A Novel Beta-Glucosidase Gene for Plant Type Was Identified by Genome-Wide Association Study and Gene Co-Expression Analysis in Widespread Bermudagrass
Abstract
1. Introduction
2. Results
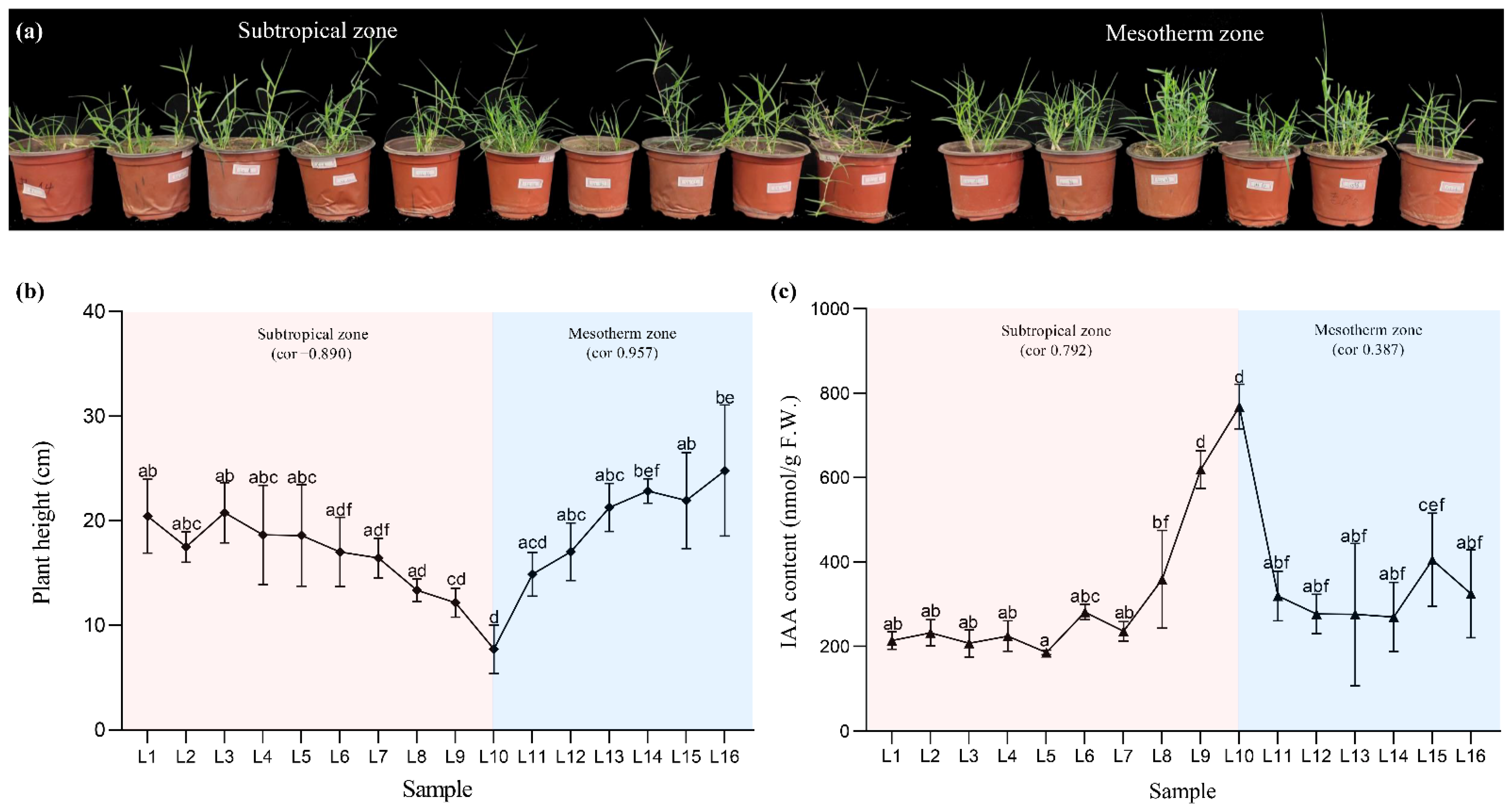
2.1. Delineating Relationship among Plant Height and IAA Content with the Latitudinal Gradient in Diversity Bermudagrass
2.2. Application of GWAS to Identify the Genetic Regions That Influence Plant Height and IAA Content in Diverse Bermudagrass
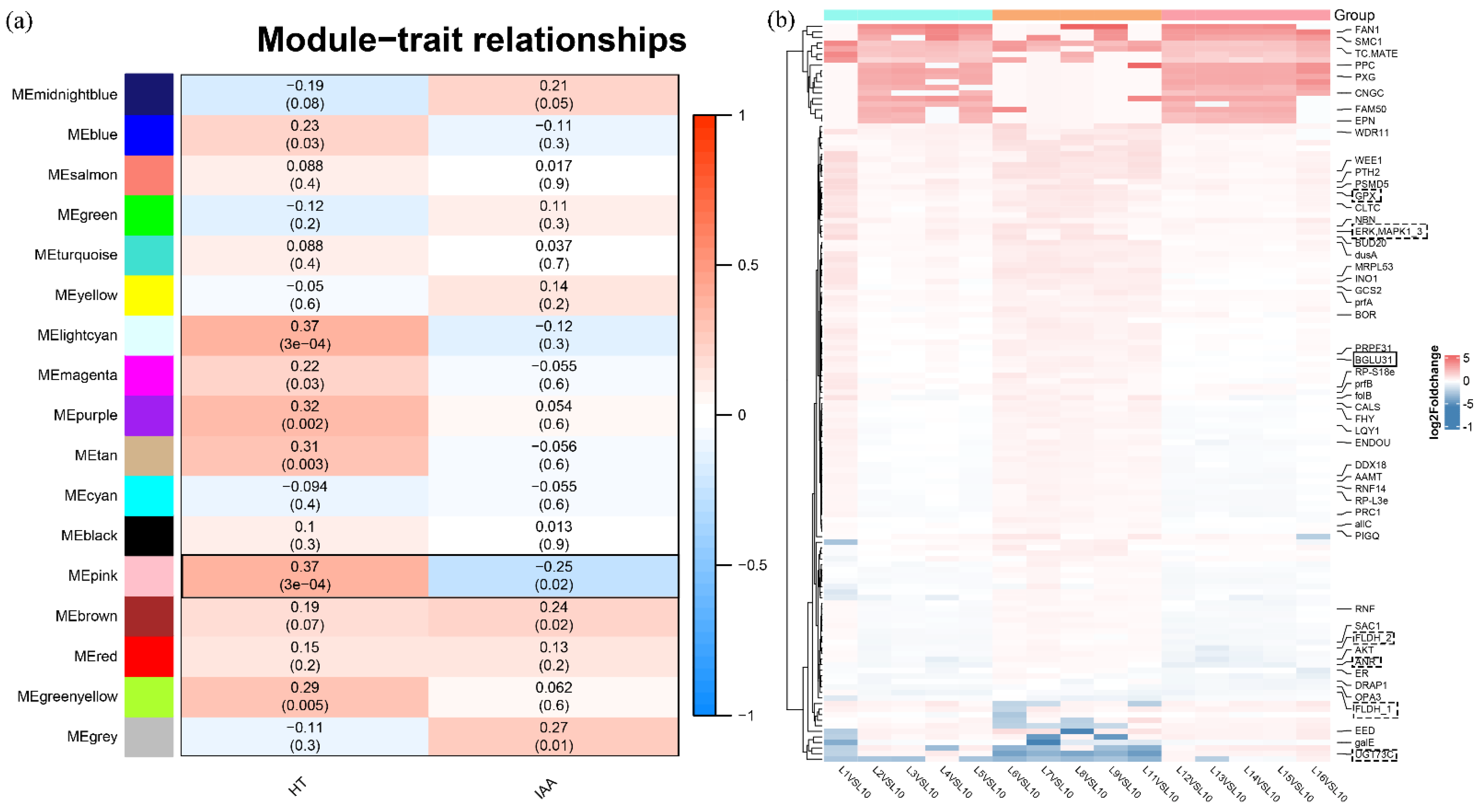
2.3. WGCNA Unraveled the Molecular Basis of the Different Phenotypes in Diverse Bermudagrass
2.4. Analysis of Candidate Gene Associated with Plant Height and IAA Content in Diverse Bermudagrass
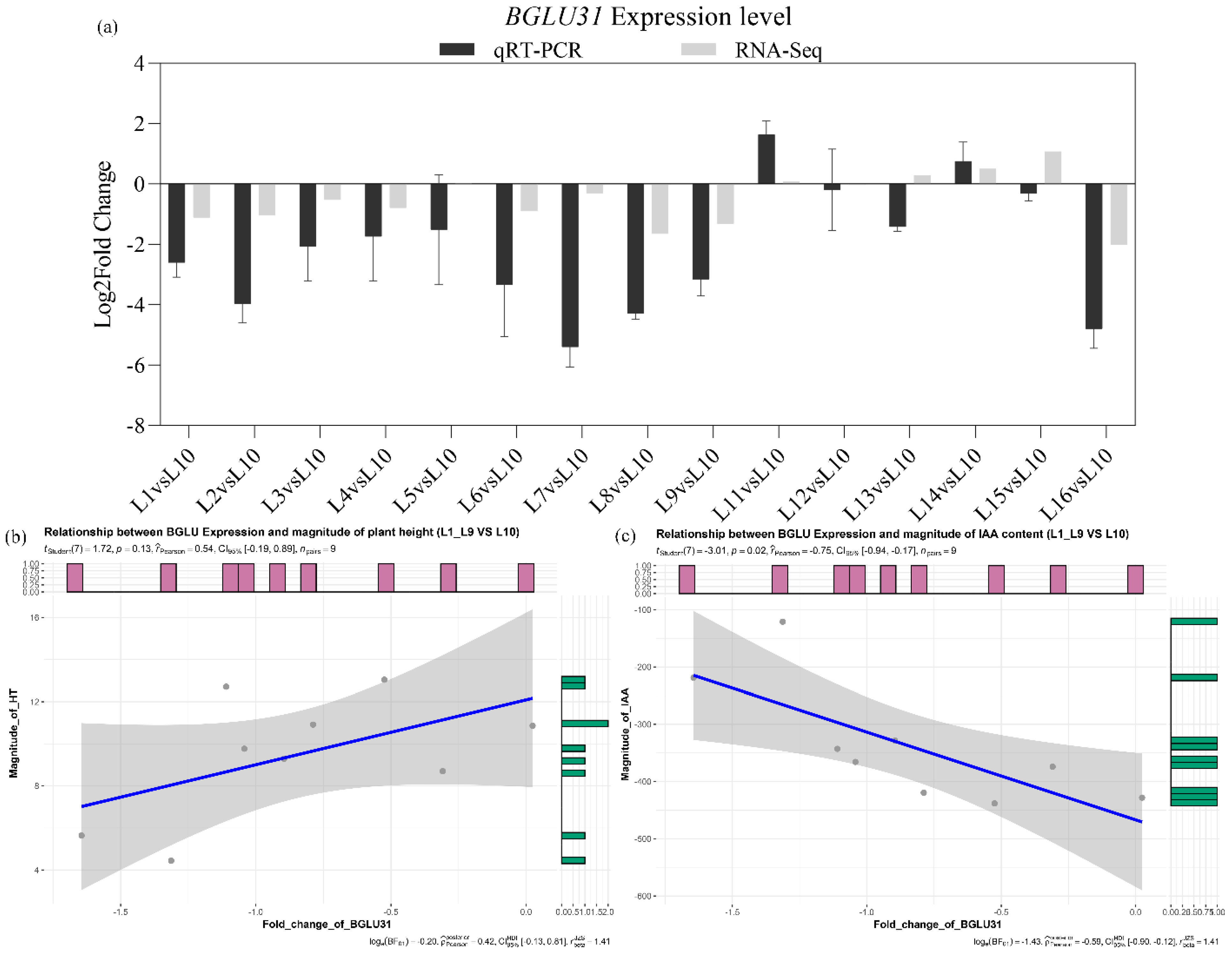
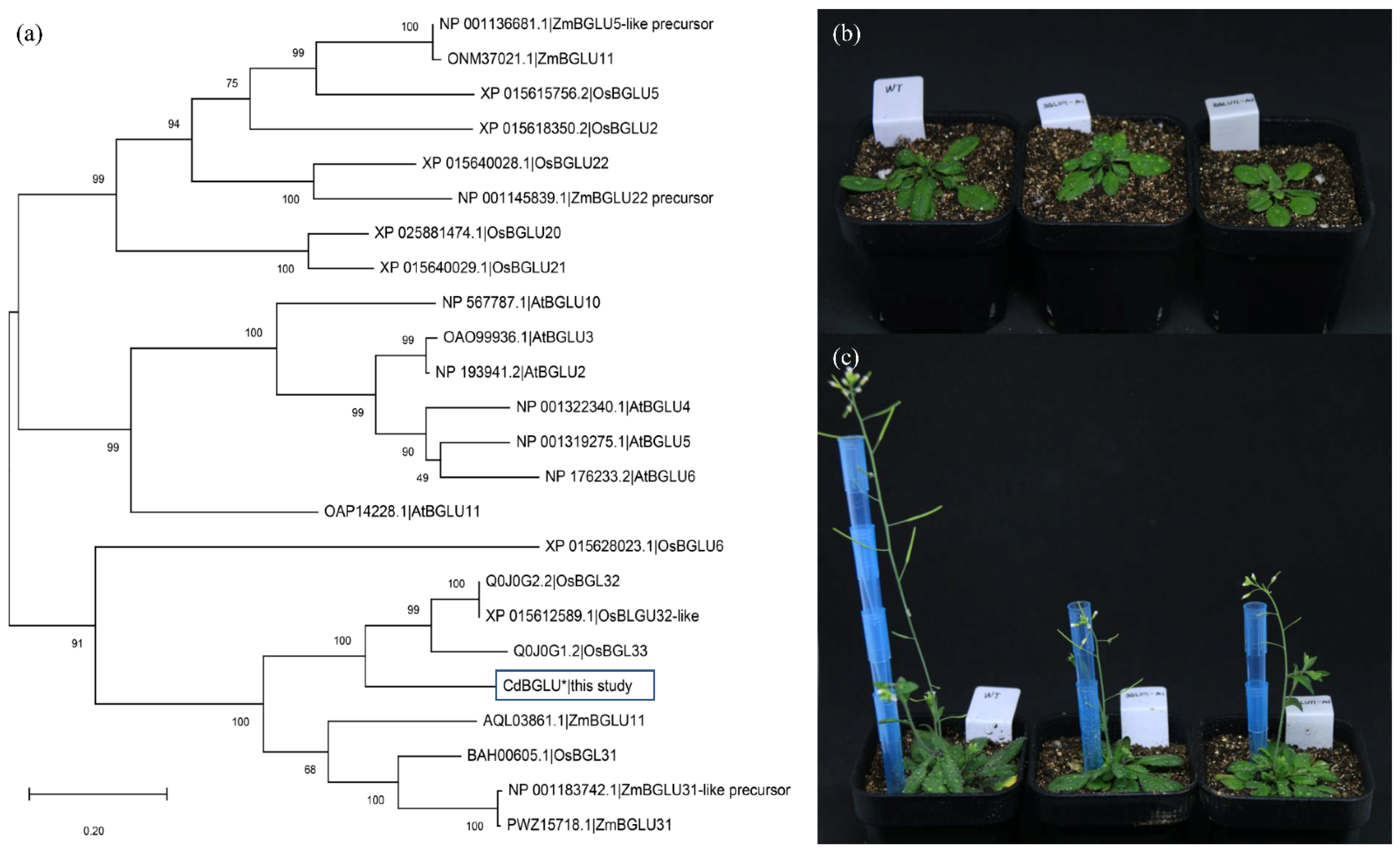
2.5. CdBGLU31 Is Likely a Key Gene for Plant Architecture and Growth Regulation in Bermudagrass
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Phenotypes
4.2. RNA Sequencing, SNP Calling, and Gene Expression
4.3. Association Analysis
4.4. Functional Analysis of Candidate Genes
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, J.; Wang, M.; Guo, Z.; Guan, Y.; Guo, Y.; Yan, X. Variations in morphological traits of bermudagrass and relationship with soil and climate along latitudinal gradients. Hereditas 2018, 155, 31. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.X.; Wang, M.L.; Guo, Z.P.; Guan, Y.Z.; Guo, Y.X.; Yan, X.B. Variation in ploidy level and genome size of Cynodon dactylon (L.) Pers. along a latitudinal gradient. Folia Geobot. 2019, 54, 267–278. [Google Scholar] [CrossRef]
