Deciphering the Basis of Molecular Biology of Selected Cardiovascular Diseases: A View on Network Medicine
Abstract
1. Introduction
1.1. Molecular Basis of Cardiac Development
1.2. Cardiac Metabolism in Health
1.3. Cardiac Metabolism in Exercise
1.4. Cardiac Metabolism in Disease
1.4.1. Obesity
1.4.2. Diabetes
2. Molecular Biology of Cardiovascular Diseases
2.1. Cardiomyopathies
- Restrictive.
- Dilated.
- Hypertrophic.
2.2. Restrictive Cardiomyopathy
Molecular and Genetic Basis of Restrictive Cardiomyopathy
- cTnC: a highly conserved Ca2+ binding subunit;
- cTnI: an actomyosin ATPase inhibitory subunit that functions to prevent actin and myosin interaction in the absence of Ca2+;
- cTnT: a tropomyosin binding subunit.
2.3. Dilated Cardiomyopathy
Molecular and Genetic Basis of Dilated Cardiomyopathy
2.4. Hypertrophic Cardiomyopathy
Molecular and Genetic Basis of Hypertrophic Cardiomyopathy
3. Heart Failure
The Molecular and Genetic Basis of Heart Failure
4. Coronary Artery Disease
5. Pericarditis
The Molecular and Genetic Basis of Pericarditis
6. Myocarditis
7. Clinical Interventions Using the Molecular and Cellular Pathways to Treat Cardiovascular Diseases
7.1. Antibodies
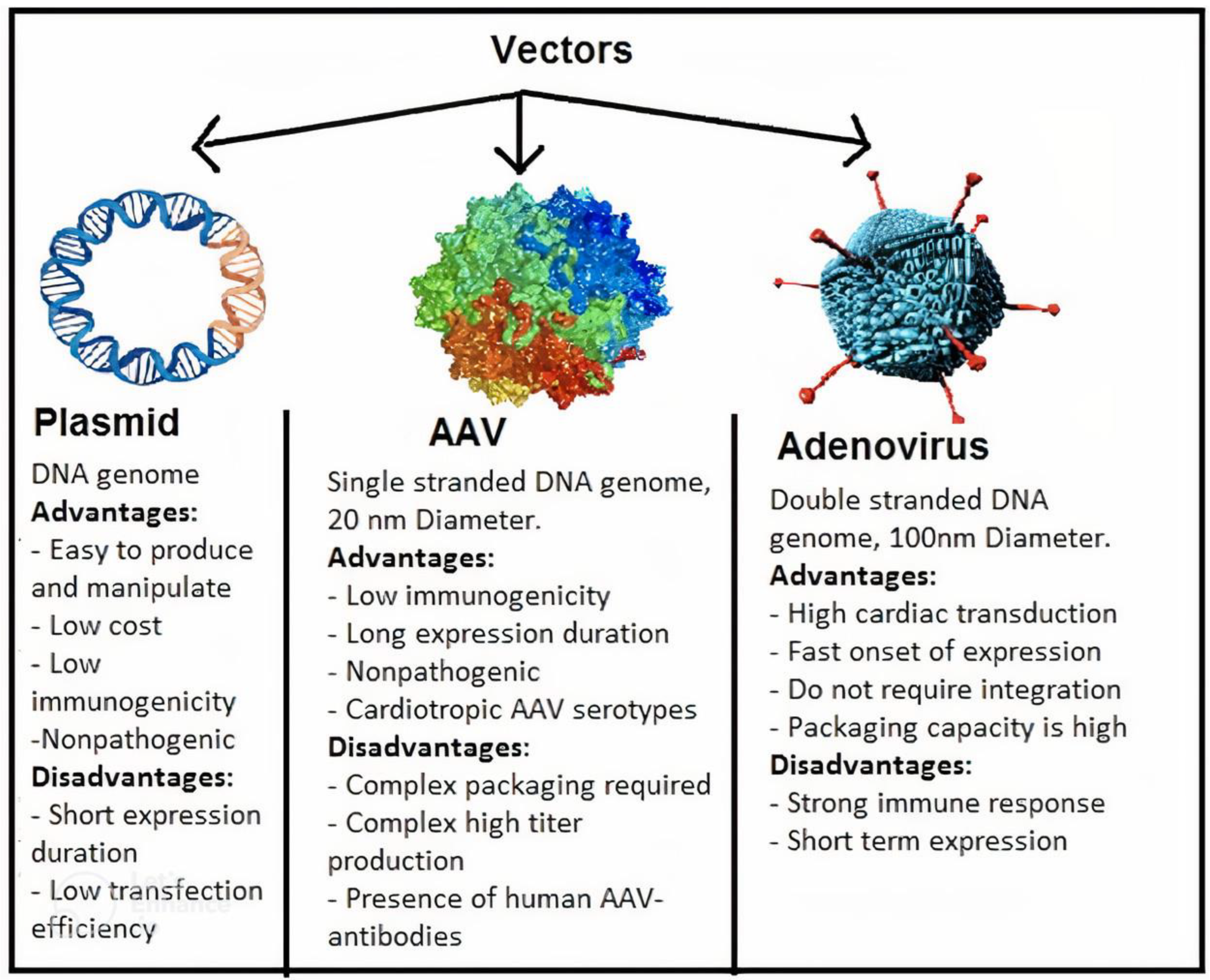
7.2. Gene Therapy
7.3. Stem Cell Therapy
7.4. Other Therapeutic Options
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Chan, S.Y.; White, K.; Loscalzo, J. Deciphering the molecular basis of human cardiovascular disease through network biology. Curr. Opin. Cardiol. 2012, 27, 202–209. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, M.W.; Kann, M.G. Chapter 4: Protein interactions and disease. PLoS Comput. Biol. 2012, 8, e1002819. [Google Scholar] [CrossRef] [PubMed]
- Lovely, C.; Rampersad, M.; Fernandes, Y.; Eberhart, J. Gene-environment interactions in development and disease. Wiley Interdiscip. Rev. Dev. Biol. 2017, 6, e247. [Google Scholar] [CrossRef]
- Sun, Z. Overview of recent advances in molecular cardiology. Can. J. Cardiol. 2006, 22, 235–240. [Google Scholar] [CrossRef]
- Stephens, J.W.; Humphries, S.E. The molecular genetics of cardiovascular disease: Clinical implications. J. Intern. Med. 2003, 253, 120–127. [Google Scholar] [CrossRef] [PubMed]
- Novelli, V.; Vatta, M. Editorial: Current Challenges in Cardiovascular Molecular Diagnostics. Front. Cardiovasc. Med. 2017, 4, 54. [Google Scholar] [CrossRef] [PubMed]
- Barabasi, A.L.; Gulbahce, N.; Loscalzo, J. Network medicine: A network-based approach to human disease. Nat. Rev. Genet. 2011, 12, 56–68. [Google Scholar] [CrossRef] [PubMed]
- Roth, G.A.; Mensah, G.A.; Johnson, C.O.; Addolorato, G.; Ammirati, E.; Baddour, L.M.; Barengo, N.C.; Beaton, A.Z.; Benjamin, E.J.; Benziger, C.P.; et al. Global Burden of Cardiovascular Diseases and Risk Factors, 1990–2019: Update From the GBD 2019 Study. J. Am. Coll. Cardiol. 2020, 76, 2982–3021. [Google Scholar] [CrossRef]
- Munoz-Chapuli, R.; Perez-Pomares, J.M. Cardiogenesis: An embryological perspective. J. Cardiovasc. Transl. Res. 2010, 3, 37–48. [Google Scholar] [CrossRef]
- Brade, T.; Pane, L.S.; Moretti, A.; Chien, K.R.; Laugwitz, K.L. Embryonic heart progenitors and cardiogenesis. Cold Spring Harb. Perspect. Med. 2013, 3, a013847. [Google Scholar] [CrossRef]
- Mirza, A.; Khan, I.; Qazi, R.E.; Salim, A.; Husain, M.; Herzig, J.W. Role of Wnt/beta-catenin pathway in cardiac lineage commitment of human umbilical cord mesenchymal stem cells by zebularine and 2′-deoxycytidine. Tissue Cell 2022, 77, 101850. [Google Scholar] [CrossRef] [PubMed]
- Steimle, J.D.; Moskowitz, I.P. TBX5: A Key Regulator of Heart Development. Curr. Top. Dev. Biol. 2017, 122, 195–221. [Google Scholar] [PubMed]
- Kim, J.S.; Seo, J.W.; Lee, Y.M.; Chi, J.G. Cardiac laterality and ventricular looping in retinoic acid-treated rat embryos. J. Korean Med. Sci. 1999, 14, 138–146. [Google Scholar] [CrossRef] [PubMed]
- Granzier, H.L.; Labeit, S. The giant protein titin: A major player in myocardial mechanics, signaling, and disease. Circ. Res. 2004, 94, 284–295. [Google Scholar] [CrossRef]
- Woodcock, E.A.; Matkovich, S.J. Cardiomyocytes structure, function and associated pathologies. Int. J. Biochem. Cell Biol. 2005, 37, 1746–1751. [Google Scholar] [CrossRef]
- Wentz, A.E.; d’Avignon, D.A.; Weber, M.L.; Cotter, D.G.; Doherty, J.M.; Kerns, R.; Nagarajan, R.; Reddy, N.; Sambandam, N.; Crawford, P.A. Adaptation of myocardial substrate metabolism to a ketogenic nutrient environment. J. Biol. Chem. 2010, 285, 24447–24456. [Google Scholar] [CrossRef]
- Gertz, E.W.; Wisneski, J.A.; Stanley, W.C.; Neese, R.A. Myocardial substrate utilization during exercise in humans. Dual carbon-labeled carbohydrate isotope experiments. J. Clin. Investig. 1988, 82, 2017–2025. [Google Scholar] [CrossRef]
- Kaijser, L.; Berglund, B. Myocardial lactate extraction and release at rest and during heavy exercise in healthy men. Acta Physiol. Scand. 1992, 144, 39–45. [Google Scholar] [CrossRef]
- De Groot, M.J.; Coumans, W.A.; Willemsen, P.H.; Van der Vusse, G.J. Substrate-induced changes in the lipid content of ischemic and reperfused myocardium. Its relation to hemodynamic recovery. Circ. Res. 1993, 72, 176–186. [Google Scholar] [CrossRef]
- Strøm, C.C.; Aplin, M.; Ploug, T.; Christoffersen, T.E.; Langfort, J.; Viese, M.; Galbo, H.; Haunsø, S.; Sheikh, S.P. Expression profiling reveals differences in metabolic gene expression between exercise-induced cardiac effects and maladaptive cardiac hypertrophy. FEBS J. 2005, 272, 2684–2695. [Google Scholar] [CrossRef]
