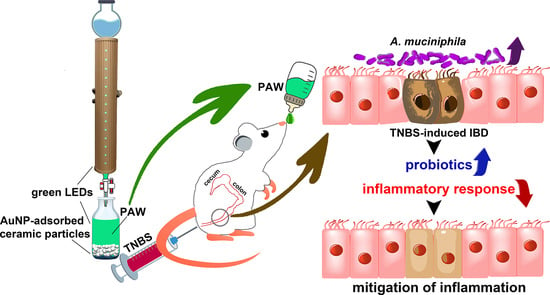
Functional Plasmon-Activated Water Increases Akkermansia muciniphila Abundance in Gut Microbiota to Ameliorate Inflammatory Bowel Disease
Abstract
1. Introduction
2. Results
2.1. Scavenging Ability of PAW against DPPH Radicals and Response of PAW in Colonic Epithelial Cells
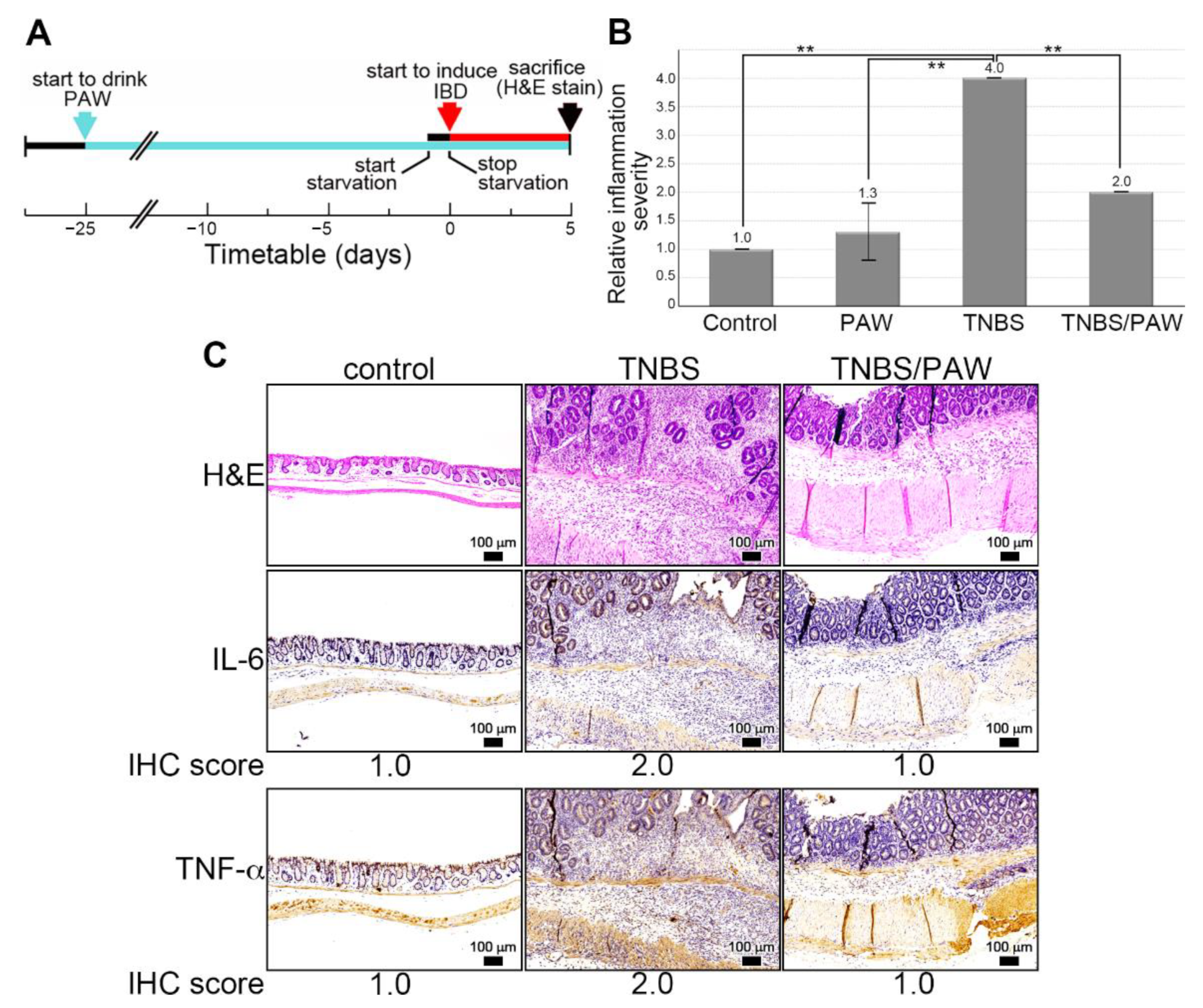
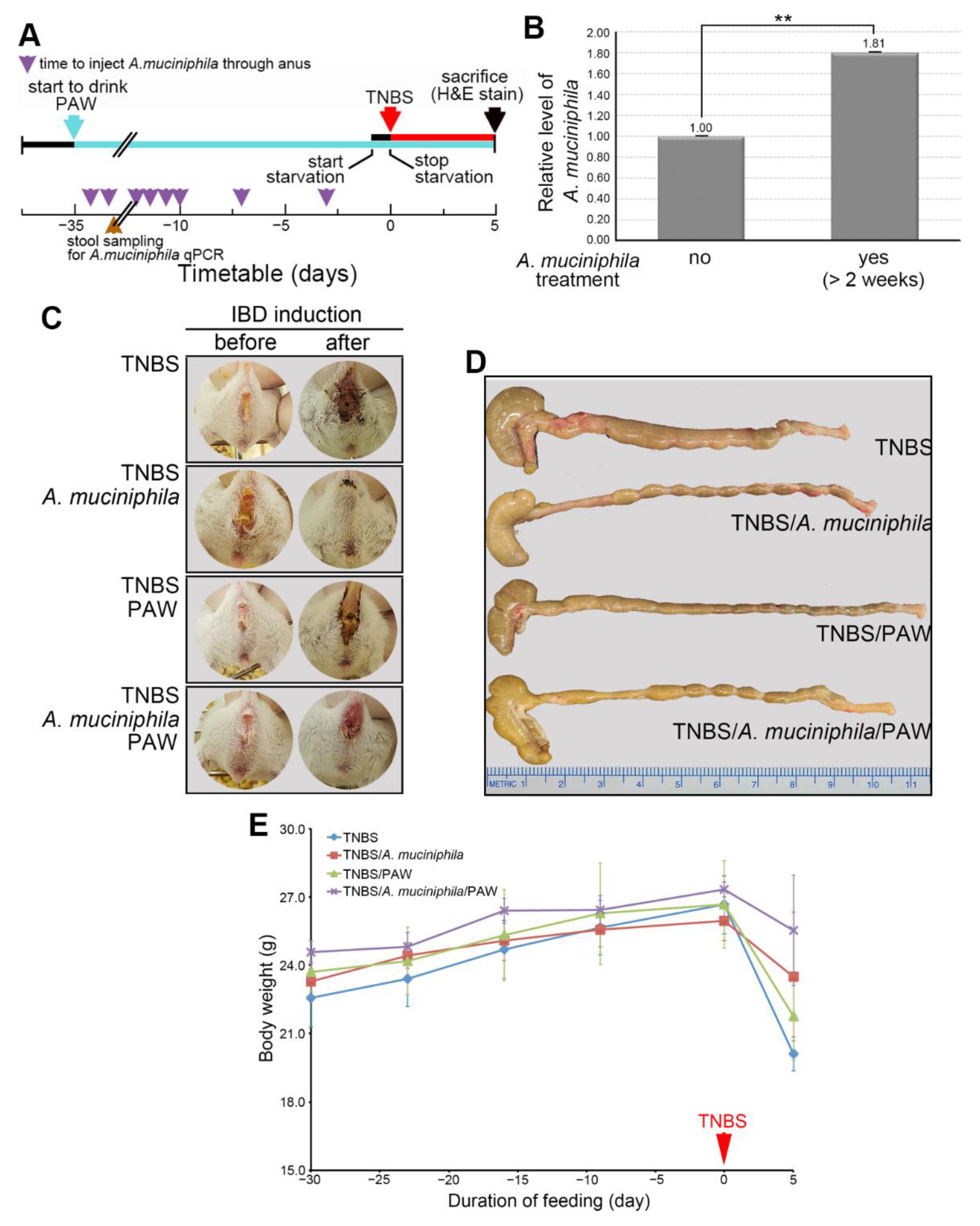
2.2. Induction of IBD in PAW-Drinking Mice with TNBS
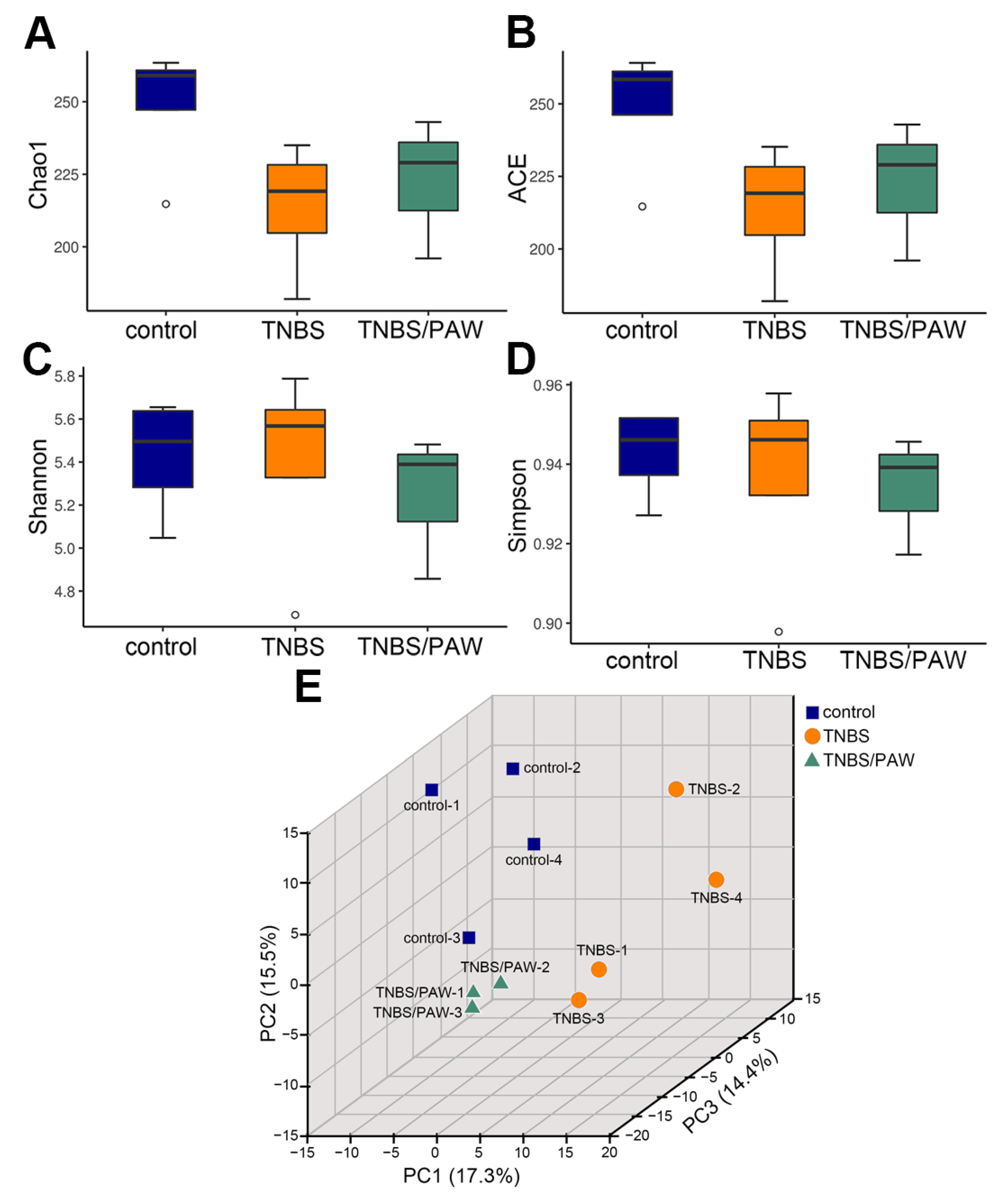
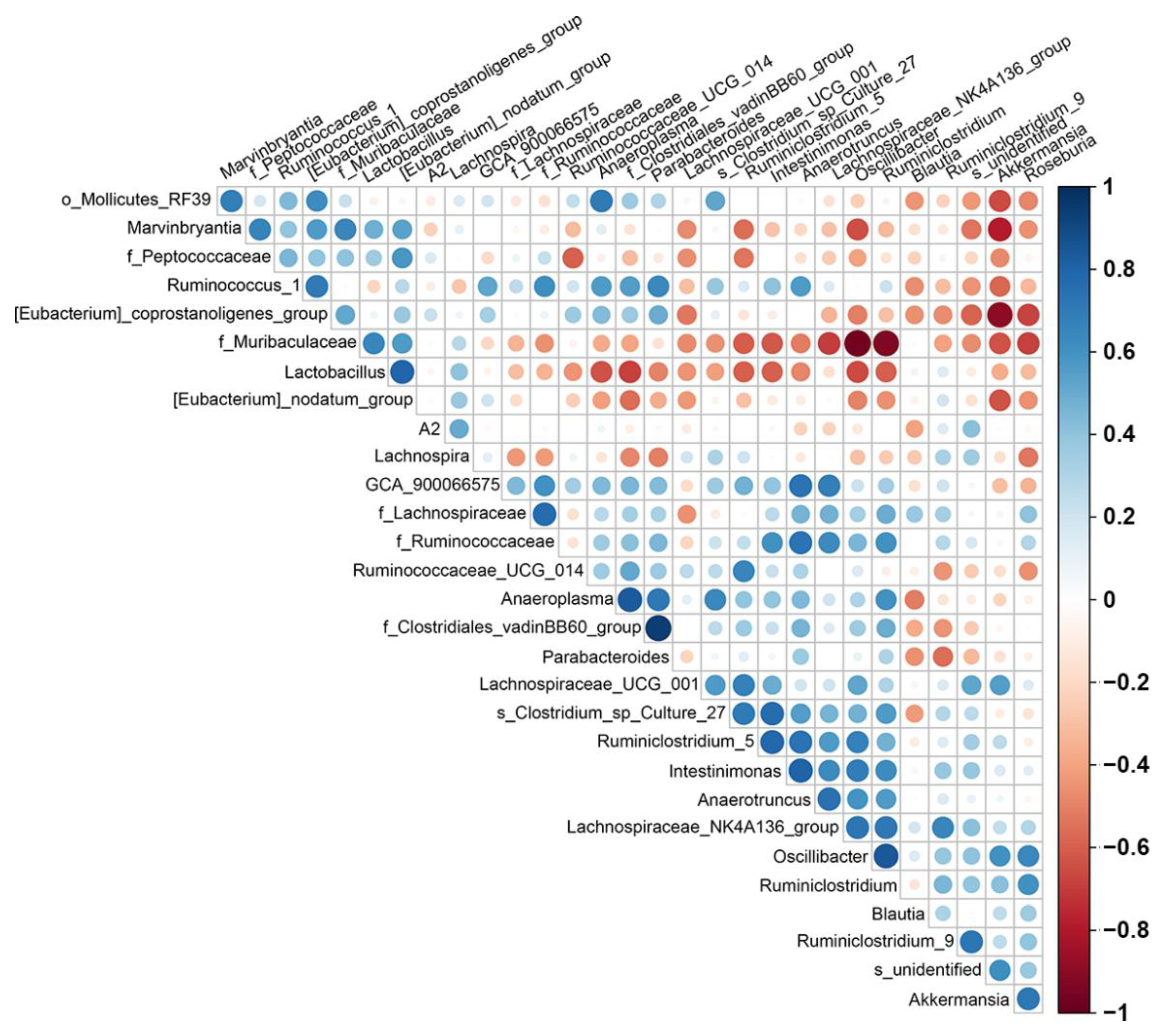
2.3. Changes in Species Richness, Diversity, and Similarity after PAW Consumption
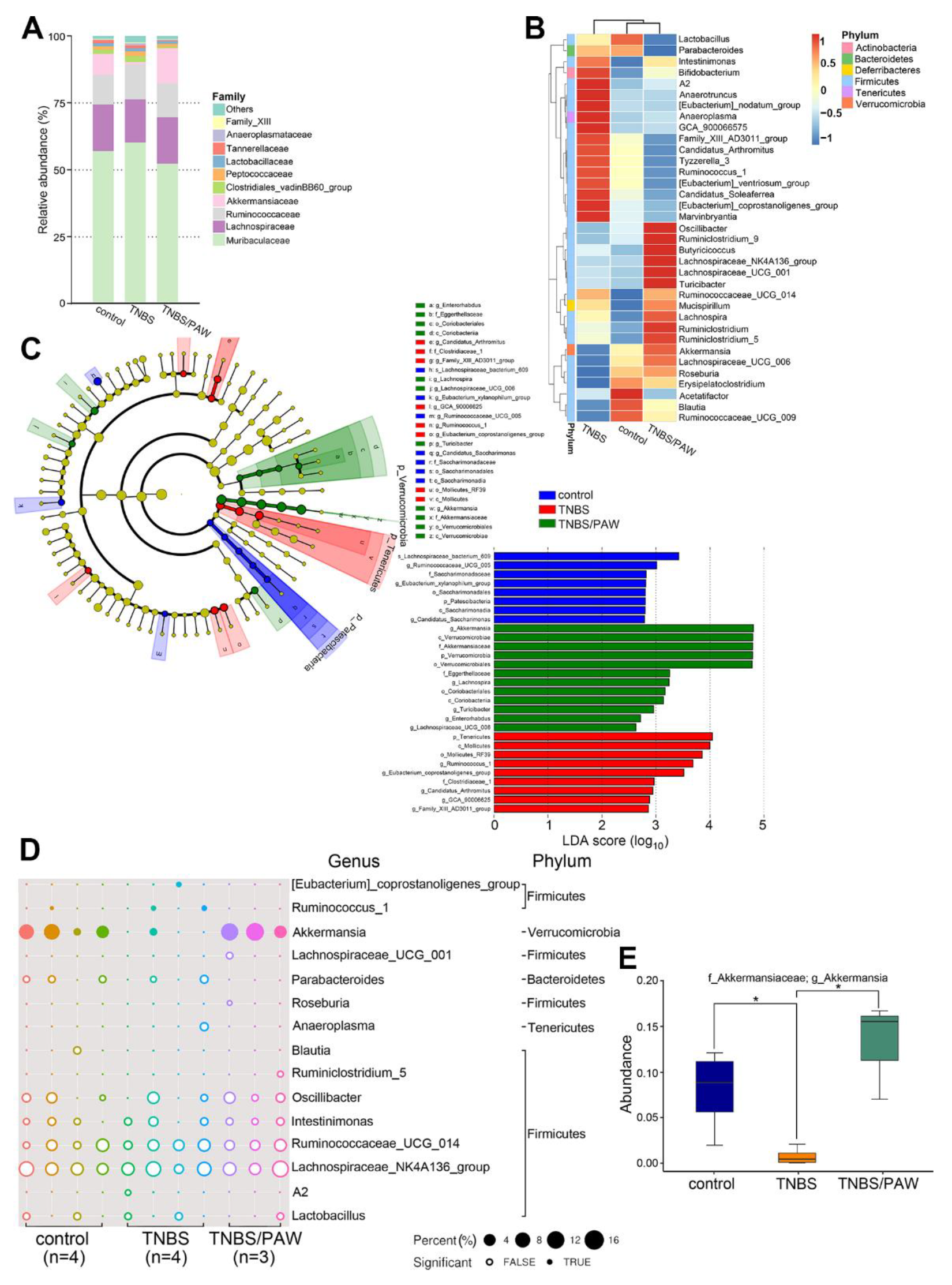
2.4. PAW Consumption Increased the Abundance of Akkermansia spp. in the Gut Microbial Community Composition
2.5. Phenotypes of A. muciniphila and PAW to IBD
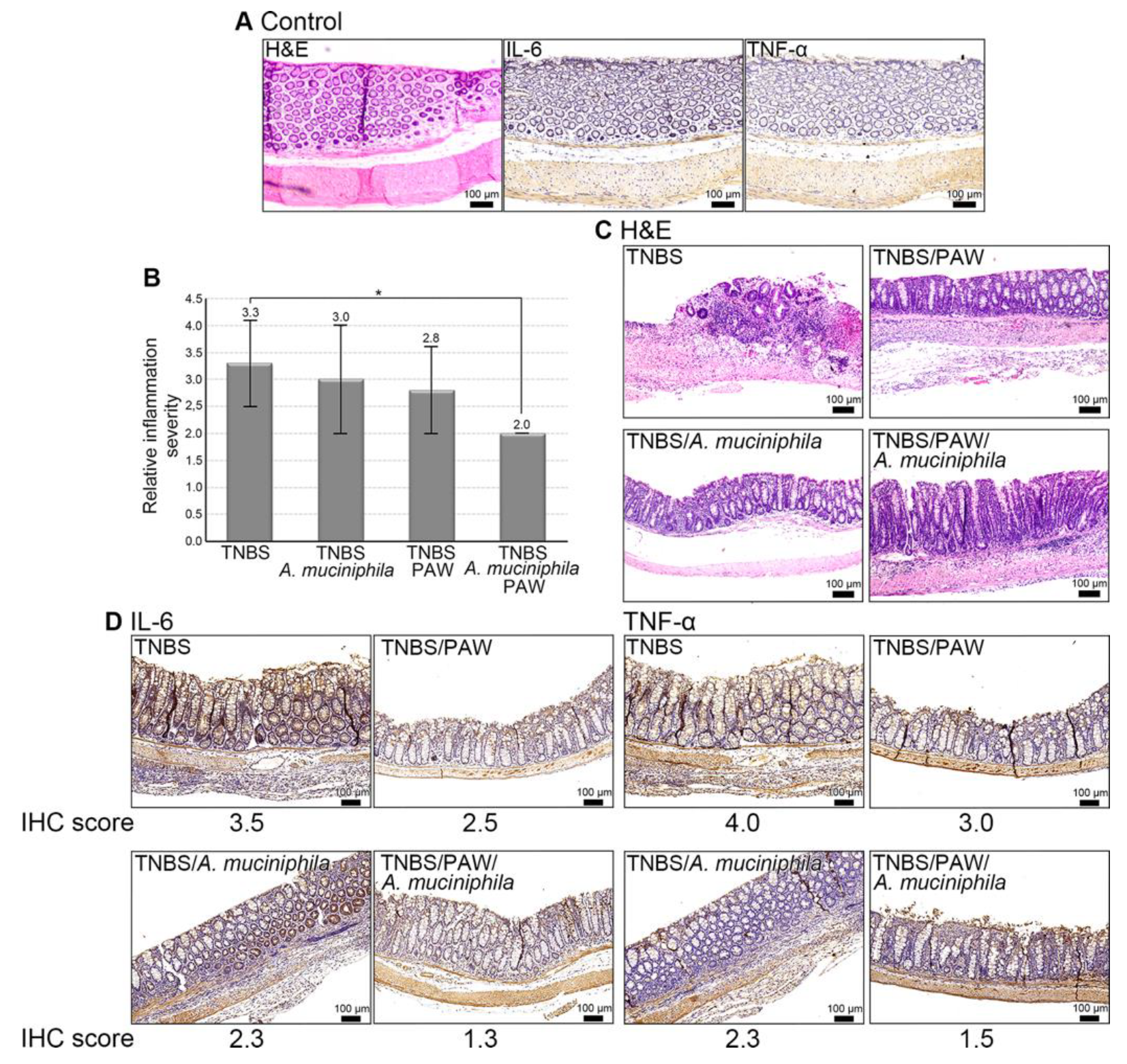
2.6. Mitigation of IBD in Mice That Consumed PAW and Received A. muciniphila Treatment
3. Discussion
4. Materials and Methods
4.1. Mouse Models of TNBS-Induced IBD and Stool Collection
4.2. Microbiota by 16S rDNA Next Generation Sequencing
4.3. PAW and A. muciniphila Preparation
4.4. Inflammation Characterization of Colon Tissue in Mice with Induced IBD
4.5. Sample Preparation for Measuring DPPH Free Radicals
4.6. Measurement of Free Radicals by Electron Spin Resonance Spectroscopy
4.7. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Le Berre, C.; Ananthakrishnan, A.N.; Danese, S.; Singh, S.; Peyrin-Biroulet, L. Ulcerative Colitis and Crohn’s Disease Have Similar Burden and Goals for Treatment. Clin. Gastroenterol. Hepatol. Off. Clin. Pract. J. Am. Gastroenterol. Assoc. 2020, 18, 14–23. [Google Scholar] [CrossRef] [PubMed]
- Li, X.X.; Liu, Y.; Luo, J.; Huang, Z.D.; Zhang, C.; Fu, Y. Vitamin D deficiency associated with Crohn’s disease and ulcerative colitis: A meta-analysis of 55 observational studies. J. Transl. Med. 2019, 17, 323. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.C. Microbiota dysbiosis and barrier dysfunction in inflammatory bowel disease and colorectal cancers: Exploring a common ground hypothesis. J. Biomed. Sci. 2018, 25, 79. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.; Ullah, N.; Zha, L.; Bai, Y.; Khan, A.; Zhao, T.; Che, T.; Zhang, C. Alteration of Gut Microbiota in Inflammatory Bowel Disease (IBD): Cause or Consequence? IBD Treatment Targeting the Gut Microbiome. Pathogens 2019, 8, 126. [Google Scholar] [CrossRef] [PubMed]
- Larabi, A.; Barnich, N.; Nguyen, H.T.T. New insights into the interplay between autophagy, gut microbiota and inflammatory responses in IBD. Autophagy 2020, 16, 38–51. [Google Scholar] [CrossRef] [PubMed]
