONE 2013, 8, e79467. [Google Scholar] [CrossRef] [PubMed]
- Place, R.F.; Li, L.-C.; Pookot, D.; Noonan, E.J.; Dahiya, R. MicroRNA-373 Induces Expression of Genes with Complementary Promoter Sequences. Proc. Natl. Acad. Sci. USA 2008, 105, 1608–1613. [Google Scholar] [CrossRef] [PubMed]
- Saliminejad, K.; Khorram Khorshid, H.R.; Soleymani Fard, S.; Ghaffari, S.H. An Overview of MicroRNAs: Biology, Functions, Therapeutics, and Analysis Methods. J. Cell. Physiol. 2019, 234, 5451–5465. [Google Scholar] [CrossRef] [PubMed]
- Frankel, D.; Delecourt, V.; Harhouri, K.; De Sandre-Giovannoli, A.; Lévy, N.; Kaspi, E.; Roll, P. MicroRNAs in Hereditary and Sporadic Premature Aging Syndromes and Other Laminopathies. Aging Cell 2018, 17, e12766. [Google Scholar] [CrossRef]
- Pathania, A.S.; Prathipati, P.; Pandey, M.K.; Byrareddy, S.N.; Coulter, D.W.; Gupta, S.C.; Challagundla, K.B. The Emerging Role of Non-Coding RNAs in the Epigenetic Regulation of Pediatric Cancers. Semin. Cancer Biol. 2022, 83, 227–241. [Google Scholar] [CrossRef]
- Beilerli, A.; Gareev, I.; Beylerli, O.; Yang, G.; Pavlov, V.; Aliev, G.; Ahmad, A. Circular RNAs as Biomarkers and Therapeutic Targets in Cancer. Semin. Cancer Biol. 2022, 83, 242–252. [Google Scholar] [CrossRef]
- Mirzaei, S.; Paskeh, M.D.A.; Okina, E.; Gholami, M.H.; Hushmandi, K.; Hashemi, M.; Kalu, A.; Zarrabi, A.; Nabavi, N.; Rabiee, N.; et al. Molecular Landscape of LncRNAs in Prostate Cancer: A Focus on Pathways and Therapeutic Targets for Intervention. J. Exp. Clin. Cancer Res. 2022, 41, 214. [Google Scholar] [CrossRef]
- Tardif, G.; Pelletier, J.-P.; Fahmi, H.; Hum, D.; Zhang, Y.; Kapoor, M.; Martel-Pelletier, J. NFAT3 and TGF-β/SMAD3 Regulate the Expression of MiR-140 in Osteoarthritis. Arthritis Res. Ther. 2013, 15, R197. [Google Scholar] [CrossRef]
- Tuddenham, L.; Wheeler, G.; Ntounia-Fousara, S.; Waters, J.; Hajihosseini, M.K.; Clark, I.; Dalmay, T. The Cartilage Specific MicroRNA-140 Targets Histone Deacetylase 4 in Mouse Cells. FEBS Lett. 2006, 580, 4214–4217. [Google Scholar] [CrossRef]
- Ghafouri-Fard, S.; Bahroudi, Z.; Shoorei, H.; Abak, A.; Ahin, M.; Taheri, M. MicroRNA-140: A MiRNA with Diverse Roles in Human Diseases. Biomed. Pharmacother. 2021, 135, 111256. [Google Scholar] [CrossRef]
- Wang, P.; Lv, C.; Zhang, T.; Liu, J.; Yang, J.; Guan, F.; Hong, T. FOXQ1 Regulates Senescence-Associated Inflammation via Activation of SIRT1 Expression. Cell Death Dis. 2017, 8, e2946. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, J.; Wang, X.; Wang, K. MiR-140-3p Inhibits Bladder Cancer Cell Proliferation and Invasion by Targeting FOXQ1. Aging 2020, 12, 20366–20379. [Google Scholar] [CrossRef]