- Bonen, A.; Luiken, J.J.; Arumugam, Y.; Glatz, J.F.; Tandon, N.N. Acute regulation of fatty acid uptake involves the cellular redistribution of fatty acid translocase. J. Biol. Chem. 2000, 275, 14501–14508. [Google Scholar] [CrossRef] [PubMed]
- Duncan, J.G.; Bharadwaj, K.G.; Fong, J.L.; Mitra, R.; Sambandam, N.; Courtois, M.R.; Lavine, K.J.; Goldberg, I.J.; Kelly, D.P. Rescue of cardiomyopathy in peroxisome proliferator-activated receptor-alpha transgenic mice by deletion of lipoprotein lipase identifies sources of cardiac lipids and peroxisome proliferator-activated receptor-alpha activators. Circulation 2010, 121, 426–435. [Google Scholar] [CrossRef] [PubMed]
- Zlobine, I.; Gopal, K.; Ussher, J.R. Lipotoxicity in obesity and diabetes-related cardiac dysfunction. Biochim. Biophys. Acta 2016, 1861, 1555–1568. [Google Scholar] [CrossRef]
- Fukushima, A.; Lopaschuk, G.D. Cardiac fatty acid oxidation in heart failure associated with obesity and diabetes. Biochim. Biophys. Acta 2016, 1861, 1525–1534. [Google Scholar] [CrossRef]
- Lopaschuk, G.D.; Karwi, Q.G.; Tian, R.; Wende, A.R.; Abel, E.D. Cardiac Energy Metabolism in Heart Failure. Circ. Res. 2021, 128, 1487–1513. [Google Scholar] [CrossRef] [PubMed]
- D’Souza, K.; Nzirorera, C.; Kienesberger, P.C. Lipid metabolism and signaling in cardiac lipotoxicity. Biochim. Biophys. Acta 2016, 1861, 1513–1524. [Google Scholar] [CrossRef]
- Unger, R.H. Lipotoxic diseases. Annu. Rev. Med. 2002, 53, 319–336. [Google Scholar] [CrossRef]
- Lopaschuk, G.D.; Folmes, C.D.; Stanley, W.C. Cardiac energy metabolism in obesity. Circ. Res. 2007, 101, 335–347. [Google Scholar] [CrossRef]
- Young, M.E.; Guthrie, P.H.; Razeghi, P.; Leighton, B.; Abbasi, S.; Patil, S.; Youker, K.A.; Taegtmeyer, H. Impaired long-chain fatty acid oxidation and contractile dysfunction in the obese Zucker rat heart. Diabetes 2002, 51, 2587–2595. [Google Scholar] [CrossRef]
- Hayat, S.A.; Patel, B.; Khattar, R.S.; Malik, R.A. Diabetic cardiomyopathy: Mechanisms, diagnosis and treatment. Clin. Sci. 2004, 107, 539–557. [Google Scholar] [CrossRef]
- Singal, P.K.; Belló-Klein, A.; Farahmand, F.; Sandhawalia, V. Oxidative stress and functional deficit in diabetic cardiomyopathy. Adv. Exp. Med. Biol. 2001, 498, 213–220. [Google Scholar] [PubMed]
- Way, K.J.; Katai, N.; King, G.L. Protein kinase C and the development of diabetic vascular complications. Diabet. Med. 2001, 18, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Semsarian, C.; Wu, M.J.; Ju, Y.K.; Marciniec, T.; Yeoh, T.; Allen, D.G.; Harvey, R.P.; Graham, R.M. Skeletal muscle hypertrophy is mediated by a Ca2+-dependent calcineurin signalling pathway. Nature 1999, 400, 576–581. [Google Scholar] [CrossRef]
- Musarò, A.; McCullagh, K.J.; Naya, F.J.; Olson, E.N.; Rosenthal, N. IGF-1 induces skeletal myocyte hypertrophy through calcineurin in association with GATA-2 and NF-ATc1. Nature 1999, 400, 581–585. [Google Scholar] [CrossRef] [PubMed]
- Qin, D.; Huang, B.; Deng, L.; El-Adawi, H.; Ganguly, K.; Sowers, J.R.; El-Sherif, N. Downregulation of K(+) channel genes expression in type I diabetic cardiomyopathy. Biochem. Biophys. Res. Commun. 2001, 283, 549–553. [Google Scholar] [CrossRef] [PubMed]
- Fein, F.S.; Sonnenblick, E.H. Diabetic cardiomyopathy. Prog. Cardiovasc. Dis. 1985, 27, 255–270. [Google Scholar] [CrossRef]
- Jordan, S.D.; Krüger, M.; Willmes, D.M.; Redemann, N.; Wunderlich, F.T.; Brönneke, H.S.; Merkwirth, C.; Kashkar, H.; Olkkonen, V.M.; Böttger, T.; et al. Obesity-induced overexpression of miRNA-143 inhibits insulin-stimulated AKT activation and impairs glucose metabolism. Nat. Cell Biol. 2011, 13, 434–446. [Google Scholar] [CrossRef]
- Zhou, B.; Li, C.; Qi, W.; Zhang, Y.; Zhang, F.; Wu, J.X.; Hu, Y.N.; Wu, D.M.; Liu, Y.; Yan, T.; et al. Downregulation of miR-181a upregulates sirtuin-1 (SIRT1) and improves hepatic insulin sensitivity. Diabetologia 2012, 55, 2032–2043. [Google Scholar] [CrossRef]
- Trajkovski, M.; Hausser, J.; Soutschek, J.; Bhat, B.; Akin, A.; Zavolan, M.; Heim, M.H.; Stoffel, M. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature 2011, 474, 649–653. [Google Scholar] [CrossRef]
- Limongelli, G.; Adorisio, R.; Baggio, C.; Bauce, B.; Biagini, E.; Castelletti, S.; Favilli, S.; Imazio, M.; Lioncino, M.; Merlo, M. Diagnosis and Management of Rare Cardiomyopathies in Adult and Paediatric Patients. A Position Paper of the Italian Society of Cardiology (SIC) and Italian Society of Paediatric Cardiology (SICP). Int. J. Cardiol. 2022, 357, 55–71. [Google Scholar] [CrossRef]
- Braunwald, E. Cardiomyopathies. Circ. Res. 2017, 121, 711–721. [Google Scholar] [CrossRef] [PubMed]
- Precone, V.; Krasi, G.; Guerri, G.; Madureri, A.; Piazzani, M.; Michelini, S.; Barati, S.; Maniscalchi, T.; Bressan, S.; Bertelli, M. Cardiomyopathies. Acta Biomed. 2019, 90, 32–43. [Google Scholar]
- Saini, H. Chapter 6—Pathophysiology of Cardiomyopathies. In Cellular and Molecular Pathobiology of Cardiovascular Disease; Willis, M.S., Homeister, J.W., Stone, J.R., Eds.; Academic Press: San Diego, CA, USA, 2014; pp. 101–119. [Google Scholar]
- Rambhatla, T.; Perk, G. The Echocardiography Companion: Study Guide and Review; Springer: Berlin/Heidelberg, Germany, 2020. [Google Scholar]
- Braam, R.L.; Post, J.G. Restrictive Cardiomyopathy. In Clinical Cardiogenetics; Baars, H.F., Doevendans, P.A.F.M., van der Smagt, J.J., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 139–149. [Google Scholar]