- Frank, D.N.; St Amand, A.L.; Feldman, R.A.; Boedeker, E.C.; Harpaz, N.; Pace, N.R. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. USA 2007, 104, 13780–13785. [Google Scholar] [CrossRef]
- Ni, J.; Wu, G.D.; Albenberg, L.; Tomov, V.T. Gut microbiota and IBD: Causation or correlation? Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 573–584. [Google Scholar] [CrossRef] [PubMed]
- Weingarden, A.R.; Vaughn, B.P. Intestinal microbiota, fecal microbiota transplantation, and inflammatory bowel disease. Gut Microbes 2017, 8, 238–252. [Google Scholar] [CrossRef] [PubMed]
- Aldars-Garcia, L.; Chaparro, M.; Gisbert, J.P. Systematic Review: The Gut Microbiome and Its Potential Clinical Application in Inflammatory Bowel Disease. Microorganisms 2021, 9, 977. [Google Scholar] [CrossRef]
- Chen, H.C.; Mai, F.D.; Hwang, B.J.; Lee, M.J.; Chen, C.H.; Wang, S.H.; Tsai, H.Y.; Yang, C.P.; Liu, Y.C. Creation of Electron-doping Liquid Water with Reduced Hydrogen Bonds. Sci. Rep. 2016, 6, 22166. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.K.; Chen, H.C.; Fang, S.U.; Ho, C.W.; Tai, C.J.; Yang, C.P.; Liu, Y.C. Innovatively Therapeutic Strategy on Lung Cancer by Daily Drinking Antioxidative Plasmon-Induced Activated Water. Sci. Rep. 2018, 8, 6316. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.P.; Liu, Y.C. Therapeutics for Inflammatory-Related Diseases Based on Plasmon-Activated Water: A Review. Int. J. Mol. Sci. 2018, 19, 1589. [Google Scholar] [CrossRef]
- Cheng, C.H.; Lin, K.J.; Hong, C.T.; Wu, D.; Chang, H.M.; Liu, C.H.; Hsiao, I.T.; Yang, C.P.; Liu, Y.C.; Hu, C.J. Plasmon-Activated Water Reduces Amyloid Burden and Improves Memory in Animals with Alzheimer’s Disease. Sci. Rep. 2019, 9, 13252. [Google Scholar] [CrossRef] [PubMed]
- Chassaing, B.; Aitken, J.D.; Malleshappa, M.; Vijay-Kumar, M. Dextran sulfate sodium (DSS)-induced colitis in mice. Curr. Protoc. Immunol. 2014, 104, Unit 15.25. [Google Scholar] [CrossRef] [PubMed]
- Shin, M.R.; Kim, K.J.; Kim, S.H.; Kim, S.J.; Seo, B.I.; An, H.J.; Roh, S.S. Comparative Evaluation between Sulfasalazine Alone and in Combination with Herbal Medicine on DSS-Induced Ulcerative Colitis Mice. BioMed Res. Int. 2017, 2017, 6742652. [Google Scholar] [CrossRef] [PubMed]
- Low, D.; Nguyen, D.D.; Mizoguchi, E. Animal models of ulcerative colitis and their application in drug research. Drug Des. Dev. Ther. 2013, 7, 1341–1357. [Google Scholar] [CrossRef]
- Yang, C.P.; Tsai, H.Y.; Tseng, C.L.; Hao, P.J.; Liu, Y.C. Strategy on Persisting in Distinct Activity of Plasmon-Activated Water. ACS Omega 2019, 4, 21197–21203. [Google Scholar] [CrossRef] [PubMed]