- Chen, L.; Liu, J.; Tao, X.; Wang, G.; Wang, Q.; Liu, X. The Role of Pin1 Protein in Aging of Human Tendon Stem/Progenitor Cells. Biochem. Biophys. Res. Commun. 2015, 464, 487–492. [Google Scholar] [CrossRef]
- Basak, I.; Patil, K.S.; Alves, G.; Larsen, J.P.; Møller, S.G. MicroRNAs as Neuroregulators, Biomarkers and Therapeutic Agents in Neurodegenerative Diseases. Cell Mol. Life Sci. 2016, 73, 811–827. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Mu, Y.; Tian, H.; Wang, D.; Zhang, S.; Wang, H.; Liu, Y.; Di, C. MicroRNA-140 Silencing Represses the Incidence of Alzheimer’s Disease. Neurosci. Lett. 2021, 758, 135674. [Google Scholar] [CrossRef] [PubMed]
- Akhter, R.; Shao, Y.; Shaw, M.; Formica, S.; Khrestian, M.; Leverenz, J.B.; Bekris, L.M. Regulation of ADAM10 by MiR-140-5p and Potential Relevance for Alzheimer’s Disease. Neurobiol. Aging 2018, 63, 110–119. [Google Scholar] [CrossRef]
- Gullett, J.M.; Chen, Z.; O’Shea, A.; Akbar, M.; Bian, J.; Rani, A.; Porges, E.C.; Foster, T.C.; Woods, A.J.; Modave, F.; et al. MicroRNA Predicts Cognitive Performance in Healthy Older Adults. Neurobiol. Aging 2020, 95, 186–194. [Google Scholar] [CrossRef]
- Kayano, M.; Higaki, S.; Satoh, J.; Matsumoto, K.; Matsubara, E.; Takikawa, O.; Niida, S. Plasma MicroRNA Biomarker Detection for Mild Cognitive Impairment Using Differential Correlation Analysis. Biomark. Res. 2016, 4, 22. [Google Scholar] [CrossRef]
- Sørensen, S.S.; Nygaard, A.-B.; Nielsen, M.-Y.; Jensen, K.; Christensen, T. MiRNA Expression Profiles in Cerebrospinal Fluid and Blood of Patients with Acute Ischemic Stroke. Transl. Stroke Res. 2014, 5, 711–718. [Google Scholar] [CrossRef]
- Song, W.; Wang, T.; Shi, B.; Wu, Z.; Wang, W.; Yang, Y. Neuroprotective Effects of MicroRNA-140-5p on Ischemic Stroke in Mice via Regulation of the TLR4/NF-ΚB Axis. Brain Res. Bull. 2021, 168, 8–16. [Google Scholar] [CrossRef]
- Wang, S.; Cui, Y.; Xu, J.; Gao, H. MiR-140-5p Attenuates Neuroinflammation and Brain Injury in Rats Following Intracerebral Hemorrhage by Targeting TLR4. Inflammation 2019, 42, 1869–1877. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Tao, S.; Liu, L.; Guo, D.; Xia, Z.; Huang, M. MiR-140-5p Regulates Angiogenesis Following Ischemic Stroke by Targeting VEGFA. Mol. Med. Rep. 2016, 13, 4499–4505. [Google Scholar] [CrossRef] [PubMed]
- Yi, M.; Li, Y.; Wang, D.; Zhang, Q.; Yang, L.; Yang, C. KCNQ1OT1 Exacerbates Ischemia–Reperfusion Injury Through Targeted Inhibition of MiR-140-3P. Inflammation 2020, 43, 1832–1845. [Google Scholar] [CrossRef] [PubMed]
- Available online: Https://www.Who.Int/News-Room/Fact-Sheets/Detail/Cardiovascular-Diseases-(Cvds) (accessed on 1 September 2022).
