Glycan-Adhering Lectins and Experimental Evaluation of a Lectin FimH Inhibitor in Enterohemorrhagic Escherichia coli (EHEC) O157:H7 Strain EDL933
Abstract
:1. Introduction
2. Results
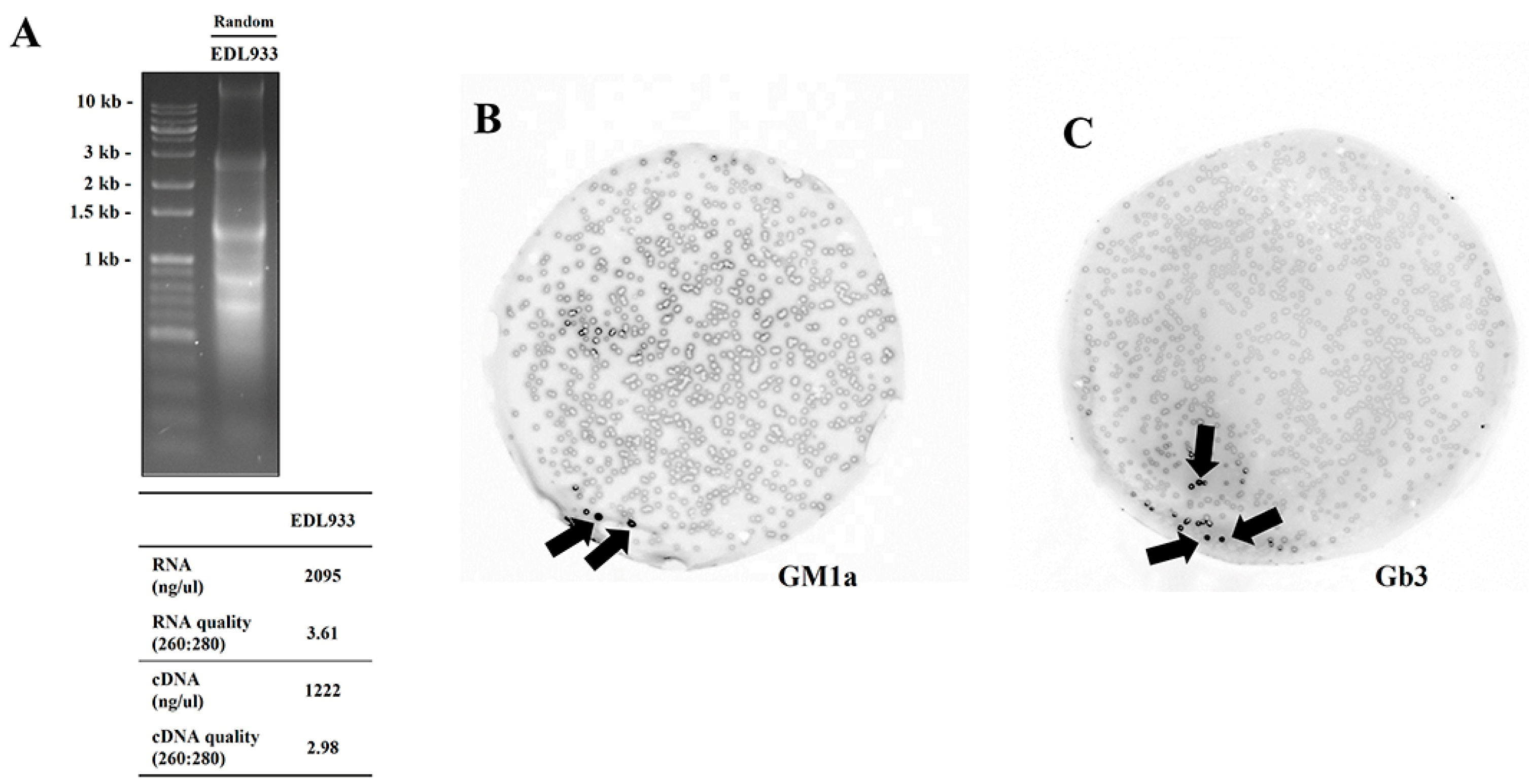
2.1. Finding Lectin Receptor of EDL933 Linked to Gb3 and GM1a Glycan Using T7 Phage Display
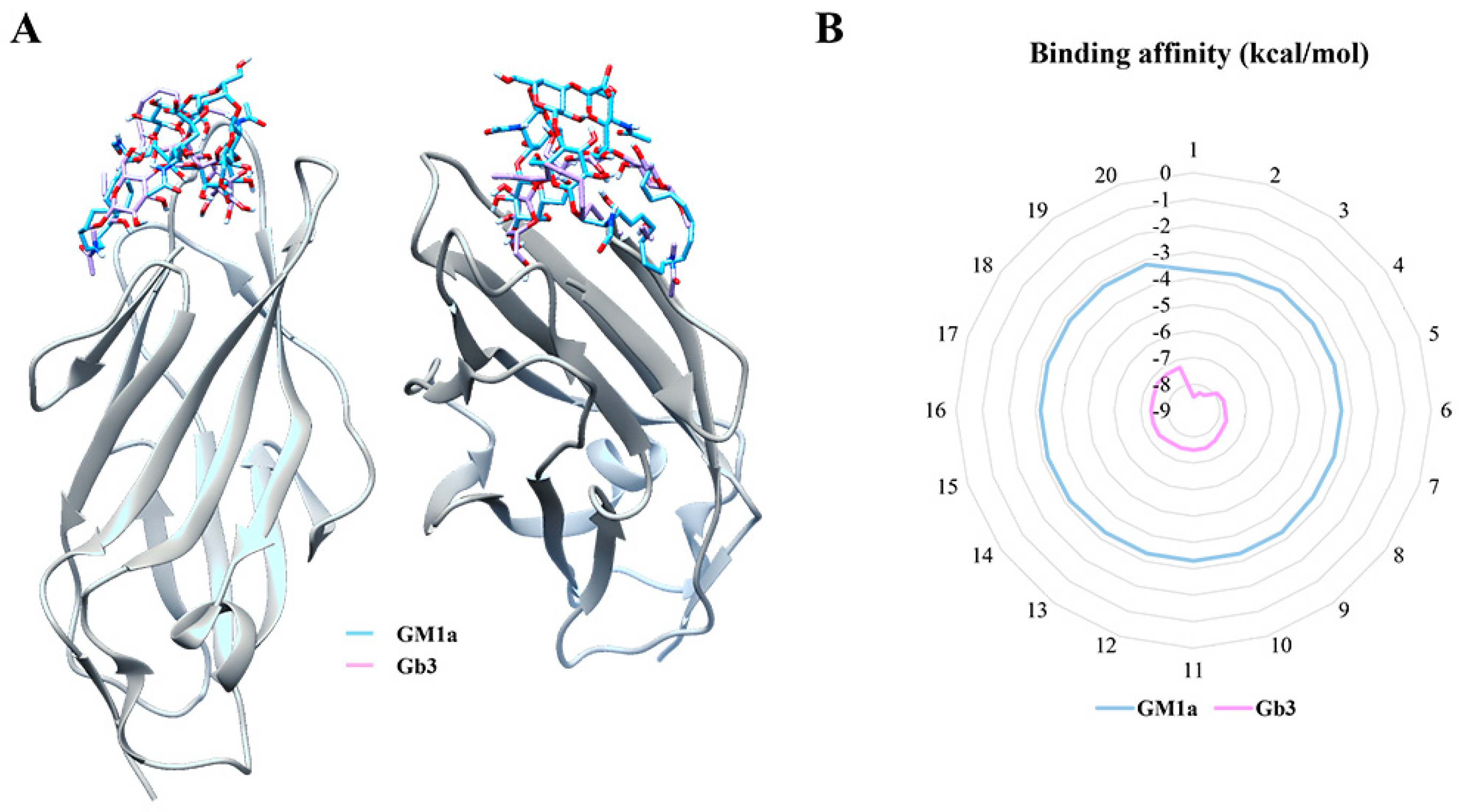
2.2. Determination of Ligand-Binding Domain (LBD) of FimH
2.3. Comparison of the Adhesion Affinity and Inhibitory Effect of the Peptides of FimH LBDs Using EDL933
2.4. Confirmation of the Inhibition Affinity of the Peptides of FimH LBDs Using the Stains Isolated from Patients
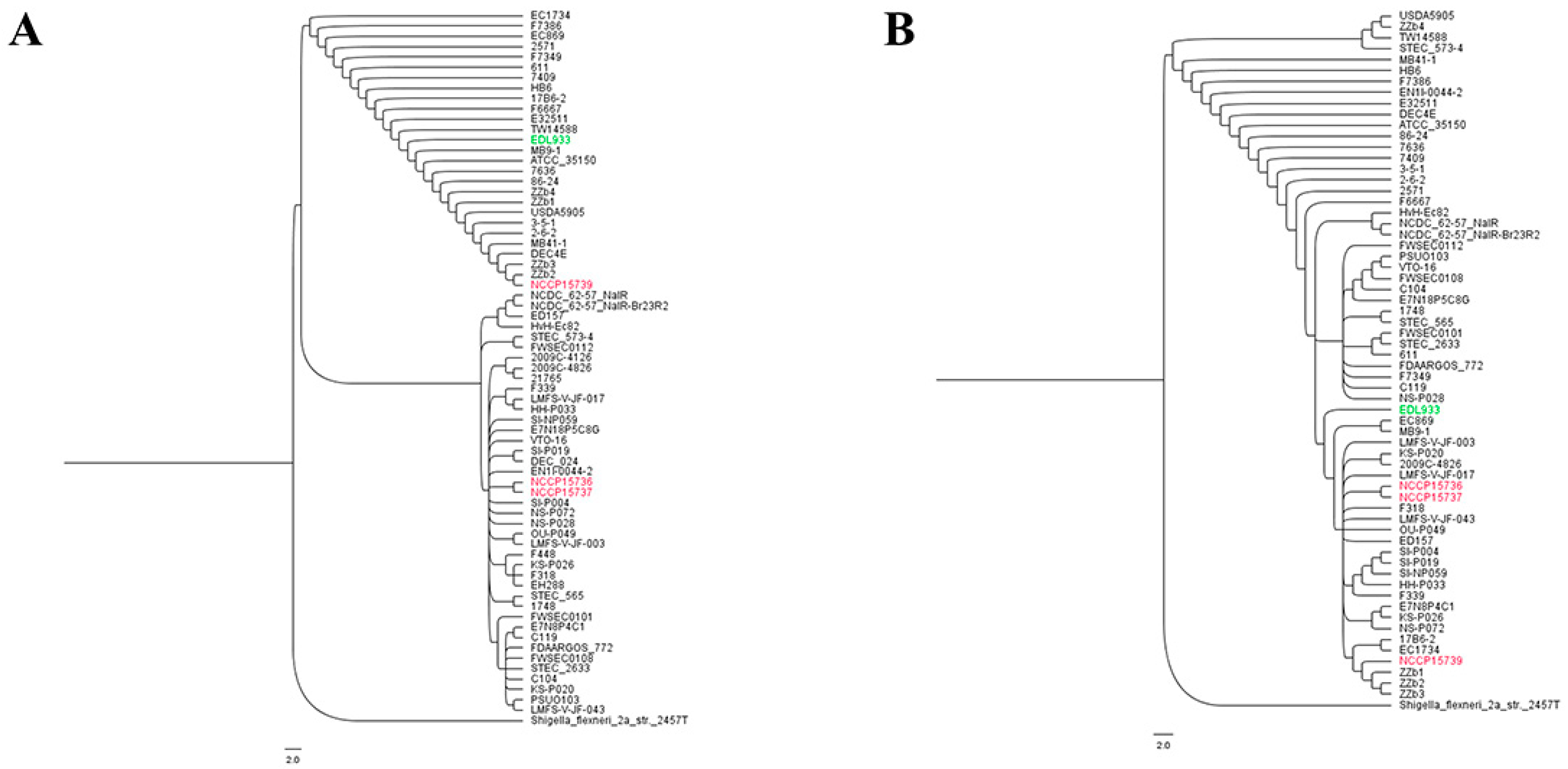
2.5. Evolutionary Analysis of EHEC Strains and Gene-Based Subtyping of EHEC Strains in Relation to LGI
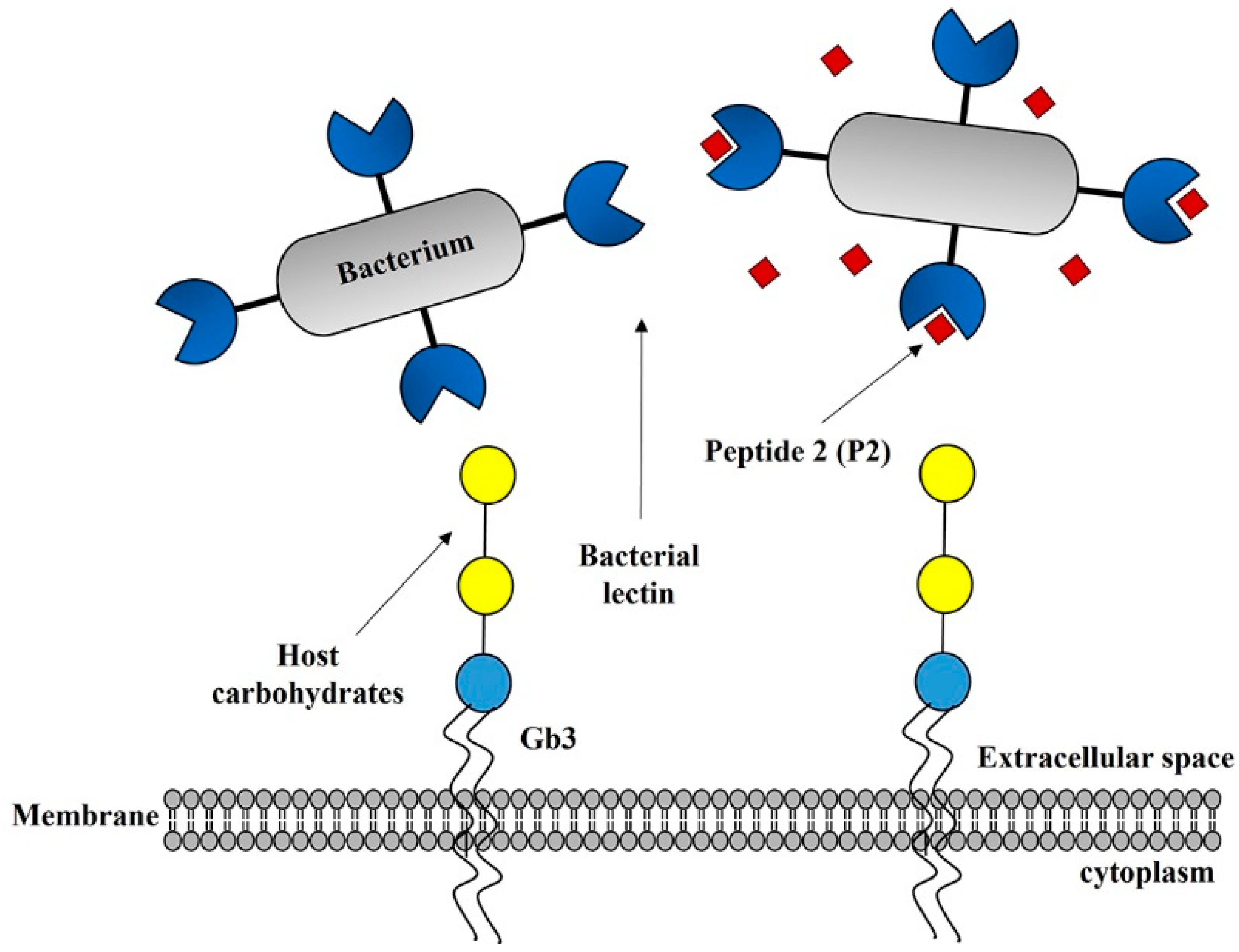
3. Discussion
4. Materials and Methods
4.1. Strain, Isolation, and Serotyping
4.2. Bacterial Growth Conditions
4.3. Total RNA Isolation
4.4. Synthesis of Double-Stranded cDNA through Reverse Transcription PCR
4.5. T7 Phage Display
4.6. Plaque Lift Assay
4.7. Docking Simulation
4.8. Solid Phase Peptide Synthesis
4.9. Peptide Binding and Inhibition Assays
4.10. Phylogenetic Analysis and Comparative Genomic Analysis
4.11. Gene-Based Subtyping of EHEC Strains in Relation to LGI
4.12. Data Accuracy Treatment
4.13. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tchesnokova, V.; Aprikian, P.; Kisiela, D.; Gowey, S.; Korotkova, N.; Thomas, W.; Sokurenko, E. Type 1 Fimbrial Adhesin FimH Elicits an Immune Response That Enhances Cell Adhesion of Escherichia coli. Infect. Immun. 2011, 79, 3895–3904. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.; Mathivanan, N.; Goyal, A. Bacterial adhesins, the pathogenic weapons to trick host defense arsenal. Biomed. Pharmacother. 2017, 93, 763–771. [Google Scholar] [CrossRef]