- Wang, M.; Zhang, J.; Guo, Z.; Guan, Y.; Qu, G.; Liu, J.; Guo, Y.; Yan, X. Morphological variation in Cynodon dactylon (L.) Pers., and its relationship with the environment along a longitudinal gradient. Hereditas 2020, 157, 4. [Google Scholar] [CrossRef] [PubMed]
- Burton, G.W. Registration of ‘Coastcross-1’ bermudagrass. Crop Sci. 1972, 12, 125. [Google Scholar] [CrossRef]
- Burton, G.W.; Gates, R.N.; Hill, G.M. Registration of ‘Tifton 85’ bermudagrass. Crop Sci. 1993, 33, 644–645. [Google Scholar] [CrossRef]
- Anderson, W.F.; Maas, A.; Ozias-Akins, P. Genetic Variability of a Forage Bermudagrass Core Collection. Crop Sci. 2009, 49, 1347–1358. [Google Scholar] [CrossRef]
- Cui, F.; Taier, G.; Li, M.; Dai, X.; Hang, N.; Zhang, X.; Wang, X.; Wang, K. The genome of the warm-season turfgrass African bermudagrass (Cynodon transvaalensis). Hortic. Res. 2021, 8, 93. [Google Scholar] [CrossRef]
- Boutigny, M.; Monnin, D.; Filali, A.E.; Carareto, C.M.; Vieira, C.; Picard, F.; Kremer, N.; Lyon, D.; Biom, L.D.; Lyon, U. De novo identification, differential analysis and functional annotation of SNPs from RNA-seq data in non-model species. bioRxiv 2015, 035238, (Preprint). [Google Scholar]
- Lopez-Maestre, H.; Brinza, L.; Marchet, C.; Kielbassa, J.; Bastien, S.; Boutigny, M.; Monnin, D.; El Filali, A.; Carareto, C.M.; Vieira, C.; et al. SNP calling from RNA-seq data without a reference genome: Identification, quantification, differential analysis and impact on the protein sequence. Nucleic Acids Res. 2016, 44, e148. [Google Scholar] [CrossRef]
- Taliaferro, C.; Hopkins, A.; Henthorn, J.; Murphy, C.; Edwards, R. Use of flow cytometry to estimate ploidy level in Cynodon species. Int. Turfgrass Soc. Res. J. 1997, 8, 385–392. [Google Scholar]