- Alkushi, A.G.; Abdelfattah-Hassan, A.; Eldoumani, H.; Elazab, S.T.; Mohamed, S.A.M.; Metwally, A.S.; El-Shetry, E.S.; Saleh, A.A.; ElSawy, N.A.; Ibrahim, D. Probiotics-loaded nanoparticles attenuated colon inflammation, oxidative stress, and apoptosis in colitis. Sci. Rep. 2022, 12, 5116. [Google Scholar] [CrossRef] [PubMed]
- Cuellar-Nunez, M.L.; Gonzalez de Mejia, E.; Loarca-Pina, G. Moringa oleifera leaves alleviated inflammation through downregulation of IL-2, IL-6, and TNF-alpha in a colitis-associated colorectal cancer model. Food Res. Int. 2021, 144, 110318. [Google Scholar] [CrossRef]
- Varshosaz, J.; Emami, J.; Fassihi, A.; Tavakoli, N.; Minaiyan, M.; Ahmadi, F.; Mahzouni, P.; Dorkoosh, F. Effectiveness of budesonide-succinate-dextran conjugate as a novel prodrug of budesonide against acetic acid-induced colitis in rats. Int. J. Colorectal Dis. 2010, 25, 1159–1165. [Google Scholar] [CrossRef] [PubMed]
- Ouwerkerk, J.P.; Aalvink, S.; Belzer, C.; de Vos, W.M. Akkermansia glycaniphila sp. nov., an anaerobic mucin-degrading bacterium isolated from reticulated python faeces. Int. J. Syst. Evol. Microbiol. 2016, 66, 4614–4620. [Google Scholar] [CrossRef] [PubMed]
- Cheng, D.; Xie, M.Z. A review of a potential and promising probiotic candidate-Akkermansia muciniphila. J. Appl. Microbiol. 2021, 130, 1813–1822. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Lv, L.; Liu, B.; Wang, S.; Zhang, S.; Wu, Z.; Yang, L.; Bian, X.; Wang, Q.; Wang, K.; et al. Akkermansia muciniphila Ameliorates Acetaminophen-Induced Liver Injury by Regulating Gut Microbial Composition and Metabolism. Microbiol. Spectr. 2022, 10, e0159621. [Google Scholar] [CrossRef] [PubMed]
- Ananthakrishnan, A.N. Epidemiology and risk factors for IBD. Nat. Rev. Gastroenterol. Hepatol. 2015, 12, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.M.; Cha, J.M.; Lee, J.L.; Jeon, J.W.; Shin, H.P.; Joo, K.R.; Yoon, J.Y.; Lee, J.I. High C-reactive protein level is associated with high-risk adenoma. Intest. Res. 2017, 15, 511–517. [Google Scholar] [CrossRef] [PubMed]
- Dignass, A.; Eliakim, R.; Magro, F.; Maaser, C.; Chowers, Y.; Geboes, K.; Mantzaris, G.; Reinisch, W.; Colombel, J.F.; Vermeire, S.; et al. Second European evidence-based consensus on the diagnosis and management of ulcerative colitis part 1: Definitions and diagnosis. J. Crohn’s Colitis 2012, 6, 965–990. [Google Scholar] [CrossRef] [PubMed]
- Kamada, N.; Seo, S.U.; Chen, G.Y.; Nunez, G. Role of the gut microbiota in immunity and inflammatory disease. Nat. Rev. Immunol. 2013, 13, 321–335. [Google Scholar] [CrossRef]