- Cheng, Y.; Regnier, M. Cardiac troponin structure-function and the influence of hypertrophic cardiomyopathy associated mutations on modulation of contractility. Arch. Biochem. Biophys. 2016, 601, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Cimiotti, D.; Budde, H.; Hassoun, R.; Jaquet, K. Genetic Restrictive Cardiomyopathy: Causes and Consequences-An Integrative Approach. Int. J. Mol. Sci. 2021, 22, 558. [Google Scholar] [CrossRef] [PubMed]
- McKenna, W.J.; Judge, D.P. Epidemiology of the inherited cardiomyopathies. Nat. Rev. Cardiol. 2021, 18, 22–36. [Google Scholar] [CrossRef] [PubMed]
- Sinagra, G.; Merlo, M.; Pinamonti, B. (Eds.) Dilated Cardiomyopathy: From Genetics to Clinical Management; Springer: Cham, Switzerland, 2019. [Google Scholar]
- Weintraub, R.G.; Semsarian, C.; Macdonald, P. Dilated cardiomyopathy. Lancet 2017, 390, 400–414. [Google Scholar] [CrossRef]
- Schultheiss, H.P.; Fairweather, D.; Caforio, A.; Escher, F.; Hershberger, R.E.; Lipshultz, S.E.; Liu, P.P.; Matsumori, A.; Mazzanti, A.; McMurray, J.; et al. Dilated cardiomyopathy. Nat. Rev. Dis. Primers 2019, 5, 32. [Google Scholar] [CrossRef]
- Mahmaljy, H.; Yelamanchili, V.S.; Singhal, M. Dilated Cardiomyopathy; StatPearls: Treasure Island, FL, USA, 2022. [Google Scholar]
- Reichart, D.; Magnussen, C.; Zeller, T.; Blankenberg, S. Dilated cardiomyopathy: From epidemiologic to genetic phenotypes: A translational review of current literature. J. Intern. Med. 2019, 286, 362–372. [Google Scholar] [CrossRef]
- Granzier, H.L.; Labeit, S. The Giant Muscle Protein Titin is an Adjustable Molecular Spring. Exerc. Sport Sci. Rev. 2006, 34, 50–53. [Google Scholar] [CrossRef]
- Chen, S.N.; Mestroni, L.; Taylor, M.R.G. Genetics of dilated cardiomyopathy. Curr. Opin. Cardiol. 2021, 36, 288–294. [Google Scholar] [CrossRef]
- Dittmer, T.A.; Misteli, T. The lamin protein family. Genome Biol. 2011, 12, 222. [Google Scholar] [CrossRef] [PubMed]
- Maron, B.J.; Maron, M.S. Hypertrophic cardiomyopathy. Lancet 2013, 381, 242–255. [Google Scholar] [CrossRef]
- Marsiglia, J.D.; Pereira, A.C. Hypertrophic cardiomyopathy: How do mutations lead to disease? Arq. Bras. Cardiol. 2014, 102, 295–304. [Google Scholar] [CrossRef] [PubMed]
- Papadakis, M.; Basu, J.; Sharma, S. Mavacamten: Treatment aspirations in hypertrophic cardiomyopathy. Lancet 2020, 396, 736–737. [Google Scholar] [CrossRef]
- Semsarian, C.; Ingles, J.; Maron, M.S.; Maron, B.J. New perspectives on the prevalence of hypertrophic cardiomyopathy. J. Am. Coll. Cardiol. 2015, 65, 1249–1254. [Google Scholar] [CrossRef]
- Geske, J.B.; Ommen, S.R.; Gersh, B.J. Hypertrophic Cardiomyopathy: Clinical Update. JACC Heart Fail. 2018, 6, 364–375. [Google Scholar] [CrossRef]
- Maron, B.J.; Maron, M.S.; Semsarian, C. Genetics of hypertrophic cardiomyopathy after 20 years: Clinical perspectives. J. Am. Coll. Cardiol. 2012, 60, 705–715. [Google Scholar] [CrossRef]
- Akhtar, M.; Elliott, P. The genetics of hypertrophic cardiomyopathy. Glob. Cardiol. Sci. Pract. 2018, 2018, 36. [Google Scholar]
- Marian, A.J.; Roberts, R. The molecular genetic basis for hypertrophic cardiomyopathy. J. Mol. Cell Cardiol. 2001, 33, 655–670. [Google Scholar] [CrossRef]
- Liew, C.C.; Dzau, V.J. Molecular genetics and genomics of heart failure. Nat. Rev. Genet. 2004, 5, 811–825. [Google Scholar] [CrossRef]
- Ibrahim, N.E.; Januzzi, J.L. Established and Emerging Roles of Biomarkers in Heart Failure. Circ. Res. 2018, 123, 614–629. [Google Scholar] [CrossRef] [PubMed]
- Lippi, G.; Sanchis-Gomar, F. Global epidemiology and future trends of heart failure. AME Med. J. 2020, 5, 15. [Google Scholar] [CrossRef]
- Truby, L.K.; Rogers, J.G. Advanced Heart Failure: Epidemiology, Diagnosis, and Therapeutic Approaches. JACC Heart Fail 2020, 8, 523–536. [Google Scholar] [CrossRef] [PubMed]
- Mirrakhimov, M.M.; Moldotashev, I.K.; Dzhaĭlobaeva, K.A.; Tenenbaum, A.M. Noninvasive assessment of the function of the lesser circulation and of the right heart in mitral stenosis. Kardiologiia 1987, 27, 45–48. [Google Scholar] [PubMed]
- Cook, C.; Cole, G.; Asaria, P.; Jabbour, R.; Francis, D.P. The annual global economic burden of heart failure. Int. J. Cardiol. 2014, 171, 368–376. [Google Scholar] [CrossRef]
- Urbich, M.; Globe, G.; Pantiri, K.; Heisen, M.; Bennison, C.; Wirtz, H.S.; Di Tanna, G.L. A Systematic Review of Medical Costs Associated with Heart Failure in the USA (2014–2020). Pharmacoeconomics 2020, 38, 1219–1236. [Google Scholar] [CrossRef]
- Malik, A.; Brito, D.; Vaqar, S.; Chhabra, L. Congestive Heart Failure; StatPearls: Treasure Island, FL, USA, 2022. [Google Scholar]
- Simmonds, S.J.; Cuijpers, I.; Heymans, S.; Jones, E. Cellular and Molecular Differences between HFpEF and HFrEF: A Step Ahead in an Improved Pathological Understanding. Cells 2020, 9, 242. [Google Scholar] [CrossRef]
- Zhu, S.; Han, Z.; Luo, Y.; Chen, Y.; Zeng, Q.; Wu, X.; Yuan, W. Molecular mechanisms of heart failure: Insights from Drosophila. Heart Fail. Rev. 2017, 22, 91–98. [Google Scholar] [CrossRef]
- Bozkurt, B.; Colvin, M.; Cook, J.; Cooper, L.T.; Deswal, A.; Fonarow, G.C.; Francis, G.S.; Lenihan, D.; Lewis, E.F.; McNamara, D.M.; et al. Current Diagnostic and Treatment Strategies for Specific Dilated Cardiomyopathies: A Scientific Statement From the American Heart Association. Circulation 2016, 134, e579–e646. [Google Scholar] [CrossRef]