- Grossman, A.Y.; Andrade, M.H.M.L.; Chaves, A.L.A.; Mendes Ferreira, M.T.; Techio, V.H.; Lopez, Y.; Begcy, K.; Kenworthy, K.E.; Rios, E.F. Ploidy Level and Genetic Parameters for Phenotypic Traits in Bermudagrass (Cynodon spp.) Germplasm. Agronomy 2021, 11, 912. [Google Scholar] [CrossRef]
- Willemoës, J.G.; Beltrano, J.; Montaldi, E.R. Stolon differentiation in Cynodon dactylon (L.) Pers. mediated by phytochrome. Environ. Exp. Bot. 1987, 27, 15–20. [Google Scholar] [CrossRef]
- Dong, M.; Kroon, H. Plasticity in morphology and biomass allocation in Cynodon dactylon, a grass species forming stolons and rhizomes. Oikos 1994, 70, 99–106. [Google Scholar] [CrossRef]
- Zhang, B.; Xiao, X.; Zong, J.; Chen, J.; Li, J.; Guo, H.; Liu, J. Comparative transcriptome analysis provides new insights into erect and prostrate growth in bermudagrass (Cynodon dactylon L.). Plant Physiol. Biochem. 2017, 121, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Fan, J.; Liu, J. Comparative proteomic analysis provides new insights into the specialization of shoots and stolons in bermudagrass (Cynodon dactylon L.). BMC Genom. 2019, 20, 708. [Google Scholar] [CrossRef] [PubMed]
- Beaty, E.; Engel, J. Forage quality measurements and forage research—A review, critique and interpretation. J. Range Manag. 1980, 33, 49–54. [Google Scholar] [CrossRef]
- Pornaro, C.; Menegon, A.; Macolino, S. Stolon Development in Four Turf-Type Perennial Ryegrass Cultivars. Agron. J. 2018, 110, 2159–2164. [Google Scholar] [CrossRef]
- Balatti, P.A.; Willemoes, J.G. Role of ethylene in the geotropic response of bermudagrass (Cynodon dactylon L. Pers.) stolons. Plant Physiol. 1989, 91, 1251–1254. [Google Scholar] [CrossRef]
