Loss of Function of the RRMF Domain in OsROS1a Causes Sterility in Rice (Oryza sativa L.)
Abstract
1. Introduction
2. Results
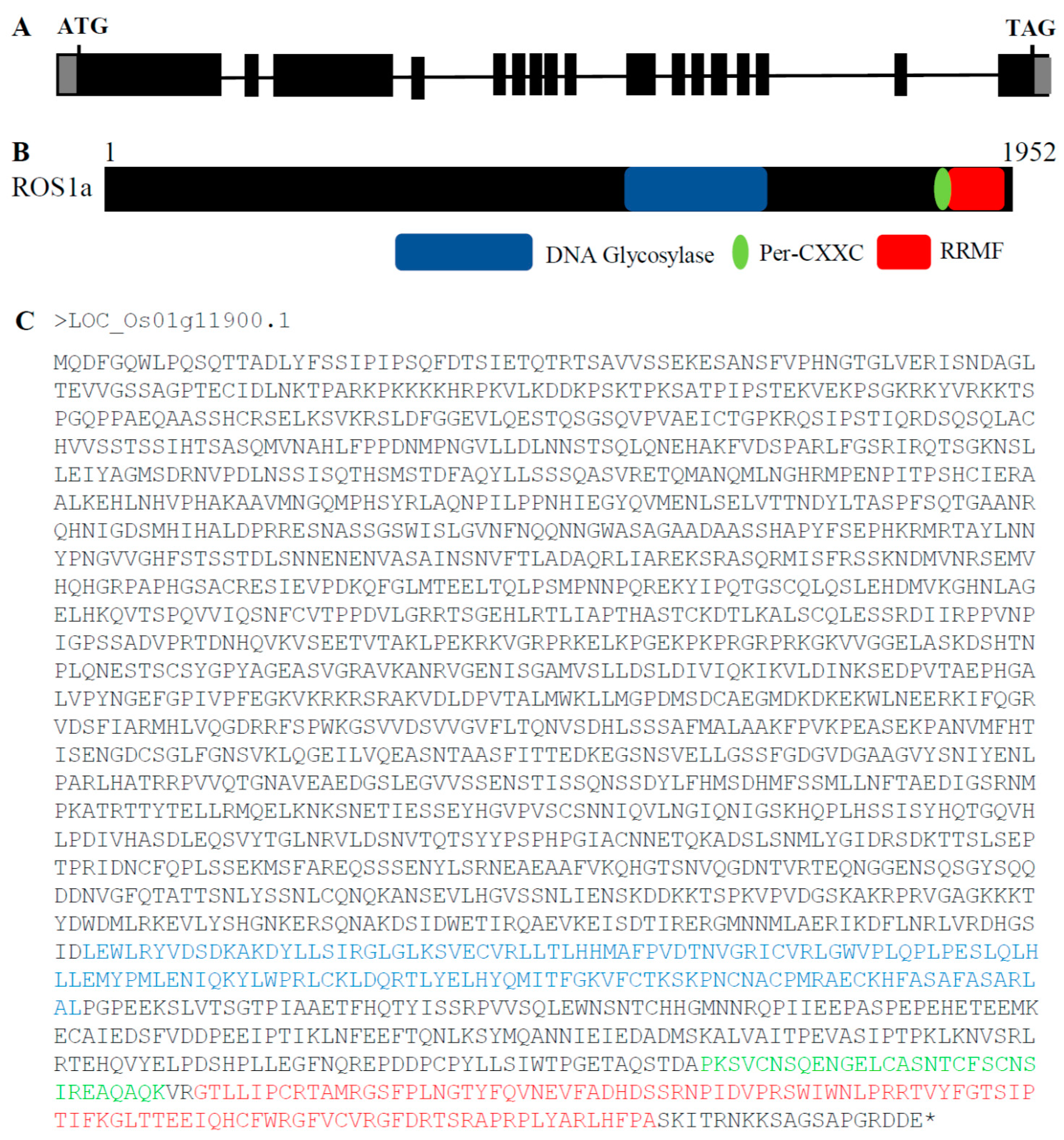
2.1. Phylogenetic Analysis of OsROS1a Glycosylase Domain
2.2. Generation of OsROS1a Gene Editing Mutants by CRISPR/Cas9
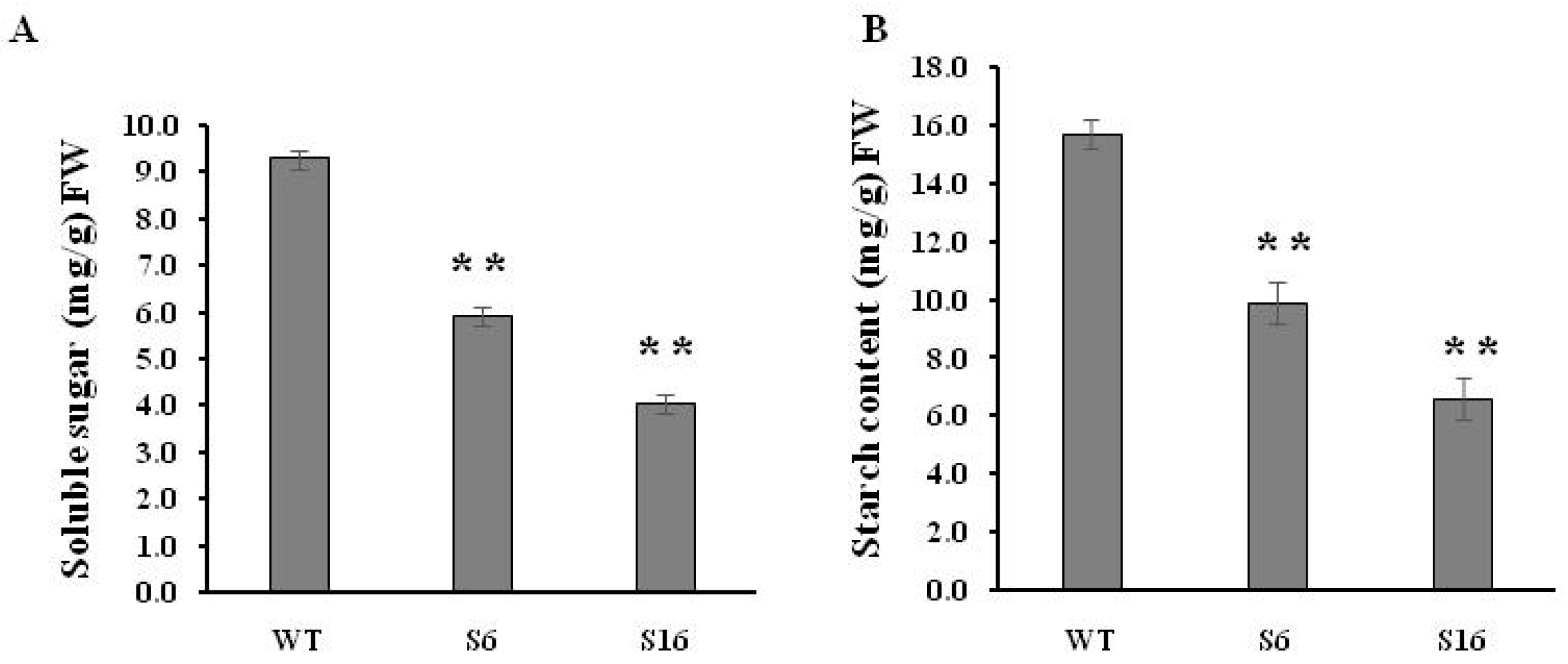
2.3. The osros1a Mutations Cause Complete Male Sterility
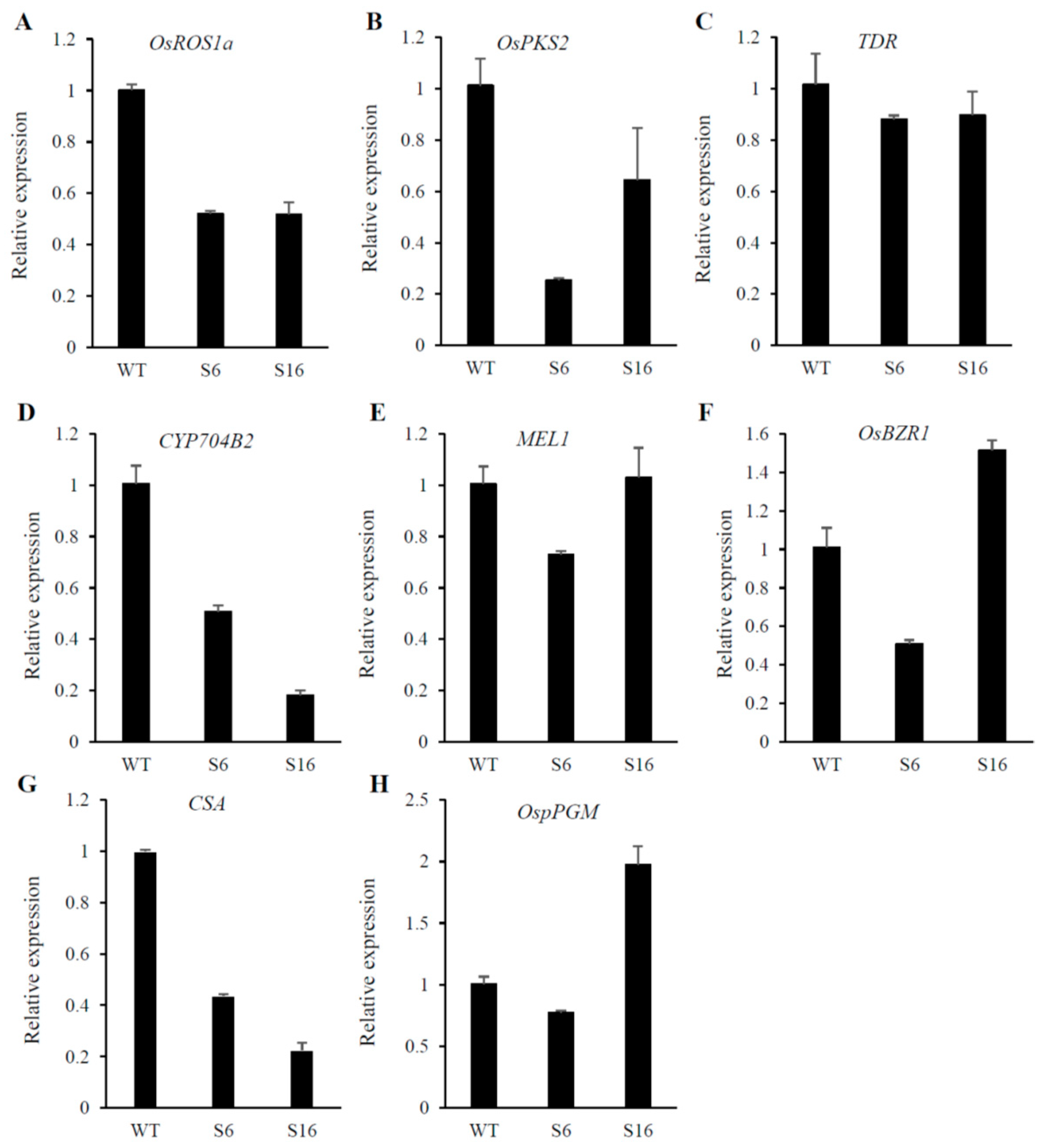
2.4. The Reduction Expression of Genes Involved in Anther and Pollen Development in Osros1a Mutants
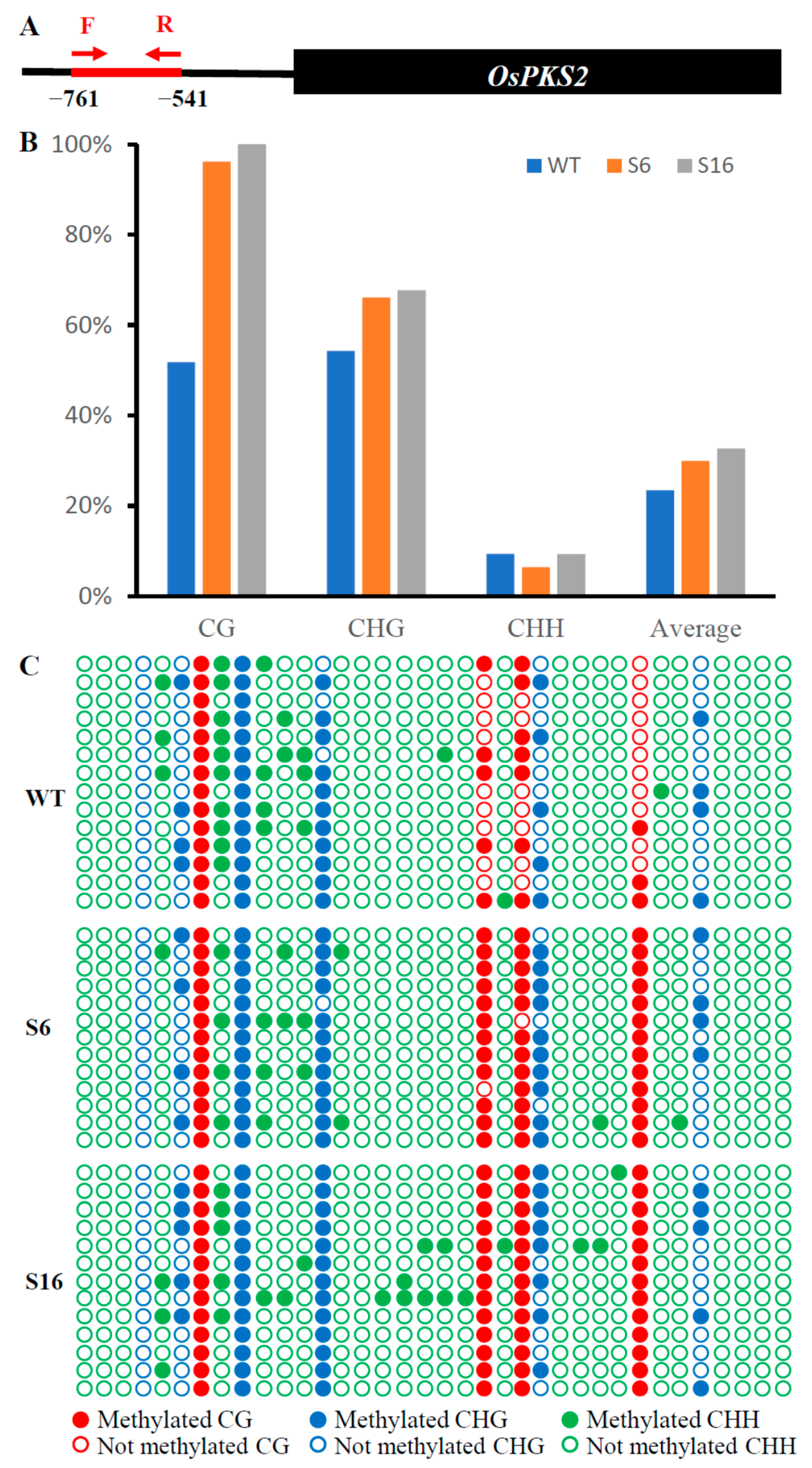
2.5. DNA Hypermethylation of OsPKS2 Gene Promoter in Osros1a Mutants
3. Discussion
4. Materials and Methods
4.1. Identification of OsROS1a Homologous Genes and Phylogenetic Analysis
4.2. OsROS1a Gene Editing Mutants Generated by CRISPR/Cas9
4.3. Pollen Fertility Examination and Histochemical Assay
4.4. Total Soluble Sugar and Starch Contents Analysis
4.5. Gene Expression by RT-PCR Analysis
4.6. DNA Methylation Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, H.; Lang, Z.; Zhu, J.K. Dynamics and function of DNA methylation in plants. Nat. Rev. Mol. Cell Biol. 2018, 19, 489–506. [Google Scholar] [CrossRef] [PubMed]
- Nash, H.M.; Bruner, S.D.; Scharer, O.D.; Kawate, T.; Addona, T.A.; Spooner, E.; Lane, W.S.; Verdine, G.L. Cloning of a yeast 8-oxoguanine DNA glycosylase reveals the existence of a base-excision DNA-repair protein superfamily. Curr. Biol. 1996, 6, 968–980. [Google Scholar] [CrossRef]