- Nakagawa, I.; Inaba, H.; Yamamura, T.; Kato, T.; Kawai, S.; Ooshima, T.; Amano, A. Invasion of Epithelial Cells and Proteolysis of Cellular Focal Adhesion Components by Distinct Types of Porphyromonas gingivalis Fimbriae. Infect. Immun. 2006, 74, 3773–3782. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, S.; Inaba, H.; Nomura, R.; Murakami, M.; Yasuda, H.; Nakano, K.; Matsumoto-Nakano, M. Efficacy of FimA antibody and clindamycin in silkworm larvae stimulated with Porphyromonas gulae. J. Oral Microbiol. 2021, 13, 1914499. [Google Scholar] [CrossRef]
- Ambrosi, C.; Scribano, D.; Sarshar, M.; Zagaglia, C.; Singer, B.B.; Palamara, A.T. Acinetobacter baumannii Targets Human Carcinoembryonic Antigen-Related Cell Adhesion Molecules (CEACAMs) for Invasion of Pneumocytes. mSystems 2020, 5, e00604-20. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.-H.; Lee, K.M.; Kim, C.-H.; Kim, S.S. Construction of a Lectin–Glycan Interaction Network from Enterohemorrhagic Escherichia coli Strains by Multi-omics Analysis. Int. J. Mol. Sci. 2020, 21, 2681. [Google Scholar] [CrossRef]
- Cho, S.H.; Park, J.Y.; Kim, C.-H. Systemic Lectin-Glycan Interaction of Pathogenic Enteric Bacteria in the Gastrointestinal Tract. Int. J. Mol. Sci. 2022, 23, 1451. [Google Scholar] [CrossRef]
- Tenaillon, O.; Skurnik, D.; Picard, B.; Denamur, E. The population genetics of commensal Escherichia coli. Nat. Rev. Microbiol. 2010, 8, 207–217. [Google Scholar]
- Nataro, J.P.; Kaper, J.B. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 1998, 11, 142–201. [Google Scholar] [CrossRef]