- Yun, L.; Larson, S.R.; Mott, I.W.; Jensen, K.B.; Staub, J.E. Genetic control of rhizomes and genomic localization of a major-effect growth habit QTL in perennial wildrye. Mol. Genet. Genom. 2014, 289, 383–397. [Google Scholar] [CrossRef]
- Kenworthy, K.E.; Martin, D.L.; Taliaferro, C.M. Growth habit determination of genotypes of African bermudagrass. HortScience 2007, 42, 1513–1516. [Google Scholar] [CrossRef]
- Sever Mutlu, S.; Mutlu, N.; Tokgöz, S.; Çakır, M.; Selim, C. Development of vegetative triploid turf-type bermudagrass [Cynodon dactylon × C. transvaalensis (C. × mangennisii Hurcombe)]. Genet. Resour. Crop Evol. 2020, 67, 177–189. [Google Scholar] [CrossRef]
- Nakatsui, M.S. Water Conservation with Soil Moisture Sensor (SMS) and Irrigation Scheduling on Bermudagrass [Cynodon dactylon × C. traansvalensis (L.)] Fairways. Master’s Thesis, California State Polytechnic University, Pomona, CA, USA, 2021. [Google Scholar]
- Zhang, J.X.; Chen, M.H.; Gan, L.; Zhang, C.J.; Shen, Y.; Qian, J.; Han, M.L.; Guo, Y.X.; Yan, X.B. Diversity Patterns of Bermuda Grass along Latitudinal Gradients at Different Temperatures in Southeastern China. Plants 2020, 9, 1778. [Google Scholar] [CrossRef] [PubMed]
- Renaut, S.; Nolte, A.W.; Bernatchez, L. Mining transcriptome sequences towards identifying adaptive single nucleotide polymorphisms in lake whitefish species pairs (Coregonus spp. Salmonidae). Mol. Ecol. 2010, 19, 115–131. [Google Scholar] [CrossRef] [PubMed]
- Rogier, O.; Chateigner, A.; Amanzougarene, S.; Lesage-Descauses, M.-C.; Balzergue, S.; Brunaud, V.; Caius, J.; Soubigou-Taconnat, L.; Jorge, V.; Segura, V. Accuracy of RNAseq based SNP discovery and genotyping in Populusnigra. BMC Genom. 2018, 19, 909. [Google Scholar] [CrossRef]