- Zhu, J.; Kapoor, A.; Sridhar, V.V.; Agius, F.; Zhu, J.K. The DNA glycosylase/lyase ROS1 functions in pruning DNA methylation patterns in Arabidopsis. Curr. Biol. 2007, 17, 54–59. [Google Scholar] [CrossRef]
- Agius, F.; Kapoor, A.; Zhu, J.K. Role of the Arabidopsis DNA glycosylase/lyase ROS1 in active DNA demethylation. Proc. Natl. Acad. Sci. USA 2006, 103, 11796–11801. [Google Scholar] [CrossRef]
- Choi, Y.; Gehring, M.; Johnson, L.; Hannon, M.; Harada, J.J.; Goldberg, R.B.; Jacobsen, S.E.; Fischer, R.L. DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in Arabidopsis. Cell 2002, 110, 33–42. [Google Scholar] [CrossRef]
- Penterman, J.; Zilberman, D.; Huh, J.H.; Ballinger, T.; Henikoff, S.; Fischer, R.L. DNA demethylation in the Arabidopsis genome. Proc. Natl. Acad. Sci. USA 2007, 104, 6752–6757. [Google Scholar] [CrossRef]
- Schoft, V.K.; Chumak, N.; Choi, Y.; Hannon, M.; Garcia-Aguilar, M.; Machlicova, A.; Slusarz, L.; Mosiolek, M.; Park, J.S.; Park, G.T.; et al. Function of the DEMETER DNA glycosylase in the Arabidopsis thaliana male gametophyte. Proc. Natl. Acad. Sci. USA 2011, 108, 8042–8047. [Google Scholar] [CrossRef]
- Gong, Z.; Morales-Ruiz, T.; Ariza, R.R.; Roldan-Arjona, T.; David, L.; Zhu, J.K. ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell 2002, 111, 803–814. [Google Scholar] [CrossRef]
- Zemach, A.; Kim, M.Y.; Silva, P.; Rodrigues, J.A.; Dotson, B.; Brooks, M.D.; Zilberman, D. Local DNA hypomethylation activates genes in rice endosperm. Proc. Natl. Acad. Sci. USA 2010, 107, 18729–18734. [Google Scholar] [CrossRef]