- Cader, M.Z.; Kaser, A. Recent advances in inflammatory bowel disease: Mucosal immune cells in intestinal inflammation. Gut 2013, 62, 1653–1664. [Google Scholar] [CrossRef]
- Cario, E. Commensal-innate immune miscommunication in IBD pathogenesis. Dig. Dis. 2012, 30, 334–340. [Google Scholar] [CrossRef]
- Wills, E.S.; Jonkers, D.M.; Savelkoul, P.H.; Masclee, A.A.; Pierik, M.J.; Penders, J. Fecal microbial composition of ulcerative colitis and Crohn’s disease patients in remission and subsequent exacerbation. PLoS ONE 2014, 9, e90981. [Google Scholar] [CrossRef]
- Papa, E.; Docktor, M.; Smillie, C.; Weber, S.; Preheim, S.P.; Gevers, D.; Giannoukos, G.; Ciulla, D.; Tabbaa, D.; Ingram, J.; et al. Non-invasive mapping of the gastrointestinal microbiota identifies children with inflammatory bowel disease. PLoS ONE 2012, 7, e39242. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, K.; Fujita, Y. Determination of the discriminant score of intestinal microbiota as a biomarker of disease activity in patients with ulcerative colitis. BMC Gastroenterol. 2014, 14, 49. [Google Scholar] [CrossRef] [PubMed]
- Gupta, V.; Rodrigues, R.; Nguyen, D.; Sauk, J.; Khalili, H.; Yajnik, V.; Ananthakrishnan, A.N. Adjuvant use of antibiotics with corticosteroids in inflammatory bowel disease exacerbations requiring hospitalisation: A retrospective cohort study and meta-analysis. Aliment. Pharmacol. Ther. 2016, 43, 52–60. [Google Scholar] [CrossRef] [PubMed]
- McIlroy, J.; Ianiro, G.; Mukhopadhya, I.; Hansen, R.; Hold, G.L. Review article: The gut microbiome in inflammatory bowel disease-avenues for microbial management. Aliment. Pharmacol. Ther. 2018, 47, 26–42. [Google Scholar] [CrossRef]
- Chen, H.C.; Cheng, C.Y.; Lin, H.C.; Chen, H.H.; Chen, C.H.; Yang, C.P.; Yang, K.H.; Lin, C.M.; Lin, T.Y.; Shih, C.M.; et al. Multifunctions of Excited Gold Nanoparticles Decorated Artificial Kidney with Efficient Hemodialysis and Therapeutic Potential. ACS Appl. Mater. Interfaces 2016, 8, 19691–19700. [Google Scholar] [CrossRef]
- Chen, H.C.; Hwang, B.J.; Mai, F.D.; Liu, Y.C.; Lin, C.M.; Kuo, H.S.; Chou, D.S.; Lee, M.J.; Yang, K.H.; Yu, C.C.; et al. Active and stable liquid water innovatively prepared using resonantly illuminated gold nanoparticles. ACS Nano 2014, 8, 2704–2713. [Google Scholar] [CrossRef]
- Tsai, C.S.; Lin, F.Y.; Liu, Y.C.; Lin, Y.W.; Tsai, Y.T.; Huang, C.Y.; Lin, S.J.; Li, C.Y.; Lin, C.Y.; Tseng, H.T.; et al. Transferring Plasmon Effect on a Biological System: Expression of Biological Polymers in Chronic Rejection and Inflammatory Rat Model. Polymers 2021, 13, 1827. [Google Scholar] [CrossRef]