- Chen, M.; Gan, L.; Zhang, J.; Shen, Y.; Qian, J.; Han, M.; Zhang, C.; Fan, J.; Sun, S.; Yan, X. A Regulatory Network of Heat Shock Modules-Photosynthesis-Redox Systems in Response to Cold Stress Across a Latitudinal Gradient in Bermudagrass. Front. Plant Sci. 2021, 12, 751901. [Google Scholar] [CrossRef]
- Slavov, G.T.; Nipper, R.; Robson, P.; Farrar, K.; Allison, G.G.; Bosch, M.; Clifton-Brown, J.C.; Donnison, I.S.; Jensen, E. Genome-wide association studies and prediction of 17 traits related to phenology, biomass and cell wall composition in the energy grass Miscanthus sinensis. New Phytol. 2014, 201, 1227–1239. [Google Scholar] [CrossRef]
- Tyler, L.; Lee, S.J.; Young, N.D.; DeIulio, G.A.; Benavente, E.; Reagon, M.; Sysopha, J.; Baldini, R.M.; Troìa, A.; Hazen, S.P. Population structure in the model grass Brachypodium distachyon is highly correlated with flowering differences across broad geographic areas. Plant Genome 2016, 9, 74. [Google Scholar] [CrossRef]
- Jaškūnė, K.; Aleliūnas, A.; Statkevičiūtė, G.; Kemešytė, V.; Studer, B.; Yates, S. Genome-wide association study to identify candidate loci for biomass formation under water deficit in perennial ryegrass. Front. Plant Sci. 2020, 11, 1923. [Google Scholar] [CrossRef]
- Henrissat, B.; Davies, G.J. Glycoside hydrolases and glycosyltransferases. Families, modules, and implications for genomics. Plant Physiol. 2000, 124, 1515–1519. [Google Scholar] [CrossRef]
- Opassiri, R.; Pomthong, B.; Onkoksoong, T.; Akiyama, T.; Esen, A.; Ketudat Cairns, J.R. Analysis of rice glycosyl hydrolase family 1 and expression of Os4bglu12 β-glucosidase. BMC Plant Biol. 2006, 6, 33. [Google Scholar] [CrossRef]