- Borén, J.; Chapman, M.J.; Krauss, R.M.; Packard, C.J.; Bentzon, J.F.; Binder, C.J.; Daemen, M.J.; Demer, L.L.; Hegele, R.A.; Nicholls, S.J.; et al. Low-Density Lipoproteins Cause Atherosclerotic Cardiovascular Disease: Pathophysiological, Genetic, and Therapeutic Insights: A Consensus Statement from the European Atherosclerosis Society Consensus Panel. Eur. Heart J. 2020, 41, 2313–2330. [Google Scholar] [CrossRef] [PubMed]
- Weber, C.; Noels, H. Atherosclerosis: Current Pathogenesis and Therapeutic Options. Nat. Med. 2011, 17, 1410–1422. [Google Scholar] [CrossRef]
- Liu, H.; Mao, Z.; Zhu, J.; Shen, M.; Chen, F. MiR-140-5p Inhibits Oxidized Low-Density Lipoprotein-Induced Oxidative Stress and Cell Apoptosis via Targeting Toll-like Receptor 4. Gene Ther. 2021, 28, 413–421. [Google Scholar] [CrossRef]
- Bennett, M.R.; Sinha, S.; Owens, G.K. Vascular Smooth Muscle Cells in Atherosclerosis. Circ. Res. 2016, 118, 692–702. [Google Scholar] [CrossRef]
- Zhao, Z.-W.; Zhang, M.; Liao, L.-X.; Zou, J.; Wang, G.; Wan, X.-J.; Zhou, L.; Li, H.; Qin, Y.-S.; Yu, X.-H.; et al. Long Non-Coding RNA PCA3 Inhibits Lipid Accumulation and Atherosclerosis through the MiR-140-5p/RFX7/ABCA1 Axis. Biochim. Et Biophys. Acta (BBA)—Mol. Cell Biol. Lipids 2021, 1866, 158904. [Google Scholar] [CrossRef]
- Yu, X.-H.; Zhang, D.-W.; Zheng, X.-L.; Tang, C.-K. Cholesterol Transport System: An Integrated Cholesterol Transport Model Involved in Atherosclerosis. Prog. Lipid Res. 2019, 73, 65–91. [Google Scholar] [CrossRef]
- Liu, Q.; Ren, K.; Liu, S.; Li, W.; Huang, C.; Yang, X. MicroRNA-140-5p Aggravates Hypertension and Oxidative Stress of Atherosclerosis via Targeting Nrf2 and Sirt2. Int. J. Mol. Med. 2018, 43, 839–849. [Google Scholar] [CrossRef]
- Kaitsuka, T.; Matsushita, M.; Matsushita, N. Regulation of Hypoxic Signaling and Oxidative Stress via the MicroRNA–SIRT2 Axis and Its Relationship with Aging-Related Diseases. Cells 2021, 10, 3316. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Qi, Y.; Xu, L.; Tao, X.; Han, X.; Yin, L.; Peng, J. MicroRNA-140-5p Aggravates Doxorubicin-Induced Cardiotoxicity by Promoting Myocardial Oxidative Stress via Targeting Nrf2 and Sirt2. Redox Biol. 2018, 15, 284–296. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Li, Y.; Peng, H.; Zhao, Y. MiR-140-5p Regulates Vascular Smooth Muscle Cell Viability, Migration and Apoptosis by Targeting ROBO4 Gene Expression in Atherosclerosis. Mol. Med. Rep. 2021, 23, 213. [Google Scholar] [CrossRef]
- Xu, Y.; Gao, J.; Gong, Y.; Chen, M.; Chen, J.; Zhao, W.; Tan, S. Hsa-MiR-140-5p down-Regulates LDL Receptor and Attenuates LDL-C Uptake in Human Hepatocytes. Atherosclerosis 2020, 297, 111–119. [Google Scholar] [CrossRef]