- Yu, S.H.; Kao, W.Y.; Mai, F.D.; Tsai, H.Y.; Liu, Y.C. Comprehensively active condensed water from vapors of plasmon-activated water. Chem. Eng. J. 2021, 426, 130875–130884. [Google Scholar] [CrossRef]
- Zhang, T.; Li, Q.; Cheng, L.; Buch, H.; Zhang, F. Akkermansia muciniphila is a promising probiotic. Microb. Biotechnol. 2019, 12, 1109–1125. [Google Scholar] [CrossRef]
- Fujisaka, S.; Usui, I.; Nawaz, A.; Igarashi, Y.; Okabe, K.; Furusawa, Y.; Watanabe, S.; Yamamoto, S.; Sasahara, M.; Watanabe, Y.; et al. Bofutsushosan improves gut barrier function with a bloom of Akkermansia muciniphila and improves glucose metabolism in mice with diet-induced obesity. Sci. Rep. 2020, 10, 5544. [Google Scholar] [CrossRef]
- Shin, N.R.; Lee, J.C.; Lee, H.Y.; Kim, M.S.; Whon, T.W.; Lee, M.S.; Bae, J.W. An increase in the Akkermansia spp. population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice. Gut 2014, 63, 727–735. [Google Scholar] [CrossRef] [PubMed]
- Zhai, R.; Xue, X.; Zhang, L.; Yang, X.; Zhao, L.; Zhang, C. Strain-Specific Anti-inflammatory Properties of Two Akkermansia muciniphila Strains on Chronic Colitis in Mice. Front. Cell. Infect. Microbiol. 2019, 9, 239. [Google Scholar] [CrossRef] [PubMed]
- Bian, X.; Wu, W.; Yang, L.; Lv, L.; Wang, Q.; Li, Y.; Ye, J.; Fang, D.; Wu, J.; Jiang, X.; et al. Administration of Akkermansia muciniphila Ameliorates Dextran Sulfate Sodium-Induced Ulcerative Colitis in Mice. Front. Microbiol. 2019, 10, 2259. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Huang, S.; Li, T.; Li, N.; Han, D.; Zhang, B.; Xu, Z.Z.; Zhang, S.; Pang, J.; Wang, S.; et al. Gut microbiota from green tea polyphenol-dosed mice improves intestinal epithelial homeostasis and ameliorates experimental colitis. Microbiome 2021, 9, 184. [Google Scholar] [CrossRef] [PubMed]
- Dong, C.; Yu, J.; Yang, Y.; Zhang, F.; Su, W.; Fan, Q.; Wu, C.; Wu, S. Berberine, a potential prebiotic to indirectly promote Akkermansia growth through stimulating gut mucin secretion. Biomed. Pharmacother. 2021, 139, 111595. [Google Scholar] [CrossRef] [PubMed]
- Depommier, C.; Everard, A.; Druart, C.; Plovier, H.; Van Hul, M.; Vieira-Silva, S.; Falony, G.; Raes, J.; Maiter, D.; Delzenne, N.M.; et al. Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: A proof-of-concept exploratory study. Nat. Med. 2019, 25, 1096–1103. [Google Scholar] [CrossRef]
- Duncan, S.H.; Hold, G.L.; Barcenilla, A.; Stewart, C.S.; Flint, H.J. Roseburia intestinalis sp. nov., a novel saccharolytic, butyrate-producing bacterium from human faeces. Int. J. Syst. Evol. Microbiol. 2002, 52, 1615–1620. [Google Scholar] [CrossRef]