- Ono, A.; Yamaguchi, K.; Fukada-Tanaka, S.; Terada, R.; Mitsui, T.; Iida, S. A null mutation of ROS1a for DNA demethylation in rice is not transmittable to progeny. Plant J. 2012, 71, 564–574. [Google Scholar] [CrossRef]
- Xu, Y.; Wang, F.; Chen, Z.; Wang, J.; Li, W.; Fan, F.; Tao, Y.; Jiang, Y.; Zhu, Q.H.; Yang, J. CRISPR/Cas9-targeted mutagenesis of the OsROS1 gene induces pollen and embryo sac defects in rice. Plant Biotechnol. J. 2020, 18, 1999–2001. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.Y.; Ono, A.; Scholten, S.; Kinoshita, T.; Zilberman, D.; Okamoto, T.; Fischer, R.L. DNA demethylation by ROS1a in rice vegetative cells promotes methylation in sperm. Proc. Natl. Acad. Sci. USA 2019, 116, 9652–9657. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wu, X.; Yao, X.; Yu, R.; Larkin, P.J.; Liu, C.M. Mutations in the DNA demethylase OsROS1 result in a thickened aleurone and improved nutritional value in rice grains. Proc. Natl. Acad. Sci. USA 2018, 115, 11327–11332. [Google Scholar] [CrossRef] [PubMed]
- Lang, Z.; Gong, Z. A role of OsROS1 in aleurone development and nutrient improvement in rice. Proc. Natl. Acad. Sci. USA 2018, 115, 11659–11660. [Google Scholar] [CrossRef]
- La, H.; Ding, B.; Mishra, G.P.; Zhou, B.; Yang, H.; Bellizzi Mdel, R.; Chen, S.; Meyers, B.C.; Peng, Z.; Zhu, J.K.; et al. A 5-methylcytosine DNA glycosylase/lyase demethylates the retrotransposon Tos17 and promotes its transposition in rice. Proc. Natl. Acad. Sci. USA 2011, 108, 15498–15503. [Google Scholar] [CrossRef]