- Czepluch, F.S.; Wollnik, B.; Hasenfuss, G. Genetic determinants of heart failure: Facts and numbers. ESC Heart Fail. 2018, 5, 211–217. [Google Scholar] [CrossRef]
- Kaviarasan, V.; Mohammed, V.; Veerabathiran, R. Genetic predisposition study of heart failure and its association with cardiomyopathy. Egypt Heart J. 2022, 74, 5. [Google Scholar] [CrossRef] [PubMed]
- Cahill, T.J.; Ashrafian, H.; Watkins, H. Genetic cardiomyopathies causing heart failure. Circ. Res. 2013, 113, 660–675. [Google Scholar] [CrossRef] [PubMed]
- McNally, E.M.; Barefield, D.Y.; Puckelwartz, M.J. The genetic landscape of cardiomyopathy and its role in heart failure. Cell Metab. 2015, 21, 174–182. [Google Scholar] [CrossRef] [PubMed]
- Hershberger, R.E.; Givertz, M.M.; Ho, C.Y.; Judge, D.P.; Kantor, P.F.; McBride, K.L.; Morales, A.; Taylor, M.; Vatta, M.; Ware, S.M. Genetic Evaluation of Cardiomyopathy-A Heart Failure Society of America Practice Guideline. J. Card. Fail. 2018, 24, 281–302. [Google Scholar] [CrossRef] [PubMed]
- Creemers, E.E.; Wilde, A.A.; Pinto, Y.M. Heart failure: Advances through genomics. Nat. Rev. Genet. 2011, 12, 357–362. [Google Scholar] [CrossRef]
- Khan, M.A.; Hashim, M.J.; Mustafa, H.; Baniyas, M.Y.; Al Suwaidi, S.; AlKatheeri, R.; Alblooshi, F.; Almatrooshi, M.; Alzaabi, M.; Al Darmaki, R.S.; et al. Global Epidemiology of Ischemic Heart Disease: Results from the Global Burden of Disease Study. Cureus 2020, 12, e9349. [Google Scholar] [CrossRef]
- Stewart, S.; Murphy, N.F.; Walker, A.; McGuire, A.; McMurray, J.J. The current cost of angina pectoris to the National Health Service in the UK. Heart 2003, 89, 848–853. [Google Scholar] [CrossRef]
- Wang, Q. Molecular genetics of coronary artery disease. Curr. Opin. Cardiol. 2005, 20, 182–188. [Google Scholar] [CrossRef]
- Gheorghe, A.; Griffiths, U.; Murphy, A.; Legido-Quigley, H.; Lamptey, P.; Perel, P. The economic burden of cardiovascular disease and hypertension in low- and middle-income countries: A systematic review. BMC Public Health 2018, 18, 975. [Google Scholar] [CrossRef]
- Colditz, G.A.; Stampfer, M.J.; Willett, W.C.; Rosner, B.; Speizer, F.E.; Hennekens, C.H. A prospective study of parental history of myocardial infarction and coronary heart disease in women. Am. J. Epidemiol. 1986, 123, 48–58. [Google Scholar] [CrossRef]
- Wang, Q. Molecular Genetics—Textbook of Cardiovascular Medicine, 2nd ed.; Lippincott, Williams Wilkins: Philadelphia, PA, USA, 2001. [Google Scholar]
- Marenberg, M.E.; Risch, N.; Berkman, L.F.; Floderus, B.; de Faire, U. Genetic susceptibility to death from coronary heart disease in a study of twins. N. Engl. J. Med. 1994, 330, 1041–1046. [Google Scholar] [CrossRef] [PubMed]
- Curran, M.E.; Splawski, I.; Timothy, K.W.; Vincent, G.M.; Green, E.D.; Keating, M.T. A molecular basis for cardiac arrhythmia: HERG mutations cause long QT syndrome. Cell 1995, 80, 795–803. [Google Scholar] [CrossRef]
- Wang, Q.; Shen, J.; Splawski, I.; Atkinson, D.; Li, Z.; Robinson, J.L.; Moss, A.J.; Towbin, J.A.; Keating, M.T. SCN5A mutations associated with an inherited cardiac arrhythmia, long QT syndrome. Cell 1995, 80, 805–811. [Google Scholar] [CrossRef]
- Chen, Q.; Kirsch, G.E.; Zhang, D.; Brugada, R.; Brugada, J.; Brugada, P.; Potenza, D.; Moya, A.; Borggrefe, M.; Breithardt, G.; et al. Genetic basis and molecular mechanism for idiopathic ventricular fibrillation. Nature 1998, 392, 293–296. [Google Scholar] [CrossRef] [PubMed]
- Imazio, M.; Gaita, F. Diagnosis and treatment of pericarditis. Heart 2015, 101, 1159–1168. [Google Scholar] [CrossRef] [PubMed]
- Kyto, V.; Sipila, J.; Rautava, P. Clinical profile and influences on outcomes in patients hospitalized for acute pericarditis. Circulation 2014, 130, 1601–1606. [Google Scholar] [CrossRef]
- Adler, Y.; Charron, P.; Imazio, M.; Badano, L.; Barón-Esquivias, G.; Bogaert, J.; Brucato, A.; Gueret, P.; Klingel, K.; Lionis, C.; et al. 2015 ESC Guidelines for the diagnosis and management of pericardial diseases: The Task Force for the Diagnosis and Management of Pericardial Diseases of the European Society of Cardiology (ESC) Endorsed by: The European Association for Cardio-Thoracic Surgery (EACTS). Eur. Heart J. 2015, 36, 2921–2964. [Google Scholar]
- Imazio, M.; Spodick, D.H.; Brucato, A.; Trinchero, R.; Adler, Y. Controversial issues in the management of pericardial diseases. Circulation 2010, 121, 916–928. [Google Scholar] [CrossRef]
- Buckley, L.F.; Viscusi, M.M.; Van Tassell, B.W.; Abbate, A. Interleukin-1 blockade for the treatment of pericarditis. Eur. Heart J. Cardiovasc. Pharmacother. 2018, 4, 46–53. [Google Scholar] [CrossRef]
- Bonaventura, A.; Vecchié, A.; Mauro, A.G.; Brucato, A.L.; Imazio, M.; Abbate, A. An update on the pathophysiology of acute and recurrent pericarditis. Panminerva Med. 2021, 63, 249–260. [Google Scholar] [CrossRef]
- Abbate, A.; Toldo, S.; Marchetti, C.; Kron, J.; Van Tassell, B.W.; Dinarello, C.A. Interleukin-1 and the Inflammasome as Therapeutic Targets in Cardiovascular Disease. Circ. Res. 2020, 126, 1260–1280. [Google Scholar] [CrossRef] [PubMed]
- Mauro, A.G.; Bonaventura, A.; Mezzaroma, E.; Quader, M.; Toldo, S. NLRP3 Inflammasome in Acute Myocardial Infarction. J. Cardiovasc. Pharmacol. 2019, 74, 175–187. [Google Scholar] [CrossRef] [PubMed]