- Caprioli, A.; Morabito, S.; Brugere, H.; Oswald, E. Enterohaemorrhagic Escherichia coli: Emerging issues on virulence and modes of transmission. Vet. Res. 2005, 36, 289–311. [Google Scholar] [CrossRef]
- Riley, L.W.; Remis, R.S.; Helgerson, S.D.; McGee, H.B.; Wells, J.G.; Davis, B.R.; Hebert, R.J.; Olcott, E.S.; Johnson, L.M.; Hargrett, N.T.; et al. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N. Engl. J. Med. 1983, 308, 681–685. [Google Scholar] [CrossRef] [PubMed]
- Beutin, L. Emerging enterohaemorrhagic Escherichia coli, causes and effects of the rise of a human pathogen. J. Vet. Med. B. Infect. Dis. Vet. Public Health 2006, 53, 299–305. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, A.D.; Newland, J.W.; Miller, S.F.; Holmes, R.K.; Smith, H.W.; Formal, S.B. Shiga-like toxin-converting phages from Escherichia coli strains that cause hemorrhagic colitis or infantile diarrhea. Science 1984, 226, 694–696. [Google Scholar] [CrossRef] [PubMed]
- Karmali, M.A. Infection by Shiga toxin-producing Escherichia coli: An overview. Mol. Biotechnol. 2004, 26, 117–122. [Google Scholar] [CrossRef]
- Sandvig, K.; Bergan, J.; Dyve, A.B.; Skotland, T.; Torgersen, M.L. Endocytosis and retrograde transport of Shiga toxin. Toxicon 2010, 56, 1181–1185. [Google Scholar] [CrossRef]
- Langermann, S.; Palaszynski, S.; Barnhart, M. Prevention of mucosal Escherichia coli infection by FimH-adhesin-based systemic vaccination. Science 1997, 276, 607–611. [Google Scholar] [CrossRef]
- Perna, N.T.; Plunkett, G., 3rd; Burland, V.; Mau, B.; Glasner, J.D.; Rose, D.J.; Mayhew, G.F.; Evans, P.S.; Gregor, J.; Kirkpatrick, H.A.; et al. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 2001, 409, 529–533. [Google Scholar] [CrossRef]