- Wen, S.; Wen, N.; Pang, J.; Langen, G.; Brew-Appiah, R.A.; Mejias, J.H.; Osorio, C.; Yang, M.; Gemini, R.; Moehs, C.P.; et al. Structural genes of wheat and barley 5-methylcytosine DNA glycosylases and their potential applications for human health. Proc. Natl. Acad. Sci. USA 2012, 109, 20543–20548. [Google Scholar] [CrossRef]
- Guo, J.X.; Liu, Y.G. Molecular control of male reproductive development and pollen fertility in rice. J. Integr. Plant Biol. 2012, 54, 967–978. [Google Scholar] [CrossRef]
- Scott, R.J.; Spielman, M.; Dickinson, H.G. Stamen structure and function. Plant Cell 2004, 16 (Suppl. 1), S46–S60. [Google Scholar] [CrossRef]
- McCormick, S. Control of male gametophyte development. Plant Cell 2004, 16 (Suppl. 1), S142–S153. [Google Scholar] [CrossRef]
- Ma, H. Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plants. Annu. Rev. Plant Biol. 2005, 56, 393–434. [Google Scholar] [CrossRef]
- Wilson, Z.A.; Zhang, D.B. From Arabidopsis to rice: Pathways in pollen development. J. Exp. Bot. 2009, 60, 1479–1492. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, G. Recent progress in plant reproduction research: The story of the male gametophyte through to successful fertilization. Plant Cell Physiol. 2009, 50, 1857–1864. [Google Scholar] [CrossRef] [PubMed]
- Zou, T.; Liu, M.; Xiao, Q.; Wang, T.; Chen, D.; Luo, T.; Yuan, G.; Li, Q.; Zhu, J.; Liang, Y.; et al. OsPKS2 is required for rice male fertility by participating in pollen wall formation. Plant Cell Rep. 2018, 37, 759–773. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.S.; Liang, W.Q.; Yuan, Z.; Li, N.; Shi, J.; Wang, J.; Liu, Y.M.; Yu, W.J.; Zhang, D.B. Tapetum degeneration retardation is critical for aliphatic metabolism and gene regulation during rice pollen development. Mol. Plant 2008, 1, 599–610. [Google Scholar] [CrossRef]