- Teng, L.; Meng, R. Long Non-Coding RNA MALAT1 Promotes Acute Cerebral Infarction Through MiRNAs-Mediated Hs-CRP Regulation. J. Mol. Neurosci. 2019, 69, 494–504. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Liu, X.; Sun, N.; Wang, T.; Zhu, J.; Yang, S.; Song, X.; Wang, R.; Wang, X.; Zhao, Y.; et al. Differentially Expressed Circular Non-Coding RNAs in Atherosclerotic Aortic Vessels and Their Potential Functions in Endothelial Injury. Front. Cardiovasc. Med. 2021, 8, 657544. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, M.; Shimizu, I.; Nagasawa, A.; Yoshida, Y.; Katsuumi, G.; Wakasugi, T.; Hayashi, Y.; Ikegami, R.; Suda, M.; Ota, Y.; et al. P53 Plays a Crucial Role in Endothelial Dysfunction Associated with Hyperglycemia and Ischemia. J. Mol. Cell. Cardiol. 2019, 129, 105–117. [Google Scholar] [CrossRef]
- Zhang, H.; Ji, N.; Gong, X.; Ni, S.; Wang, Y. NEAT1/MiR-140-3p/MAPK1 Mediates the Viability and Survival of Coronary Endothelial Cells and Affects Coronary Atherosclerotic Heart Disease. Acta Biochim. Et Biophys. Sin. 2020, 52, 967–974. [Google Scholar] [CrossRef]
- Yang, L.; Chu, Y.; Wang, Y.; Zhao, X.; Xu, W.; Zhang, P.; Liu, X.; Dong, S.; He, W.; Gao, C. SiRNA-Mediated Silencing of Wnt5a Regulates Inflammatory Responses in Atherosclerosis through the MAPK/NF-ΚB Pathways. Int. J. Mol. Med. 2014, 34, 1147–1152. [Google Scholar] [CrossRef][Green Version]
- Zhu, Z.-R.; He, Q.; Wu, W.-B.; Chang, G.-Q.; Yao, C.; Zhao, Y.; Wang, M.; Wang, S.-M. MiR-140-3p Is Involved in In-Stent Restenosis by Targeting C-Myb and BCL-2 in Peripheral Artery Disease. J. Atheroscler. Thromb. 2018, 25, 1168–1181. [Google Scholar] [CrossRef]
- Glatz, M.; Riedl, R.; Glatz, W.; Schneider, M.; Wedrich, A.; Bolz, M.; Strauss, R.W. Blindness and Visual Impairment in Central Europe. PLoS ONE 2022, 17, e0261897. [Google Scholar] [CrossRef] [PubMed]
- Velez-Montoya, R.; Oliver, S.C.N.; Olson, J.L.; Fine, S.L.; Quiroz-Mercado, H.; Mandava, N. Current knowledge and trends in age-related macular degeneration: Genetics, Epidemiology, and Prevention. Retina 2014, 34, 423–441. [Google Scholar] [CrossRef] [PubMed]
- Ertekin, S.; Yıldırım, O.; Dinç, E.; Ayaz, L.; Fidancı, S.B.; Tamer, L. Evaluation of Circulating MiRNAs in Wet Age-Related Macular Degeneration. Mol. Vis. 2014, 20, 1057–1066. [Google Scholar] [PubMed]
- Yu, X.; Zheng, H.; Chan, M.T.; Wu, W.K.K. MicroRNAs: New Players in Cataract. Am. J. Transl. Res. 2017, 9, 3896–3903. [Google Scholar] [PubMed]
- Drake, M.T.; Clarke, B.L.; Lewiecki, E.M. The Pathophysiology and Treatment of Osteoporosis. Clin. Ther. 2015, 37, 1837–1850. [Google Scholar] [CrossRef]
- Benedetti, M.G.; Furlini, G.; Zati, A.; Letizia Mauro, G. The Effectiveness of Physical Exercise on Bone Density in Osteoporotic Patients. BioMed Res. Int. 2018, 2018, 4840531. [Google Scholar] [CrossRef]
- Zhu, G.; Zeng, C.; Qian, Y.; Yuan, S.; Ye, Z.; Zhao, S.; Li, R. Tensile Strain Promotes Osteogenic Differentiation of Bone Marrow Mesenchymal Stem Cells through Upregulating LncRNA-MEG3. Histol. Histopathol. 2021, 36, 939–946. [Google Scholar] [CrossRef]
- Kocijan, R.; Muschitz, C.; Geiger, E.; Skalicky, S.; Baierl, A.; Dormann, R.; Plachel, F.; Feichtinger, X.; Heimel, P.; Fahrleitner-Pammer, A.; et al. Circulating MicroRNA Signatures in Patients With Idiopathic and Postmenopausal Osteoporosis and Fragility Fractures. J. Clin. Endocrinol. Metab. 2016, 101, 4125–4134. [Google Scholar] [CrossRef]