- Proulx, F.; Turgeon, J.P.; Delage, G.; Lafleur, L.; Chicoine, L. Randomized, controlled trial of antibiotic therapy for Escherichia coli O157:H7 enteritis. J. Pediatr. 1992, 121, 299–303. [Google Scholar] [CrossRef]
- Smith, K.E.; Wilker, P.R.; Reiter, P.L.; Hedican, E.B.; Bender, J.B.; Hedberg, C.W. Antibiotic Treatment of Escherichia coli O157 Infection and the Risk of Hemolytic Uremic Syndrome, Minnesota. Pediatr. Infect. Dis. J. 2012, 31, 37–41. [Google Scholar] [CrossRef]
- Wong, C.S.; Jelacic, S.; Habeeb, R.L.; Watkins, S.L.; Tarr, P.I. The Risk of the Hemolytic–Uremic Syndrome after Antibiotic Treatment of Escherichia coli O157:H7 Infections. N. Engl. J. Med. 2000, 342, 1930–1936. [Google Scholar] [CrossRef]
- Asadi, K.; Oloomi, M.; Habibi, M.; Bouzari, S. Cloning of fimH and fliC and expression of the fusion protein FimH/FliC from Uropathogenic Escherichia coli (UPEC) isolated in Iran. Iran. J. Microbiol. 2012, 4, 55–62. [Google Scholar] [PubMed]
- Schwartz, D.J.; Kalas, V.; Pinkner, J.S.; Chen, S.L.; Spaulding, C.N.; Dodson, K.W.; Hultgren, S.J. Positively selected FimH residues enhance virulence during urinary tract infection by altering FimH conformation. Proc. Natl. Acad. Sci. USA 2013, 110, 15530–15537. [Google Scholar] [CrossRef] [PubMed]
- Aprikian, P.; Tchesnokova, V.; Kidd, B.; Yakovenko, O.; Yarov-Yarovoy, V.; Trinchina, E.; Vogel, V.; Thomas, W.; Sokurenko, E. Interdomain Interaction in the FimH Adhesin of Escherichia coli Regulates the Affinity to Mannose. J. Biol. Chem. 2007, 282, 23437–23446. [Google Scholar] [CrossRef] [PubMed]