- Machiels, K.; Joossens, M.; Sabino, J.; De Preter, V.; Arijs, I.; Eeckhaut, V.; Ballet, V.; Claes, K.; Van Immerseel, F.; Verbeke, K.; et al. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut 2014, 63, 1275–1283. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xie, Q.; Sun, S.; Huang, B.; Zhang, Y.; Xu, Y.; Zhang, S.; Xiang, H. Probiotics-fermented Massa Medicata Fermentata ameliorates weaning stress in piglets related to improving intestinal homeostasis. Appl. Microbiol. Biotechnol. 2018, 102, 10713–10727. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Qi, W.; Song, G.; Pang, S.; Peng, Z.; Li, Y.; Wang, P. High-Fructose Diet Increases Inflammatory Cytokines and Alters Gut Microbiota Composition in Rats. Mediat. Inflamm. 2020, 2020, 6672636. [Google Scholar] [CrossRef] [PubMed]
- Everard, A.; Belzer, C.; Geurts, L.; Ouwerkerk, J.P.; Druart, C.; Bindels, L.B.; Guiot, Y.; Derrien, M.; Muccioli, G.G.; Delzenne, N.M.; et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. USA 2013, 110, 9066–9071. [Google Scholar] [CrossRef]
- Liu, X.; Zhao, F.; Liu, H.; Xie, Y.; Zhao, D.; Li, C. Transcriptomics and metabolomics reveal the adaption of Akkermansia muciniphila to high mucin by regulating energy homeostasis. Sci. Rep. 2021, 11, 9073. [Google Scholar] [CrossRef] [PubMed]
- Ou, Z.; Deng, L.; Lu, Z.; Wu, F.; Liu, W.; Huang, D.; Peng, Y. Protective effects of Akkermansia muciniphila on cognitive deficits and amyloid pathology in a mouse model of Alzheimer’s disease. Nutr. Diabetes 2020, 10, 12. [Google Scholar] [CrossRef] [PubMed]
- Earley, H.; Lennon, G.; Balfe, A.; Coffey, J.C.; Winter, D.C.; O’Connell, P.R. The abundance of Akkermansia muciniphila and its relationship with sulphated colonic mucins in health and ulcerative colitis. Sci. Rep. 2019, 9, 15683. [Google Scholar] [CrossRef] [PubMed]
- Varum, F.J.; Veiga, F.; Sousa, J.S.; Basit, A.W. Mucus thickness in the gastrointestinal tract of laboratory animals. J. Pharm. Pharm. 2012, 64, 218–227. [Google Scholar] [CrossRef] [PubMed]
- Hung, C.S.; Wang, Y.C.; Guo, J.W.; Yang, R.N.; Lee, C.L.; Shen, M.H.; Huang, C.C.; Huang, C.J.; Yang, J.Y.; Liu, C.Y. Expression pattern of placenta specific 8 and keratin 20 in different types of gastrointestinal cancer. Mol. Med. Rep. 2020, 21, 659–666. [Google Scholar] [CrossRef] [PubMed]
- Negi, R.R.; Rana, S.V.; Gupta, V.; Gupta, R.; Chadha, V.D.; Prasad, K.K.; Dhawan, D.K. Over-Expression of Cyclooxygenase-2 in Colorectal Cancer Patients. Asian Pac. J. Cancer Prev. 2019, 20, 1675–1681. [Google Scholar] [CrossRef]