- Cantarini, L.; Lucherini, O.M.; Cimaz, R.; Baldari, C.T.; Bellisai, F.; Rossi Paccani, S.; Laghi Pasini, F.; Capecchi, P.L.; Sebastiani, G.D.; Galeazzi, M. Idiopathic recurrent pericarditis refractory to colchicine treatment can reveal tumor necrosis factor receptor-associated periodic syndrome. Int. J. Immunopathol. Pharmacol. 2009, 22, 1051–1058. [Google Scholar] [CrossRef] [PubMed]
- Peet, C.J.; Rowczenio, D.; Omoyinmi, E.; Papadopoulou, C.; Mapalo, B.; Wood, M.R.; Capon, F.; Lachmann, H.J. Pericarditis and Autoinflammation: A Clinical and Genetic Analysis of Patients with Idiopathic Recurrent Pericarditis and Monogenic Autoinflammatory Diseases at a National Referral Center. J. Am. Heart Assoc. 2022, 11, e024931. [Google Scholar] [CrossRef] [PubMed]
- Dodé, C.; André, M.; Bienvenu, T.; Hausfater, P.; Pêcheux, C.; Bienvenu, J.; Lecron, J.C.; Reinert, P.; Cattan, D.; Piette, J.C.; et al. The enlarging clinical, genetic, and population spectrum of tumor necrosis factor receptor-associated periodic syndrome. Arthritis Rheum. 2002, 46, 2181–2188. [Google Scholar] [CrossRef] [PubMed]
- Vos, T.; Barber, R.M.; Bell, B.; Bertozzi-Villa, A.; Biryukov, S.; Bolliger, I.; Charlson, F.; Davis, A.; Degenhardt, L.; Dicker, D.; et al. Global, regional, and national incidence, prevalence, and years lived with disability for 301 acute and chronic diseases and injuries in 188 countries, 1990–2013: A systematic analysis for the Global Burden of Disease Study 2013. Lancet 2015, 386, 743–800. [Google Scholar] [CrossRef]
- Dionne, A.; Dahdah, N. Myocarditis and Kawasaki disease. Int. J. Rheum. Dis. 2018, 21, 45–49. [Google Scholar] [CrossRef]
- Cooper, L.T., Jr. Myocarditis. N. Engl. J. Med. 2009, 360, 1526–1538. [Google Scholar] [CrossRef]
- Takeuchi, S.; Kawada, J.I.; Okuno, Y.; Horiba, K.; Suzuki, T.; Torii, Y.; Yasuda, K.; Numaguchi, A.; Kato, T.; Takahashi, Y.; et al. Identification of potential pathogenic viruses in patients with acute myocarditis using next-generation sequencing. J. Med. Virol. 2018, 90, 1814–1821. [Google Scholar] [CrossRef]
- Chapman, S.J.; Hill, A.V. Human genetic susceptibility to infectious disease. Nat. Rev. Genet. 2012, 13, 175–188. [Google Scholar] [CrossRef]
- Campuzano, O.; Fernández-Falgueras, A.; Sarquella-Brugada, G.; Sanchez, O.; Cesar, S.; Mademont, I.; Allegue, C.; Mates, J.; Pérez-Serra, A.; Coll, M.; et al. A Genetically Vulnerable Myocardium May Predispose to Myocarditis. J. Am. Coll. Cardiol. 2015, 66, 2913–2914. [Google Scholar] [CrossRef] [PubMed]
- Baggio, C.; Gagno, G.; Porcari, A.; Paldino, A.; Artico, J.; Castrichini, M.; Dal Ferro, M.; Bussani, R.; Merlo, M. Myocarditis: Which Role for Genetics? Curr. Cardiol. Rep. 2021, 23, 58. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Song, J. Targeted Therapy in Cardiovascular Disease: A Precision Therapy Era. Front. Pharmacol. 2021, 12, 623674. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, G.G.; Steg, P.G.; Szarek, M.; Bhatt, D.L.; Bittner, V.A.; Diaz, R.; Edelberg, J.M.; Goodman, S.G.; Hanotin, C.; Harrington, R.A.; et al. Alirocumab and Cardiovascular Outcomes after Acute Coronary Syndrome. N. Engl. J. Med. 2018, 379, 2097–2107. [Google Scholar] [CrossRef] [PubMed]
- Ikonomidis, I.; Tzortzis, S.; Andreadou, I.; Paraskevaidis, I.; Katseli, C.; Katsimbri, P.; Pavlidis, G.; Parissis, J.; Kremastinos, D.; Anastasiou-Nana, M.; et al. Increased benefit of interleukin-1 inhibition on vascular function, myocardial deformation, and twisting in patients with coronary artery disease and coexisting rheumatoid arthritis. Circ. Cardiovasc. Imaging 2014, 7, 619–628. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.J.; Fang, Q.; Davol, P.A.; Gu, Y.; Sievers, R.E.; Grabert, R.C.; Gall, J.M.; Tsang, E.; Yee, M.S.; Fok, H.; et al. Antibody targeting of stem cells to infarcted myocardium. Stem Cells 2007, 25, 712–717. [Google Scholar] [CrossRef]
- Gundlach, C.W., IV; Caivano, A.; Cabreira-Hansen, M.; Gahremanpour, A.; Brown, W.S.; Zheng, Y.; McIntyre, B.W.; Willerson, J.T.; Dixon, R.A.; Perin, E.C.; et al. Synthesis and evaluation of an anti-MLC1 x anti-CD90 bispecific antibody for targeting and retaining bone-marrow-derived multipotent stromal cells in infarcted myocardium. Bioconjug. Chem. 2011, 22, 1706–1714. [Google Scholar] [CrossRef]
- Lum, L.G.; Fok, H.; Sievers, R.; Abedi, M.; Quesenberry, P.J.; Lee, R.J. Targeting of Lin-Sca+ hematopoietic stem cells with bispecific antibodies to injured myocardium. Blood Cells Mol. Dis. 2004, 32, 82–87. [Google Scholar] [CrossRef]
- Helas, S.; Goettsch, C.; Schoppet, M.; Zeitz, U.; Hempel, U.; Morawietz, H.; Kostenuik, P.J.; Erben, R.G.; Hofbauer, L.C. Inhibition of receptor activator of NF-kappaB ligand by denosumab attenuates vascular calcium deposition in mice. Am. J. Pathol. 2009, 175, 473–478. [Google Scholar] [CrossRef]
- Lerman, D.A.; Prasad, S.; Alotti, N. Denosumab could be a Potential Inhibitor of Valvular Interstitial Cells Calcification in vitro. Int. J. Cardiovasc. Res. 2016, 5, 1000249. [Google Scholar] [CrossRef]
- Imazio, M.; Brucato, A.; Cemin, R.; Ferrua, S.; Maggiolini, S.; Beqaraj, F.; Demarie, D.; Forno, D.; Ferro, S.; Maestroni, S.; et al. A randomized trial of colchicine for acute pericarditis. N. Engl. J. Med. 2013, 369, 1522–1528. [Google Scholar] [CrossRef] [PubMed]