- Price, A.L.; Patterson, N.J.; Plenge, R.M.; Weinblatt, M.E.; Shadick, N.A.; Reich, D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006, 38, 904–909. [Google Scholar] [CrossRef] [PubMed]
- Luang, S.; Cho, J.-I.; Mahong, B.; Opassiri, R.; Akiyama, T.; Phasai, K.; Komvongsa, J.; Sasaki, N.; Hua, Y.-l.; Matsuba, Y. Rice Os9BGlu31 is a transglucosidase with the capacity to equilibrate phenylpropanoid, flavonoid, and phytohormone glycoconjugates. J. Biol. Chem. 2013, 288, 10111–10123. [Google Scholar] [CrossRef] [PubMed]
- Wolters, H.; Jürgens, G. Survival of the flexible: Hormonal growth control and adaptation in plant development. Nat. Rev. Genet. 2009, 10, 305–317. [Google Scholar] [CrossRef] [PubMed]
- Shkolnik-Inbar, D.; Bar-Zvi, D. ABI4 mediates abscisic acid and cytokinin inhibition of lateral root formation by reducing polar auxin transport in Arabidopsis. Plant Cell 2010, 22, 3560–3573. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Escamilla-Trevino, L.; Zeng, L.; Lalgondar, M.; Bevan, D.; Winkel, B.; Mohamed, A.; Cheng, C.L.; Shih, M.C.; Poulton, J.; et al. Functional genomic analysis of Arabidopsis thaliana glycoside hydrolase family 1. Plant Mol. Biol. 2004, 55, 343–367. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Yu, J.; Pressoir, G.; Briggs, W.H.; Vroh Bi, I.; Yamasaki, M.; Doebley, J.F.; McMullen, M.D.; Gaut, B.S.; Nielsen, D.M.; Holland, J.B. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 2006, 38, 203–208. [Google Scholar] [CrossRef]
- Liu, X.; Huang, M.; Fan, B.; Buckler, E.S.; Zhang, Z. Iterative usage of fixed and random effect models for powerful and efficient genome-wide association studies. PLoS Genet. 2016, 12, e1005767. [Google Scholar] [CrossRef]