- Li, N.; Zhang, D.S.; Liu, H.S.; Yin, C.S.; Li, X.X.; Liang, W.Q.; Yuan, Z.; Xu, B.; Chu, H.W.; Wang, J.; et al. The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell 2006, 18, 2999–3014. [Google Scholar] [CrossRef] [PubMed]
- Lian, J.P.; Yang, Y.W.; He, R.R.; Yang, L.; Zhou, Y.F.; Lei, M.Q.; Zhang, Z.; Huang, J.H.; Cheng, Y.; Liu, Y.W.; et al. Ubiquitin-dependent Argonauteprotein MEL1 degradation is essential for rice sporogenesis and phasiRNA target regulation. Plant Cell 2021, 33, 2685–2700. [Google Scholar] [CrossRef]
- Li, H.; Pinot, F.; Sauveplane, V.; Werck-Reichhart, D.; Diehl, P.; Schreiber, L.; Franke, R.; Zhang, P.; Chen, L.; Gao, Y.; et al. Cytochrome P450 family member CYP704B2 catalyzes the {omega}-hydroxylation of fatty acids and is required for anther cutin biosynthesis and pollen exine formation in rice. Plant Cell 2010, 22, 173–190. [Google Scholar] [CrossRef]
- Zhang, D.B.; Wilson, Z.A. Stamen specification and anther development in rice. Chin. Sci. Bull. 2009, 54, 2342–2353. [Google Scholar] [CrossRef]
- Zhang, D.; Luo, X.; Zhu, L. Cytological analysis and genetic control of rice anther development. J. Genet. Genomics 2011, 38, 379–390. [Google Scholar] [CrossRef]
- Zhu, X.; Liang, W.; Cui, X.; Chen, M.; Yin, C.; Luo, Z.; Zhu, J.; Lucas, W.J.; Wang, Z.; Zhang, D. Brassinosteroids promote development of rice pollen grains and seeds by triggering expression of Carbon Starved Anther, a MYB domain protein. Plant J. 2015, 82, 570–581. [Google Scholar] [CrossRef]