- Ramírez-Salazar, E.G.; Carrillo-Patiño, S.; Hidalgo-Bravo, A.; Rivera-Paredez, B.; Quiterio, M.; Ramírez-Palacios, P.; Patiño, N.; Valdés-Flores, M.; Salmerón, J.; Velázquez-Cruz, R. Serum MiRNAs MiR-140-3p and MiR-23b-3p as Potential Biomarkers for Osteoporosis and Osteoporotic Fracture in Postmenopausal Mexican-Mestizo Women. Gene 2018, 679, 19–27. [Google Scholar] [CrossRef]
- Yin, R.; Jiang, J.; Deng, H.; Wang, Z.; Gu, R.; Wang, F. MiR-140-3p Aggregates Osteoporosis by Targeting PTEN and Activating PTEN/PI3K/AKT Signaling Pathway. Hum. Cell 2020, 33, 569–581. [Google Scholar] [CrossRef]
- Li, K.-C.; Chang, Y.-H.; Yeh, C.-L.; Hu, Y.-C. Healing of Osteoporotic Bone Defects by Baculovirus-Engineered Bone Marrow-Derived MSCs Expressing MicroRNA Sponges. Biomaterials 2016, 74, 155–166. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.-Z.; Huang, H.-T.; Cheng, T.-L.; Lu, Y.-M.; Lin, S.-Y.; Ho, C.-J.; Lee, T.-C.; Hsu, C.-H.; Huang, P.-J.; Huang, H.H.; et al. Application of MicroRNA in Human Osteoporosis and Fragility Fracture: A Systemic Review of Literatures. Int. J. Mol. Sci. 2021, 22, 5232. [Google Scholar] [CrossRef] [PubMed]
- Fushimi, S.; Nohno, T.; Nagatsuka, H.; Katsuyama, H. Involvement of MiR-140-3p in Wnt3a and TGFβ3 Signaling Pathways during Osteoblast Differentiation in MC3T3-E1 Cells. Genes Cells 2018, 23, 517–527. [Google Scholar] [CrossRef] [PubMed]
- Mao, J.-H.; Sui, Y.-X.; Ao, S.; Wang, Y.; Liu, Y.; Leng, H. MiR-140-3p Exhibits Repressive Functions on Preosteoblast Viability and Differentiation by Downregulating MCF2L in Osteoporosis. In Vitro Cell Dev. Biol. Anim. 2020, 56, 49–58. [Google Scholar] [CrossRef]
- Loeser, R.F.; Collins, J.A.; Diekman, B.O. Ageing and the Pathogenesis of Osteoarthritis. Nat. Rev. Rheumatol. 2016, 12, 412–420. [Google Scholar] [CrossRef]
- Woods, S.; Charlton, S.; Cheung, K.; Hao, Y.; Soul, J.; Reynard, L.N.; Crowe, N.; Swingler, T.E.; Skelton, A.J.; Piróg, K.A.; et al. MicroRNA-Seq of Cartilage Reveals an Overabundance of MiR-140-3p Which Contains Functional IsomiRs. RNA 2020, 26, 1575–1588. [Google Scholar] [CrossRef]
- Duan, L.; Liang, Y.; Xu, X.; Xiao, Y.; Wang, D. Recent Progress on the Role of MiR-140 in Cartilage Matrix Remodelling and Its Implications for Osteoarthritis Treatment. Arthritis Res. Ther. 2020, 22, 194. [Google Scholar] [CrossRef]
- Miyaki, S.; Nakasa, T.; Otsuki, S.; Grogan, S.P.; Higashiyama, R.; Inoue, A.; Kato, Y.; Sato, T.; Lotz, M.K.; Asahara, H. MicroRNA-140 Is Expressed in Differentiated Human Articular Chondrocytes and Modulates Interleukin-1 Responses. Arthritis Rheum. 2009, 60, 2723–2730. [Google Scholar] [CrossRef]
- Miyaki, S.; Sato, T.; Inoue, A.; Otsuki, S.; Ito, Y.; Yokoyama, S.; Kato, Y.; Takemoto, F.; Nakasa, T.; Yamashita, S.; et al. MicroRNA-140 Plays Dual Roles in Both Cartilage Development and Homeostasis. Genes Dev. 2010, 24, 1173–1185. [Google Scholar] [CrossRef]
- Castanheira, C.I.G.D.; Anderson, J.R.; Fang, Y.; Milner, P.I.; Goljanek-Whysall, K.; House, L.; Clegg, P.D.; Peffers, M.J. Mouse MicroRNA Signatures in Joint Ageing and Post-Traumatic Osteoarthritis. Osteoarthr. Cartil. Open 2021, 3, 100186. [Google Scholar] [CrossRef]