- Wang, D.; Li, Y.; Zhang, C.; Li, X.; Yu, J. MiR-216a-3p inhibits colorectal cancer cell proliferation through direct targeting COX-2 and ALOX5. J. Cell Biochem. 2018, 119, 1755–1766. [Google Scholar] [CrossRef] [PubMed]
- Bradford, E.M.; Ryu, S.H.; Singh, A.P.; Lee, G.; Goretsky, T.; Sinh, P.; Williams, D.B.; Cloud, A.L.; Gounaris, E.; Patel, V.; et al. Epithelial TNF Receptor Signaling Promotes Mucosal Repair in Inflammatory Bowel Disease. J. Immunol. (Baltimore Md. 1950) 2017, 199, 1886–1897. [Google Scholar] [CrossRef] [PubMed]
- Muhamad Rizal, N.S.; Neoh, H.M.; Ramli, R.; PR, A.L.K.P.; Hanafiah, A.; Abdul Samat, M.N.; Tan, T.L.; Wong, K.K.; Nathan, S.; Chieng, S.; et al. Advantages and Limitations of 16S rRNA Next-Generation Sequencing for Pathogen Identification in the Diagnostic Microbiology Laboratory: Perspectives from a Middle-Income Country. Diagnostics 2020, 10, 816. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.Y.; Cho, K.A.; Kang, J.L.; Kim, K.H.; Woo, S.Y. Comparison of experimental mouse models of inflammatory bowel disease. Int. J. Mol. Med. 2014, 33, 333–340. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Kechin, A.; Boyarskikh, U.; Kel, A.; Filipenko, M. cutPrimers: A New Tool for Accurate Cutting of Primers from Reads of Targeted Next Generation Sequencing. J. Comput. Biol. 2017, 24, 1138–1143. [Google Scholar] [CrossRef] [PubMed]
- Quin, C.; Estaki, M.; Vollman, D.M.; Barnett, J.A.; Gill, S.K.; Gibson, D.L. Probiotic supplementation and associated infant gut microbiome and health: A cautionary retrospective clinical comparison. Sci. Rep. 2018, 8, 8283. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef] [PubMed]
- McDonald, D.; Price, M.N.; Goodrich, J.; Nawrocki, E.P.; DeSantis, T.Z.; Probst, A.; Andersen, G.L.; Knight, R.; Hugenholtz, P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012, 6, 610–618. [Google Scholar] [CrossRef] [PubMed]
- Jolliffe, I.T.; Cadima, J. Principal component analysis: A review and recent developments. Philos. Trans. A Math. Phys. Eng. Sci. 2016, 374, 20150202. [Google Scholar] [CrossRef] [PubMed]
- Jami, E.; Israel, A.; Kotser, A.; Mizrahi, I. Exploring the bovine rumen bacterial community from birth to adulthood. ISME J. 2013, 7, 1069–1079. [Google Scholar] [CrossRef] [PubMed]