- Zhang, H.; Xu, C.; He, Y.; Zong, J.; Yang, X.; Si, H.; Sun, Z.; Hu, J.; Liang, W.; Zhang, D. Mutation in CSA creates a new photoperiod-sensitive genic male sterile line applicable for hybrid rice seed production. Proc. Natl. Acad. Sci. USA 2013, 110, 76–81. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liang, W.; Shi, J.; Xu, J.; Zhang, D. MYB56 encoding a R2R3 MYB transcription factor regulates seed size in Arabidopsis thaliana. J. Integr. Plant Biol. 2013, 55, 1166–1178. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Liang, W.; Yang, X.; Luo, X.; Jiang, N.; Ma, H.; Zhang, D. Carbon starved anther encodes a MYB domain protein that regulates sugar partitioning required for rice pollen development. Plant Cell 2010, 22, 672–689. [Google Scholar] [CrossRef] [PubMed]
- Dickinson, D.B. Rapid starch synthesis associated with increased respiration in germinating lily pollen. Plant Physiol. 1968, 43, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Wen, L.Y.; Chase, C.D. Mitochondrial gene expression in developing male gametophytes of male-fertile and S male-sterile maize. Sex. Plant Rep. 1999, 11, 323–330. [Google Scholar] [CrossRef]
- Datta, R.; Chamusco, K.C.; Chourey, P.S. Starch biosynthesis during pollen maturation is associated with altered patterns of gene expression in maize. Plant Physiol. 2002, 130, 1645–1656. [Google Scholar] [CrossRef]
- Wu, Y.; Fox, T.W.; Trimnell, M.R.; Wang, L.; Xu, R.J.; Cigan, A.M.; Huffman, G.A.; Garnaat, C.W.; Hershey, H.; Albertsen, M.C. Development of a novel recessive genetic male sterility system for hybrid seed production in maize and other cross-pollinating crops. Plant Biotechnol. J. 2016, 14, 1046–1054. [Google Scholar] [CrossRef]