- Scharenberg, M.; Jiang, X.; Pang, L.; Navarra, G.; Rabbani, S.; Binder, F.; Schwardt, O.; Ernst, B. Kinetic Properties of Carbohydrate-Lectin Interactions: FimH Antagonists. ChemMedChem 2013, 9, 78–83. [Google Scholar] [CrossRef] [PubMed]
- Mydock-McGrane, L.K.; Cusumano, Z.T.; Janetka, J.W. Mannose-derived FimH antagonists: A promising anti-virulence therapeutic strategy for urinary tract infections and Crohn’s disease. Expert. Opin. Ther. Pat. 2016, 26, 175–197. [Google Scholar] [CrossRef]
- Lee, K.M.; Kim, C.H.; Kim, J.H.; Kim, S.S.; Cho, S.H. E-Membranome: A Database for Genome-Wide Analysis of Escherichia coli Outer Membrane Proteins. Curr. Pharm. Biotechnol. 2021, 22, 501–507. [Google Scholar] [CrossRef]
- Danner, S.; Belasco, J.G. T7 phage display: A novel genetic selection system for cloning RNA-binding proteins from cDNA libraries. Proc. Natl. Acad. Sci. USA 2001, 98, 12954–12959. [Google Scholar] [CrossRef]
- Segundo-Acosta, P.S.; Garranzo-Asensio, M.; Oeo-Santos, C.; Montero-Calle, A.; Quiralte, J.; Cuesta-Herranz, J.; Villalba, M.; Barderas, R. High-throughput screening of T7 phage display and protein microarrays as a methodological approach for the identification of IgE-reactive components. J. Immunol. Methods 2018, 456, 44–53. [Google Scholar] [CrossRef]
- Yamamoto, M.; Kominato, Y.; Yamamoto, F. Phage display cDNA cloning of protein with carbohydrate affinity. Biochem. Biophys. Res. Commun. 1999, 255, 194–199. [Google Scholar] [CrossRef]
- Jafari, A.; Aslani, M.; Bouzari, S. Escherichia coli: A brief review of diarrheagenic pathotypes and their role in diarrheal diseases in Iran. Iran. J. Microbiol. 2012, 4, 102–117. [Google Scholar]