- Yin, L.; Zhang, H.; Tang, Z.; Xu, J.; Yin, D.; Zhang, Z.; Yuan, X.; Zhu, M.; Zhao, S.; Li, X. rMVP: A memory-efficient, visualization-enhanced, and parallel-accelerated tool for genome-wide association study. Genom. Proteom. Bioinform. 2021, 19, 619–628. [Google Scholar] [CrossRef] [PubMed]
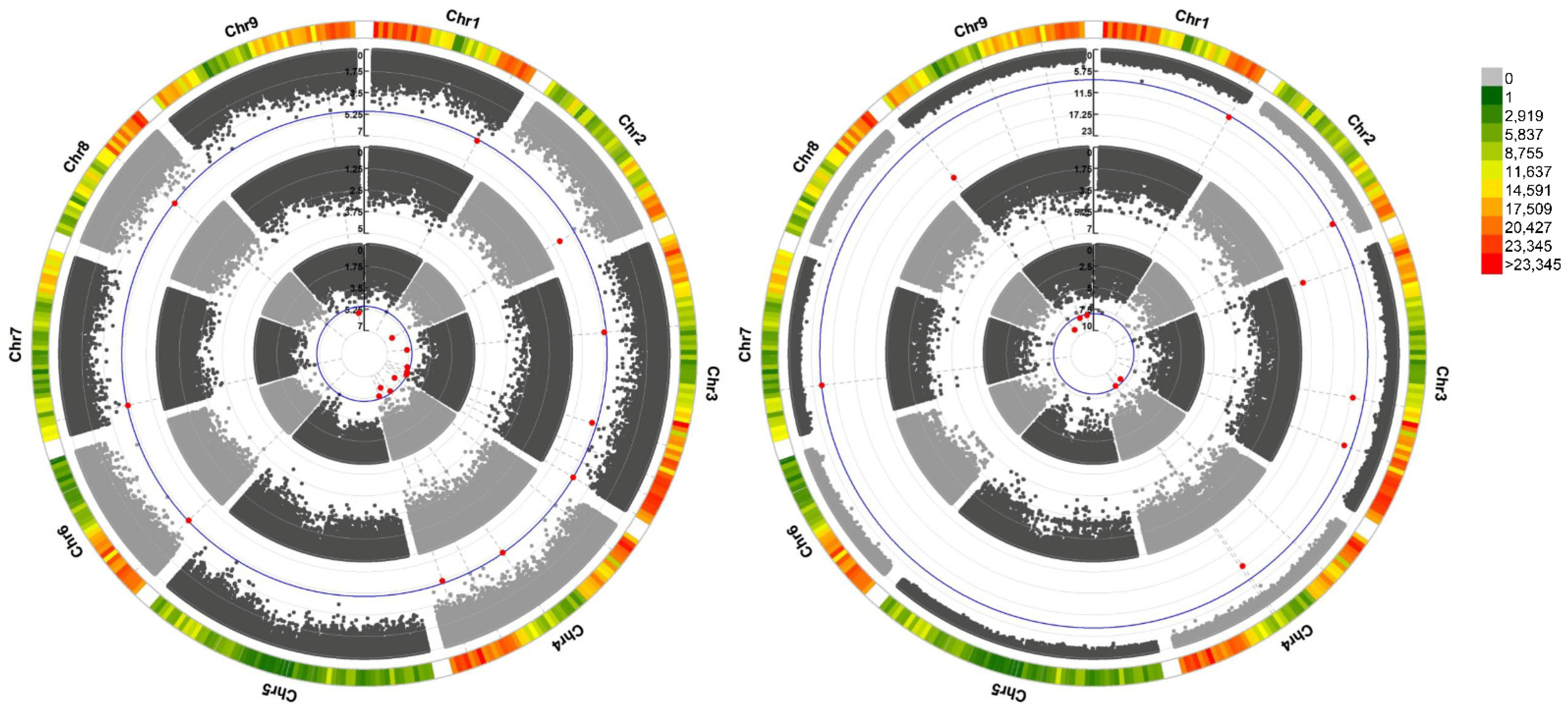

| SNP | Chr. | Position | p | Allele | Candidate Gene ID | Pfam_Annotation |
|---|---|---|---|---|---|---|
| Associate with Plant Height | ||||||
| LG01-30416755 | 1 | 30416755 | 7 × 10−6 | C/T | evm.model.LG01.2990 | Unknown |
| LG03-18657355 | 3 | 18657355 | 4.2 × 10−6 | T/C | evm.model.LG03.1638 | CytoLGome b5-like Heme/Steroid |
| LG03-44157320 | 3 | 44157320 | 6.1 × 10−6 | A/C | evm.model.LG03.3214 | TMEM33/Pom33 family |
| LG03-44157337 | 3 | 44157337 | 6.1 × 10−6 | T/G | evm.model.LG03.3214 | TMEM33/Pom33 family |
| LG03-51374819 | 3 | 51374819 | 8.2 × 10−6 | G/A | evm.model.LG03.4036 | Protein of unknown function (DUF620) |
| LG04-34820631 | 4 | 34820631 | 1.6 × 10−6 | A/G | evm.model.LG04.2874 | GTP cyclohydrolase II |
| LG04-4787029 | 4 | 4787029 | 1.8 × 10−6 | A/G | evm.model.LG04.571 | Surface antigen |
| LG04-42658778 | 4 | 42658778 | 6.1 × 10−6 | A/G | evm.model.LG04.3796 | TLD |
| LG04-24215127 | 4 | 24215127 | 6.9 × 10−6 | G/A | evm.model.LG04.1979 | Serine-threonine/tyrosine-protein kinase |
| LG06-3448481 | 6 | 3448481 | 8.8 × 10−6 | T/C | evm.model.LG06.412 | Ring finger domain98 |
| LG09-36689561 | 9 | 36689561 | 3.0 × 10−6 | G/A | evm.model.LG09.2890 | Beta-glucosidase |
| Associate with IAA Content | ||||||
| LG01-32609814 | 1 | 32609814 | 3.7 × 10−9 | G/A | evm.model.LG01.3294 | Lipase (class 3) |