- Zhu, J.K. Active DNA demethylation mediated by DNA glycosylases. Annu. Rev. Genet. 2009, 43, 143–166. [Google Scholar] [CrossRef]
- Xu, J.H.; Wang, R.; Li, X.; Miclaus, M.; Messing, J. Locus- and Site-Specific DNA Methylation of 19 kDa Zein Genes in Maize. PLoS ONE 2016, 11, e0146416. [Google Scholar] [CrossRef]
- Iyer, L.M.; Abhiman, S.; Aravind, L. Natural history of eukaryotic DNA methylation systems. Prog. Mol. Biol. Transl. Sci. 2011, 101, 25–104. [Google Scholar]
- Gehring, M.; Bubb, K.L.; Henikoff, S. Extensive demethylation of repetitive elements during seed development underlies gene imprinting. Science 2009, 324, 1447–1451. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Hung, Y.H.; Rim, H.J.; Zhang, D.; Frost, J.M.; Shin, H.; Jang, H.; Liu, F.; Xiao, W.; Iyer, L.M.; et al. The catalytic core of DEMETER guides active DNA demethylation in Arabidopsis. Proc. Natl. Acad. Sci. USA 2019, 116, 17563–17571. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Rechkoblit, O.; Bestor, T.H.; Patel, D.J. Structure of DNMT1-DNA complex reveals a role for autoinhibition in maintenance DNA methylation. Science 2011, 331, 1036–1040. [Google Scholar] [CrossRef] [PubMed]