- Piérard, D.; De Greve, H.; Haesebrouck, F.; Mainil, J. O157:H7 and O104:H4 Vero/Shiga toxin-producing Escherichia coli outbreaks: Respective role of cattle and humans. Veter. Res. 2012, 43, 13. [Google Scholar] [CrossRef] [PubMed]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M.; et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2014, 42, D206–D214. [Google Scholar] [CrossRef] [PubMed]
- Sharma, V.; Hecker, N.; Roscito, J.G.; Foerster, L.; Langer, B.E.; Hiller, M. A genomics approach reveals insights into the importance of gene losses for mammalian adaptations. Nat. Commun. 2018, 9, 1215. [Google Scholar] [CrossRef] [PubMed]
- Goldwater, P.N.; Bettelheim, K.A. Treatment of enterohemorrhagic Escherichia coli (EHEC) infection and hemolytic uremic syndrome (HUS). BMC Med. 2012, 10, 12. [Google Scholar] [CrossRef] [PubMed]
- Mohawk, K.L.; O’Brien, A.D. Mouse Models of Escherichia coli O157:H7 Infection and Shiga Toxin Injection. J. Biomed. Biotechnol. 2011, 2011, 258185. [Google Scholar] [CrossRef] [PubMed]
- Ielasi, F.S.; Alioscha-Perez, M.; Donohue, D.; Claes, S.; Sahli, H.; Schols, D.; Willaert, R.G. Lectin-Glycan Interaction Network-Based Identification of Host Receptors of Microbial Pathogenic Adhesins. mBio 2016, 7, e00584-16. [Google Scholar] [CrossRef]
- Mariethoz, J.; Khatib, K.; Alocci, D.; Campbell, M.P.; Karlsson, N.G.; Packer, N.H.; Mullen, E.H.; Lisacek, F. SugarBindDB, a resource of glycan-mediated host–pathogen interactions. Nucleic Acids Res. 2016, 44, D1243–D1250. [Google Scholar] [CrossRef]
- Kwon, T.; Bak, Y.-S.; Jung, Y.-H.; Yu, Y.-B.; Choi, J.T.; Kim, C.-H.; Kim, J.-B.; Kim, W.; Cho, S.-H. Whole-genome sequencing and comparative genomic analysis of Escherichia coli O91 strains isolated from symptomatic and asymptomatic human carriers. Gut Pathog. 2016, 8, 57. [Google Scholar] [CrossRef]