| LG04-24619228 | 4 | 24619228 | 7.0 × 10−9 | G/A | evm.model.LG04.1998 | Calcineurin-like phosphoesterase |
| LG04-23975106 | 4 | 23975106 | 1.0 × 10−12 | A/T | evm.model.LG04.1964 | Transferase family |
| LG07-10265433 | 7 | 10265433 | 7.5 × 10−9 | C/T | evm.model.LG07.712 | Unknown |
| LG09-1324439 | 9 | 1324439 | 7.8 × 10−9 | A/G | evm.model.LG09.98 | Cell division control protein 14, SIN component |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gan, L.; Chen, M.; Zhang, J.; Fan, J.; Yan, X. A Novel Beta-Glucosidase Gene for Plant Type Was Identified by Genome-Wide Association Study and Gene Co-Expression Analysis in Widespread Bermudagrass. Int. J. Mol. Sci. 2022, 23, 11432. https://doi.org/10.3390/ijms231911432
Gan L, Chen M, Zhang J, Fan J, Yan X. A Novel Beta-Glucosidase Gene for Plant Type Was Identified by Genome-Wide Association Study and Gene Co-Expression Analysis in Widespread Bermudagrass. International Journal of Molecular Sciences. 2022; 23(19):11432. https://doi.org/10.3390/ijms231911432
Chicago/Turabian StyleGan, Lu, Minghui Chen, Jingxue Zhang, Jibiao Fan, and Xuebing Yan. 2022. "A Novel Beta-Glucosidase Gene for Plant Type Was Identified by Genome-Wide Association Study and Gene Co-Expression Analysis in Widespread Bermudagrass" International Journal of Molecular Sciences 23, no. 19: 11432. https://doi.org/10.3390/ijms231911432
APA StyleGan, L., Chen, M., Zhang, J., Fan, J., & Yan, X. (2022). A Novel Beta-Glucosidase Gene for Plant Type Was Identified by Genome-Wide Association Study and Gene Co-Expression Analysis in Widespread Bermudagrass. International Journal of Molecular Sciences, 23(19), 11432. https://doi.org/10.3390/ijms231911432