- Tardif, G.; Hum, D.; Pelletier, J.-P.; Duval, N.; Martel-Pelletier, J. Regulation of the IGFBP-5 and MMP-13 Genes by the MicroRNAs MiR-140 and MiR-27a in Human Osteoarthritic Chondrocytes. BMC Musculoskelet. Disord. 2009, 10, 148. [Google Scholar] [CrossRef] [PubMed]
- Si, H.-B.; Yang, T.-M.; Li, L.; Tian, M.; Zhou, L.; Li, D.-P.; Huang, Q.; Kang, P.; Yang, J.; Zhou, Z.-K.; et al. MiR-140 Attenuates the Progression of Early-Stage Osteoarthritis by Retarding Chondrocyte Senescence. Mol. Ther. Nucleic Acids 2020, 19, 15–30. [Google Scholar] [CrossRef] [PubMed]
- Si, H.-B.; Zeng, Y.; Liu, S.-Y.; Zhou, Z.-K.; Chen, Y.-N.; Cheng, J.-Q.; Lu, Y.-R.; Shen, B. Intra-Articular Injection of MicroRNA-140 (MiRNA-140) Alleviates Osteoarthritis (OA) Progression by Modulating Extracellular Matrix (ECM) Homeostasis in Rats. Osteoarthr. Cartil. 2017, 25, 1698–1707. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, S.; Miyaki, S.; Kato, Y.; Yokoyama, S.; Sato, T.; Barrionuevo, F.; Akiyama, H.; Scherer, G.; Takada, S.; Asahara, H. L-Sox5 and Sox6 Proteins Enhance Chondrogenic MiR-140 MicroRNA Expression by Strengthening Dimeric Sox9 Activity. J. Biol. Chem. 2012, 287, 22206–22215. [Google Scholar] [CrossRef] [PubMed]
- Papathanasiou, I.; Trachana, V.; Mourmoura, E.; Tsezou, A. DNA Methylation Regulates MiR-140-5p and MiR-146a Expression in Osteoarthritis. Life Sci. 2019, 228, 274–284. [Google Scholar] [CrossRef]
- Wei, W.; He, S.; Wang, Z.; Dong, J.; Xiang, D.; Li, Y.; Ren, L.; Kou, N.; Lv, J. LINC01534 Promotes the Aberrant Metabolic Dysfunction and Inflammation in IL-1β-Simulated Osteoarthritic Chondrocytes by Targeting MiR-140-5p. Cartilage 2021, 13, 898S–907S. [Google Scholar] [CrossRef]
- Cao, F.; Chen, Y.; Wang, X.; Wu, L.-M.; Tian, M.; Li, H.-Y.; Si, H.-B.; Shen, B. Therapeutic Effect and Potential Mechanisms of Intra-Articular Injections of MiR-140-5p on Early-Stage Osteoarthritis in Rats. Int. Immunopharmacol. 2021, 96, 107786. [Google Scholar] [CrossRef]
- Ren, T.; Wei, P.; Song, Q.; Ye, Z.; Wang, Y.; Huang, L. MiR-140-3p Ameliorates the Progression of Osteoarthritis via Targeting CXCR4. Biol. Pharm. Bull. 2020, 43, 810–816. [Google Scholar] [CrossRef]
- Karlsen, T.A.; Jakobsen, R.B.; Mikkelsen, T.S.; Brinchmann, J.E. MicroRNA-140 Targets RALA and Regulates Chondrogenic Differentiation of Human Mesenchymal Stem Cells by Translational Enhancement of SOX9 and ACAN. Stem. Cells Dev. 2014, 23, 290–304. [Google Scholar] [CrossRef]
- Karlsen, T.A.; de Souza, G.A.; Ødegaard, B.; Engebretsen, L.; Brinchmann, J.E. MicroRNA-140 Inhibits Inflammation and Stimulates Chondrogenesis in a Model of Interleukin 1β-Induced Osteoarthritis. Mol. Ther. Nucleic Acids 2016, 5, e373. [Google Scholar] [CrossRef]
- Liang, Y.; Duan, L.; Xiong, J.; Zhu, W.; Liu, Q.; Wang, D.; Liu, W.; Li, Z.; Wang, D. E2 Regulates MMP-13 via Targeting MiR-140 in IL-1β-Induced Extracellular Matrix Degradation in Human Chondrocytes. Arthritis Res. Ther. 2016, 18, 105. [Google Scholar] [CrossRef]