- Pleger, S.T.; Brinks, H.; Ritterhoff, J.; Raake, P.; Koch, W.J.; Katus, H.A.; Most, P. Heart failure gene therapy: The path to clinical practice. Circ. Res. 2013, 113, 792–809. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Parmacek, M.S.; Morle, G.; Bolling, S.; Leiden, J.M. Expression of recombinant genes in myocardium in vivo after direct injection of DNA. Circulation 1990, 82, 2217–2221. [Google Scholar] [CrossRef] [PubMed]
- Pacak, C.A.; Byrne, B.J. AAV Vectors for Cardiac Gene Transfer: Experimental Tools and Clinical Opportunities. Mol. Ther. 2011, 19, 1582–1590. [Google Scholar] [CrossRef]
- Korpela, H.; Järveläinen, N.; Siimes, S.; Lampela, J.; Airaksinen, J.; Valli, K.; Turunen, M.; Pajula, J.; Nurro, J.; Ylä-Herttuala, S. Gene therapy for ischaemic heart disease and heart failure. J. Intern. Med. 2021, 290, 567–582. [Google Scholar] [CrossRef]
- Wolfram, J.A.; Donahue, J.K. Gene therapy to treat cardiovascular disease. J. Am. Heart Assoc. 2013, 2, e000119. [Google Scholar] [CrossRef]
- Hedman, M.; Muona, K.; Hedman, A.; Kivelä, A.; Syvänne, M.; Eränen, J.; Rantala, A.; Stjernvall, J.; Nieminen, M.S.; Hartikainen, J.; et al. Eight-year safety follow-up of coronary artery disease patients after local intracoronary VEGF gene transfer. Gene Ther. 2009, 16, 629–634. [Google Scholar] [CrossRef]
- Kukuła, K.; Chojnowska, L.; Dąbrowski, M.; Witkowski, A.; Chmielak, Z.; Skwarek, M.; Kądziela, J.; Teresińska, A.; Małecki, M.; Janik, P.; et al. Intramyocardial plasmid-encoding human vascular endothelial growth factor A165/basic fibroblast growth factor therapy using percutaneous transcatheter approach in patients with refractory coronary artery disease (VIF-CAD). Am. Heart J. 2011, 161, 581–589. [Google Scholar] [CrossRef]
- Li, J.; Wei, Y.; Liu, K.; Yuan, C.; Tang, Y.; Quan, Q.; Chen, P.; Wang, W.; Hu, H.; Yang, L. Synergistic effects of FGF-2 and PDGF-BB on angiogenesis and muscle regeneration in rabbit hindlimb ischemia model. Microvasc. Res. 2010, 80, 10–17. [Google Scholar] [CrossRef]
- Favaloro, L.; Diez, M.; Mendiz, O.; Janavel, G.V.; Valdivieso, L.; Ratto, R.; Garelli, G.; Salmo, F.; Criscuolo, M.; Bercovich, A.; et al. High-dose plasmid-mediated VEGF gene transfer is safe in patients with severe ischemic heart disease (Genesis-I). A phase I, open-label, two-year follow-up trial. Catheter. Cardiovasc. Interv. 2013, 82, 899–906. [Google Scholar] [CrossRef]
- Hammond, H.K.; Penny, W.F.; Traverse, J.H.; Henry, T.D.; Watkins, M.W.; Yancy, C.W.; Sweis, R.N.; Adler, E.D.; Patel, A.N.; Murray, D.R. Intracoronary Gene Transfer of Adenylyl Cyclase 6 in Patients With Heart Failure: A Randomized Clinical Trial. JAMA Cardiol. 2016, 1, 163–171. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, B.; Butler, J.; Felker, G.M.; Ponikowski, P.; Voors, A.A.; Desai, A.S.; Barnad, D.; Bouchard, A.; Jaski, B.; Lyon, A.R. Calcium upregulation by percutaneous administration of gene therapy in patients with cardiac disease (CUPID 2): A randomised, multinational, double-blind, placebo-controlled, phase 2b trial. Lancet 2016, 387, 1178–1186. [Google Scholar] [CrossRef]
- Stewart, D.J.; Kutryk, M.J.; Fitchett, D.; Freeman, M.; Camack, N.; Su, Y.; Della Siega, A.; Bilodeau, L.; Burton, J.R.; Proulx, G.; et al. VEGF gene therapy fails to improve perfusion of ischemic myocardium in patients with advanced coronary disease: Results of the NORTHERN trial. Mol Ther. 2009, 17, 1109–1115. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.J.; Zhang, Y.R.; Chen, B.; Zhang, S.L.; Jia, E.Z.; Wang, L.S.; Zhu, T.B.; Li, C.J.; Wang, H.; Huang, J.; et al. Phase I clinical trial on intracoronary administration of Ad-hHGF treating severe coronary artery disease. Mol. Biol. Rep. 2009, 36, 1323–1329. [Google Scholar] [CrossRef]
- Jin, Y.N.; Inubushi, M.; Masamoto, K.; Odaka, K.; Aoki, I.; Tsuji, A.B.; Sagara, M.; Koizumi, M.; Saga, T. Long-term effects of hepatocyte growth factor gene therapy in rat myocardial infarct model. Gene Ther. 2012, 19, 836–843. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.J.; Chen, B.; Sheng, Z.; Zhang, D.G.; Jia, E.Z.; Wang, W.; Ma, D.C.; Zhu, T.B.; Wang, L.S.; Li, C.J.; et al. Improvement of heart function in postinfarct heart failure swine models after hepatocyte growth factor gene transfer: Comparison of low-, medium- and high-dose groups. Mol. Biol. Rep. 2010, 37, 2075–2081. [Google Scholar] [CrossRef]
- Kilian, E.G.; Sadoni, S.; Vicol, C.; Kelly, R.; van Hulst, K.; Schwaiger, M.; Kupatt, C.; Boekstegers, P.; Pillai, R.; Channon, K.; et al. Myocardial transfection of hypoxia inducible factor-1alpha via an adenoviral vector during coronary artery bypass grafting—A multicenter phase I and safety study. Circ. J. 2010, 74, 916–924. [Google Scholar] [CrossRef]
- Heinl-Green, A.; Radke, P.W.; Munkonge, F.M.; Frass, O.; Zhu, J.; Vincent, K.; Geddes, D.M.; Alton, E.W. The efficacy of a ‘master switch gene’ HIF-1alpha in a porcine model of chronic myocardial ischaemia. Eur. Heart J. 2005, 26, 1327–1332. [Google Scholar] [CrossRef]
- Jessup, M.; Greenberg, B.; Mancini, D.; Cappola, T.; Pauly, D.F.; Jaski, B.; Yaroshinsky, A.; Zsebo, K.M.; Dittrich, H.; Hajjar, R.J. Calcium Upregulation by Percutaneous Administration of Gene Therapy in Cardiac Disease (CUPID): A phase 2 trial of intracoronary gene therapy of sarcoplasmic reticulum Ca2+-ATPase in patients with advanced heart failure. Circulation 2011, 124, 304–313. [Google Scholar] [CrossRef]
- Penn, M.S.; Mendelsohn, F.O.; Schaer, G.L.; Sherman, W.; Farr, M.; Pastore, J.; Rouy, D.; Clemens, R.; Aras, R.; Losordo, D.W. An open-label dose escalation study to evaluate the safety of administration of nonviral stromal cell-derived factor-1 plasmid to treat symptomatic ischemic heart failure. Circ. Res. 2013, 112, 816–825. [Google Scholar] [CrossRef]