- Kwon, T.; Cho, S.-H. Draft Genome Sequence of Enterohemorrhagic Escherichia coli O157 NCCP15739, Isolated in the Republic of Korea. Genome Announc. 2015, 3, e00522-15. [Google Scholar] [CrossRef]
- Miller, N.D.; Draughon, F.A.; D’Souza, D.H. Real-Time Reverse-Transcriptase–Polymerase Chain Reaction for Salmonella enterica Detection from Jalapeño and Serrano Peppers. Foodborne Pathog. Dis. 2010, 7, 367–373. [Google Scholar] [CrossRef]
- Rosenow, C.; Saxena, R.M.; Durst, M.; Gingeras, T.R. Prokaryotic RNA preparation methods useful for high density array analysis: Comparison of two approaches. Nucleic Acids Res. 2001, 29, E112. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Wang, L.; You, X.; Dai, P.; Zeng, Y. Advances in the T7 phage display system (Review). Mol. Med. Rep. 2017, 17, 714–720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kopp, J.; Schwede, T. The SWISS-MODEL Repository of annotated three-dimensional protein structure homology models. Nucleic Acids Res. 2004, 32, D230–D234. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. Auto Dock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [PubMed]
- Klett, J.; Núñez-Salgado, A.; Dos Santos, H.G.; Cabrera, A.C.; Perona, A.; Gil-Redondo, R.; Abia, D.; Gago, F.; Morreale, A. MM-ISMSA: An Ultrafast and Accurate Scoring Function for Protein–Protein Docking. J. Chem. Theory Comput. 2012, 8, 3395–3408. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X. Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Khan, N.H.; Ahsan, M.; Yoshizawa, S.; Hosoya, S.; Yokota, A.; Kogure, K. Multilocus sequence typing and phylogenetic analyses of Pseudomonas aeruginosa Isolates from the ocean. Appl. Environ. Microbiol. 2008, 74, 6194–6205. [Google Scholar] [CrossRef]