- Li, W.; Zhao, S.; Yang, H.; Zhang, C.; Kang, Q.; Deng, J.; Xu, Y.; Ding, Y.; Li, S. Potential Novel Prediction of TMJ-OA: MiR-140-5p Regulates Inflammation Through Smad/TGF-β Signaling. Front. Pharmacol. 2019, 10, 15. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Qiu, J.; Shi, J.; Liu, S.; Zou, H. MicroRNA-140-5p Represses Chondrocyte Pyroptosis and Relieves Cartilage Injury in Osteoarthritis by Inhibiting Cathepsin B/Nod-like Receptor Protein 3. Bioengineered 2021, 12, 9949–9964. [Google Scholar] [CrossRef] [PubMed]
- Swingler, T.E.; Niu, L.; Smith, P.; Paddy, P.; Le, L.; Barter, M.J.; Young, D.A.; Clark, I.M. The Function of MicroRNAs in Cartilage and Osteoarthritis. Clin. Exp. Rheumatol. 2019, 37 (Suppl. S120), 40–47. [Google Scholar] [PubMed]
- Wang, Z.; Huang, C.; Zhao, C.; Zhang, H.; Zhen, Z.; Xu, D. Knockdown of LINC01385 Inhibits Osteoarthritis Progression by Modulating the MicroRNA-140-3p/TLR4 Axis. Exp. Ther. Med. 2021, 22, 1244. [Google Scholar] [CrossRef] [PubMed]
- Kwak, Y.H.; Kwak, D.-K.; Moon, H.-S.; Kim, N.Y.; Yee, J.-S.; Yoo, J.-H. Significant Changes in Serum MicroRNAs after High Tibial Osteotomy in Medial Compartmental Knee Osteoarthritis: Potential Prognostic Biomarkers. Diagnostics 2021, 11, 258. [Google Scholar] [CrossRef]
- Tao, S.-C.; Yuan, T.; Zhang, Y.-L.; Yin, W.-J.; Guo, S.-C.; Zhang, C.-Q. Exosomes Derived from MiR-140-5p-Overexpressing Human Synovial Mesenchymal Stem Cells Enhance Cartilage Tissue Regeneration and Prevent Osteoarthritis of the Knee in a Rat Model. Theranostics 2017, 7, 180–195. [Google Scholar] [CrossRef]
- Chaudhry, N.; Muhammad, H.; Seidl, C.; Downes, D.; Young, D.A.; Hao, Y.; Zhu, L.; Vincent, T.L. Highly Efficient CRISPR-Cas9-Mediated Editing Identifies Novel Mechanosensitive MicroRNA-140 Targets in Primary Human Articular Chondrocytes. Osteoarthr. Cartil. 2022, 30, 596–604. [Google Scholar] [CrossRef]
- Ugalde, A.P.; Español, Y.; López-Otín, C. Micromanaging Aging with MiRNAs: New Messages from the Nuclear Envelope. Nucleus 2011, 2, 549–555. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Toury, L.; Frankel, D.; Airault, C.; Magdinier, F.; Roll, P.; Kaspi, E. miR-140-5p and miR-140-3p: Key Actors in Aging-Related Diseases? Int. J. Mol. Sci. 2022, 23, 11439. https://doi.org/10.3390/ijms231911439
Toury L, Frankel D, Airault C, Magdinier F, Roll P, Kaspi E. miR-140-5p and miR-140-3p: Key Actors in Aging-Related Diseases? International Journal of Molecular Sciences. 2022; 23(19):11439. https://doi.org/10.3390/ijms231911439
Chicago/Turabian StyleToury, Léa, Diane Frankel, Coraline Airault, Frédérique Magdinier, Patrice Roll, and Elise Kaspi. 2022. "miR-140-5p and miR-140-3p: Key Actors in Aging-Related Diseases?" International Journal of Molecular Sciences 23, no. 19: 11439. https://doi.org/10.3390/ijms231911439
APA StyleToury, L., Frankel, D., Airault, C., Magdinier, F., Roll, P., & Kaspi, E. (2022). miR-140-5p and miR-140-3p: Key Actors in Aging-Related Diseases? International Journal of Molecular Sciences, 23(19), 11439. https://doi.org/10.3390/ijms231911439