- Rebolledo, B.; Lai, N.C.; Gao, M.H.; Takahashi, T.; Roth, D.M.; Baird, S.M.; Hammond, H.K. Adenylylcyclase gene transfer increases function of the failing heart. Hum. Gene Ther. 2006, 17, 1043–1048. [Google Scholar] [CrossRef] [PubMed]
- Roth, D.M.; Gao, M.H.; Lai, N.C.; Drumm, J.; Dalton, N.; Zhou, J.Y.; Zhu, J.; Entrikin, D.; Hammond, H.K. Cardiac-directed adenylyl cyclase expression improves heart function in murine cardiomyopathy. Circulation 1999, 99, 3099–3102. [Google Scholar] [CrossRef] [PubMed]
- Roth, D.M.; Bayat, H.; Drumm, J.D.; Gao, M.H.; Swaney, J.S.; Ander, A.; Hammond, H.K. Adenylyl cyclase increases survival in cardiomyopathy. Circulation 2002, 105, 1989–1994. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, T.; Tang, T.; Lai, N.C.; Roth, D.M.; Rebolledo, B.; Saito, M.; Lew, W.Y.; Clopton, P.; Hammond, H.K. Increased cardiac adenylyl cyclase expression is associated with increased survival after myocardial infarction. Circulation 2006, 114, 388–396. [Google Scholar] [CrossRef] [PubMed]
- Lai, N.C.; Roth, D.M.; Gao, M.; Fine, S.; Head, B.P.; Zhu, J.; McKirnan, M.D.; Kwong, C.; Dalton, N.; Urasawa, K.; et al. Intracoronary delivery of adenovirus encoding adenylyl cyclase VI increases left ventricular function and cAMP-generating capacity. Circulation 2000, 102, 2396–2401. [Google Scholar] [CrossRef] [PubMed]
- Shah, A.S.; White, D.C.; Emani, S.; Kypson, A.P.; Lilly, R.E.; Wilson, K.; Glower, D.D.; Lefkowitz, R.J.; Koch, W.J. In vivo ventricular gene delivery of a beta-adrenergic receptor kinase inhibitor to the failing heart reverses cardiac dysfunction. Circulation 2001, 103, 1311–1316. [Google Scholar] [CrossRef]
- Williams, M.L.; Hata, J.A.; Schroder, J.; Rampersaud, E.; Petrofski, J.; Jakoi, A.; Milano, C.A.; Koch, W.J. Targeted beta-adrenergic receptor kinase (betaARK1) inhibition by gene transfer in failing human hearts. Circulation 2004, 109, 1590–1593. [Google Scholar] [CrossRef]
- Most, P.; Remppis, A.; Pleger, S.T.; Katus, H.A.; Koch, W.J. S100A1: A novel inotropic regulator of cardiac performance. Transition from molecular physiology to pathophysiological relevance. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2007, 293, R568–R577. [Google Scholar] [CrossRef]
- Pleger, S.T.; Most, P.; Boucher, M.; Soltys, S.; Chuprun, J.K.; Pleger, W.; Gao, E.; Dasgupta, A.; Rengo, G.; Remppis, A.; et al. Stable myocardial-specific AAV6-S100A1 gene therapy results in chronic functional heart failure rescue. Circulation 2007, 115, 2506–2515. [Google Scholar] [CrossRef] [PubMed]
- Szatkowski, M.L.; Westfall, M.V.; Gomez, C.A.; Wahr, P.A.; Michele, D.E.; DelloRusso, C.; Turner, I.I.; Hong, K.E.; Albayya, F.P.; Metzger, J.M. In vivo acceleration of heart relaxation performance by parvalbumin gene delivery. J. Clin. Investig. 2001, 107, 191–198. [Google Scholar] [CrossRef]
- Terashvili, M.; Bosnjak, Z.J. Stem Cell Therapies in Cardiovascular Disease. J. Cardiothorac. Vasc. Anesth. 2019, 33, 209–222. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Gordillo-Martinez, F.; Jiang, L.; He, P.; Hong, W.; Wei, X.; Staines, K.A.; Macrae, V.E.; Zhang, C.; Yu, D.; et al. Zinc ameliorates human aortic valve calcification through GPR39 mediated ERK1/2 signalling pathway. Cardiovasc. Res. 2021, 117, 820–835. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Han, D.; Zhou, T.; Zhang, J.; Liu, C.; Cao, F.; Dong, N. Melatonin ameliorates aortic valve calcification via the regulation of circular RNA CircRIC3/miR-204-5p/DPP4 signaling in valvular interstitial cells. J. Pineal Res. 2020, 69, e12666. [Google Scholar] [CrossRef] [PubMed]

| Gene Name and Functional Role | Gene Expression Changes in Heart Failure | Isoform Switch I the Failing Heart |
|---|---|---|
| Structural | ||
| TNNT1 *, TNNC1 *, and TNNI1 * | ↑ | No change |
| Tropomyosin * | ↑ | TMP1K |
| MYH6 | ↓ | MYH6 to MYH7 |
| MYH7 * | ↑ | MYH6 to MYH7 |
| MYBPC3 * | No change | No change |
| ACTC1 * | ↑ | No change |
| Sarcoglycan, delta * | No change | No change |
| Dystrophin * | Unknown | No change |
| Desmin * | ↑ | No change |
| Metavinculin | Unknown | No change |
| Muscle LIM protein | ↓ | No change |
| Actinin, alpha | ↑ | ACTN1 to ACTN2 |
| Titin * | No change | N2BA to N2B |
| Lamin A/C | ↓ | No change |
| Energy production | ||
| Succinate dehydrogenase complex * | Unknown | No change |
| Calcium handling ion channels | ||
| Phospholamban | ↑ | No change |
| SUR2A | Unknown | No change |
| SCN5A | ↓ | No change |
| Other | ||
| Ankyrin repeat domain 1 | ↑ | No change |
| Thymopoietin | Unknown | No change |
| RNA-binding motif 20 * | Unknown | No change |
| LIM-binding domain 3 * | Unknown | No change |
| Tafazzin * | Unknown | No change |
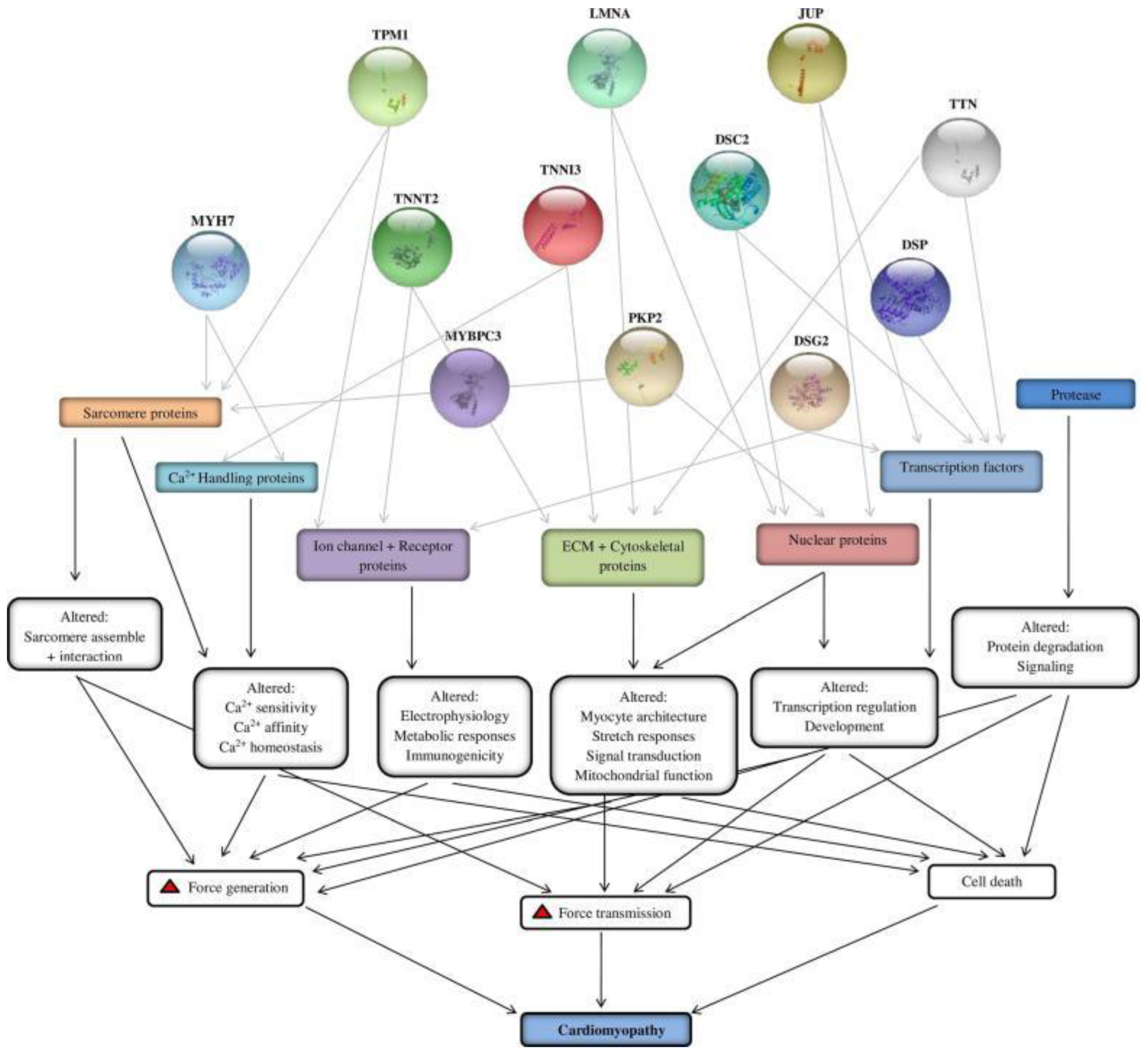
| Gene | Protein Name | Chromosome | Chromosome Location | Inheritance Type | Cardiomyopathy Form | Function |
|---|---|---|---|---|---|---|
| TNNT2 | Troponin T | 1 | 1q32.1 | AD | HCM/DCM/RCM | Ca+2 dependent regulator of muscle contraction |
| MYH7 | Beta myosin heavy chain | 14 | 14q12 | AD | HCM/DCM/RCM | Beta heavy chain subunit of cardiac myosin |
| MYBPC3 | Cardiac myosin binding protein C | 11 | 11p11.2 | AD | HCM/DCM/RCM | Cardiac isoform of myosin binding protein found in the cross-bridge zone (C area) of A bands |