- Mutiah, R.; Sari, R.A.; Firsyaradha, W.Y.; Listiyana, A.; Indrawijaya, Y.Y.A.; Wafi, A.; Suryadinata, A.; Susilowati, R.; Rahmawati, A. Activity and Toxicity of Eleutherine palmifolia (L.) Merr. Extract on BALB/c Mice Colitis-Associated Colon Cancer Model. Asian Pac. J. Cancer Prev. 2020, 21, 3579–3586. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Shang, Y.; Sun, F.; Dong, X.; Niu, J.; Li, F. Interleukin-6 Promotes Epithelial-Mesenchymal Transition and Cell Invasion through Integrin beta6 Upregulation in Colorectal Cancer. Oxid. Med. Cell. Longev. 2020, 2020, 8032187. [Google Scholar] [CrossRef] [PubMed]
- Fields, C.T.; Chassaing, B.; Paul, M.J.; Gewirtz, A.T.; de Vries, G.J. Vasopressin deletion is associated with sex-specific shifts in the gut microbiome. Gut Microbes 2018, 9, 13–25. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; King, E.; Deek, R.; Wei, Z.; Yu, Y.; Grill, D.; Ballman, K.; Stegle, O. An omnibus test for differential distribution analysis of microbiome sequencing data. Bioinformatics 2018, 34, 643–651. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.; Tian, H.; Wang, Y.; Li, Y.; Li, Y.; Xie, J.; Zeng, B.; Zhou, J.; Li, G.; Ma, T. Spatial isolation and environmental factors drive distinct bacterial and archaeal communities in different types of petroleum reservoirs in China. Sci. Rep. 2016, 6, 20174. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, C.-C.; Liu, C.-Y.; Su, I.-C.; Lee, Y.-J.; Yeh, H.-J.; Chen, W.-C.; Yu, C.-J.; Kao, W.-Y.; Liu, Y.-C.; Huang, C.-J. Functional Plasmon-Activated Water Increases Akkermansia muciniphila Abundance in Gut Microbiota to Ameliorate Inflammatory Bowel Disease. Int. J. Mol. Sci. 2022, 23, 11422. https://doi.org/10.3390/ijms231911422
Chang C-C, Liu C-Y, Su I-C, Lee Y-J, Yeh H-J, Chen W-C, Yu C-J, Kao W-Y, Liu Y-C, Huang C-J. Functional Plasmon-Activated Water Increases Akkermansia muciniphila Abundance in Gut Microbiota to Ameliorate Inflammatory Bowel Disease. International Journal of Molecular Sciences. 2022; 23(19):11422. https://doi.org/10.3390/ijms231911422
Chicago/Turabian StyleChang, Chun-Chao, Chih-Yi Liu, I-Chia Su, Yuarn-Jang Lee, Hsing-Jung Yeh, Wen-Chao Chen, Chih-Jui Yu, Wei-Yu Kao, Yu-Chuan Liu, and Chi-Jung Huang. 2022. "Functional Plasmon-Activated Water Increases Akkermansia muciniphila Abundance in Gut Microbiota to Ameliorate Inflammatory Bowel Disease" International Journal of Molecular Sciences 23, no. 19: 11422. https://doi.org/10.3390/ijms231911422
APA StyleChang, C.-C., Liu, C.-Y., Su, I.-C., Lee, Y.-J., Yeh, H.-J., Chen, W.-C., Yu, C.-J., Kao, W.-Y., Liu, Y.-C., & Huang, C.-J. (2022). Functional Plasmon-Activated Water Increases Akkermansia muciniphila Abundance in Gut Microbiota to Ameliorate Inflammatory Bowel Disease. International Journal of Molecular Sciences, 23(19), 11422. https://doi.org/10.3390/ijms231911422