- Irshad, F.; Li, C.; Wu, H.Y.; Yan, Y.; Xu, J.H. The function of DNA demethylase gene ROS1a null mutant on seed development in rice (Oryza Sativa) using the CRISPR/CAS9 system. Int. J. Mol. Sci. 2022, 23, 6357. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA-X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, Q.; Zhu, Q.; Liu, W.; Chen, Y.; Qiu, R.; Wang, B.; Yang, Z.; Li, H.; Lin, Y.; et al. A Robust CRISPR/Cas9 System for Convenient, High-Efficiency Multiplex Genome Editing in Monocot and Dicot Plants. Mol. Plant 2015, 8, 1274–1284. [Google Scholar] [CrossRef]
- Sinay, H.; Karuwa, R.L. Proline and total soluble sugar content at the vegetative phase of six corn cultivars from Kisar Island Maluku, grown under drought stress conditions. Int. J. Adv. Agric. Res. 2014, 2, 77–82. [Google Scholar]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, J.-H.; Irshad, F.; Yan, Y.; Li, C. Loss of Function of the RRMF Domain in OsROS1a Causes Sterility in Rice (Oryza sativa L.). Int. J. Mol. Sci. 2022, 23, 11349. https://doi.org/10.3390/ijms231911349
Xu J-H, Irshad F, Yan Y, Li C. Loss of Function of the RRMF Domain in OsROS1a Causes Sterility in Rice (Oryza sativa L.). International Journal of Molecular Sciences. 2022; 23(19):11349. https://doi.org/10.3390/ijms231911349
Chicago/Turabian StyleXu, Jian-Hong, Faiza Irshad, Yan Yan, and Chao Li. 2022. "Loss of Function of the RRMF Domain in OsROS1a Causes Sterility in Rice (Oryza sativa L.)" International Journal of Molecular Sciences 23, no. 19: 11349. https://doi.org/10.3390/ijms231911349
APA StyleXu, J.-H., Irshad, F., Yan, Y., & Li, C. (2022). Loss of Function of the RRMF Domain in OsROS1a Causes Sterility in Rice (Oryza sativa L.). International Journal of Molecular Sciences, 23(19), 11349. https://doi.org/10.3390/ijms231911349