| TNN13 | Troponin I | 19 | 19q13.42 | AD | HCM/DCM/RCM | Mediates striated muscle relaxation |
| TPM1 | Alpha-tropomyosin | 15 | 15q22.2 | AD | HCM/DCM/RCM | Ca+2 dependent striated muscle contraction regulator |
| LMNA | Lamin A/C | 1 | 1q22 | AD | DCM/ARVC | Cardiac homeostasis |
| PKP2 | Plakophilin 2 | 12 | 12p11.21 | AR | ARVC | Plays a role in junctional plaques |
| DSC2 | Desmocollin | 18 | 18q12.1 | AD | ARVC | Major components of Desmosome |
| DSG2 | Desmoglein 2 | 18 | 18q12.1 | AD | ARVC/DCM | Ca+2 binding transmembrane glycoprotein, component of desmosome between myocardial cells |
| DSP | Desmoplakin | 6 | 6p24.3 | AD/AR | ARVC/DCM | It is an essential component of functional desmosome |
| JUP | Plakoglobin | 17 | 17q21.2 | AD | ARVC | The common component of desmosome and intermediate junction |
| TTN | Titin | 2 | 2q31.2 | AD | ARVC/HCM/DCM | Essential for striated muscle assembly and function. Connects microfilaments. |
| Gene Therapy Target for Coronary Heart Disease | ||||
|---|---|---|---|---|
| Molecular Target | Stage in Development | Findings | Model Assessed | Reference |
| Vascular endothelial growth factor (VEGF) | Clinical trials, phase 2/3 Continued safety and efficacy | Safe but not consistently efficacious with increasing myocardial perfusion. Success with secondary end points, i.e., increased exercise capacity and reduction in ischemic area | Human | Hedman et al., Gene Ther., 2009 [124] Stewart et al., Mol. Ther., 2009 [130] |
| Fibroblast growth factor (FGF) | Clinical trials, phase 2/3 Continued safety and efficacy | Safe but most trials have not increased myocardial perfusion. Some have improved exercise capacity and symptom alleviation | Human | Kukula et al., Am. Heart J., 2011 [125] |
| Hepatocyte growth factor (HGF) | Clinical trial, phase 1 Preclinical | Safe with negligible side effects from ADs; HGF in serum not detected after 35 days Increased capillary density and end-diastolic volume Improved cardiac perfusion and reduced apoptosis | Human Rat Pig | Yang et al., Mol. Biol. Rep., 2009 [131] Jin et al., Gene Ther., 2012 [132] Yang et al., Mol. Biol. Rep., 2010 [133] |
| Platelet-derived growth factor (PDGF) | Preclinical | Increased capillary growth and collateral formation from single naked DNA injection | Rabbit | Li et al., Microvasc. Res., 2010 [126] |
| Hypoxia-inducible factor (HIF1α) | Clinical trial, phase 1 Preclinical | Preliminary safety of ADs after 1 year Increased myocardial perfusion and improved LV function but no improvement in bioactivity end points | Human Pig | Kilian et al., Circ. J., 2010 [134] Heinl-Green et al., Eur. Heart J., 2005 [135] |
| Gene therapy targets for heart failure | ||||
| Molecular target | Stage in development | Findings | Model assessed | Reference |
| Sarcoendoplasmic Reticulum calcium-ATPase 2a (SERCA2a) | Clinical trials, phase 2 | Decreased HF symptoms, increased functional status, and reversal of negative LV remodeling | Human | Jessup et al., Circulation, 2011 [136] |
| Stromal-derived factor-1 (SDF-1) | Clinical trials, phase 1/2 | Safe and improved 6-min walk test, quality of life, and NYHA class | Human | Penn et al., Circ. Res., 2013 [137] |
| Adenylyl cyclase 6 (ADCY6) | Preclinical | Increased LV function, increased cAMP levels, reversal of dysfunctional β-AR signaling, and increased survival Improved LV contractility | Mice Pig | Rebolledo et al., Hum. Gene Ther., 2006 [138] Roth et al., Circulation, 1999, 2002 [139,140] Takahashi et al., Circulation, 2006 [141] Lai et al., Circulation, 2000 [142] |
| βARKct-carboxy terminal peptide from GRK2 | Preclinical | Heart failure rescue Improved β-AR signaling and contractile dysfunction | Rabbit Human Cardiomyocytes | Shah et al., Circulation, 2001 [143] Williams et al., Circulation, 2004 [144] |
| S100A1 | Preclinical | Increased reuptake SR Ca2+, lowered Ca2+ leak, enhanced cardiac function, and reversed LV remodeling | Rat Cardiomyocytes | Most et al., J. Clin. Invest., 2004 [145] Pleger et al., Circulation, 2007 [146] |
| Parvalbumin (PVALB) | Preclinical | Increased rate of Ca2+ removal and improved relaxation rate | Rat | Szatkowski et al., J. Clin. Invest., 2001 [147] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alhajri, N.; Rustom, M.; Adegbile, A.; Ahmed, W.; Kilidar, S.; Afify, N. Deciphering the Basis of Molecular Biology of Selected Cardiovascular Diseases: A View on Network Medicine. Int. J. Mol. Sci. 2022, 23, 11421. https://doi.org/10.3390/ijms231911421
Alhajri N, Rustom M, Adegbile A, Ahmed W, Kilidar S, Afify N. Deciphering the Basis of Molecular Biology of Selected Cardiovascular Diseases: A View on Network Medicine. International Journal of Molecular Sciences. 2022; 23(19):11421. https://doi.org/10.3390/ijms231911421
Chicago/Turabian StyleAlhajri, Noora, Mohammad Rustom, Adedayo Adegbile, Weshah Ahmed, Salsabeel Kilidar, and Nariman Afify. 2022. "Deciphering the Basis of Molecular Biology of Selected Cardiovascular Diseases: A View on Network Medicine" International Journal of Molecular Sciences 23, no. 19: 11421. https://doi.org/10.3390/ijms231911421
APA StyleAlhajri, N., Rustom, M., Adegbile, A., Ahmed, W., Kilidar, S., & Afify, N. (2022). Deciphering the Basis of Molecular Biology of Selected Cardiovascular Diseases: A View on Network Medicine. International Journal of Molecular Sciences, 23(19), 11421. https://doi.org/10.3390/ijms231911421