- Glaeser, S.P.; Kampfer, P. Multilocus sequence analysis (MLSA) in prokaryotic taxonomy. Syst. Appl. Microbiol. 2015, 38, 237–245. [Google Scholar] [CrossRef]
- Patiño, L.H.; Camargo, M.; Muñoz, M.; Ríos-Chaparro, D.I.; Patarroyo, M.A.; Ramírez, J.D. Unveiling the Multilocus Sequence Typing (MLST) Schemes and Core Genome Phylogenies for Genotyping Chlamydia trachomatis. Front. Microbiol. 2018, 9, 1854. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, J.-Y.; Kim, C.-H.; Cho, S.-H. Glycan-Adhering Lectins and Experimental Evaluation of a Lectin FimH Inhibitor in Enterohemorrhagic Escherichia coli (EHEC) O157:H7 Strain EDL933. Int. J. Mol. Sci. 2022, 23, 9931. https://doi.org/10.3390/ijms23179931
Park J-Y, Kim C-H, Cho S-H. Glycan-Adhering Lectins and Experimental Evaluation of a Lectin FimH Inhibitor in Enterohemorrhagic Escherichia coli (EHEC) O157:H7 Strain EDL933. International Journal of Molecular Sciences. 2022; 23(17):9931. https://doi.org/10.3390/ijms23179931
Chicago/Turabian StylePark, Jun-Young, Cheorl-Ho Kim, and Seung-Hak Cho. 2022. "Glycan-Adhering Lectins and Experimental Evaluation of a Lectin FimH Inhibitor in Enterohemorrhagic Escherichia coli (EHEC) O157:H7 Strain EDL933" International Journal of Molecular Sciences 23, no. 17: 9931. https://doi.org/10.3390/ijms23179931
APA StylePark, J.-Y., Kim, C.-H., & Cho, S.-H. (2022). Glycan-Adhering Lectins and Experimental Evaluation of a Lectin FimH Inhibitor in Enterohemorrhagic Escherichia coli (EHEC) O157:H7 Strain EDL933. International Journal of Molecular Sciences, 23(17), 9931. https://doi.org/10.3390/ijms23179931








