Sox6 Differentially Regulates Inherited Myogenic Abilities and Muscle Fiber Types of Satellite Cells Derived from Fast- and Slow-Type Muscles
Abstract
1. Introduction
2. Results
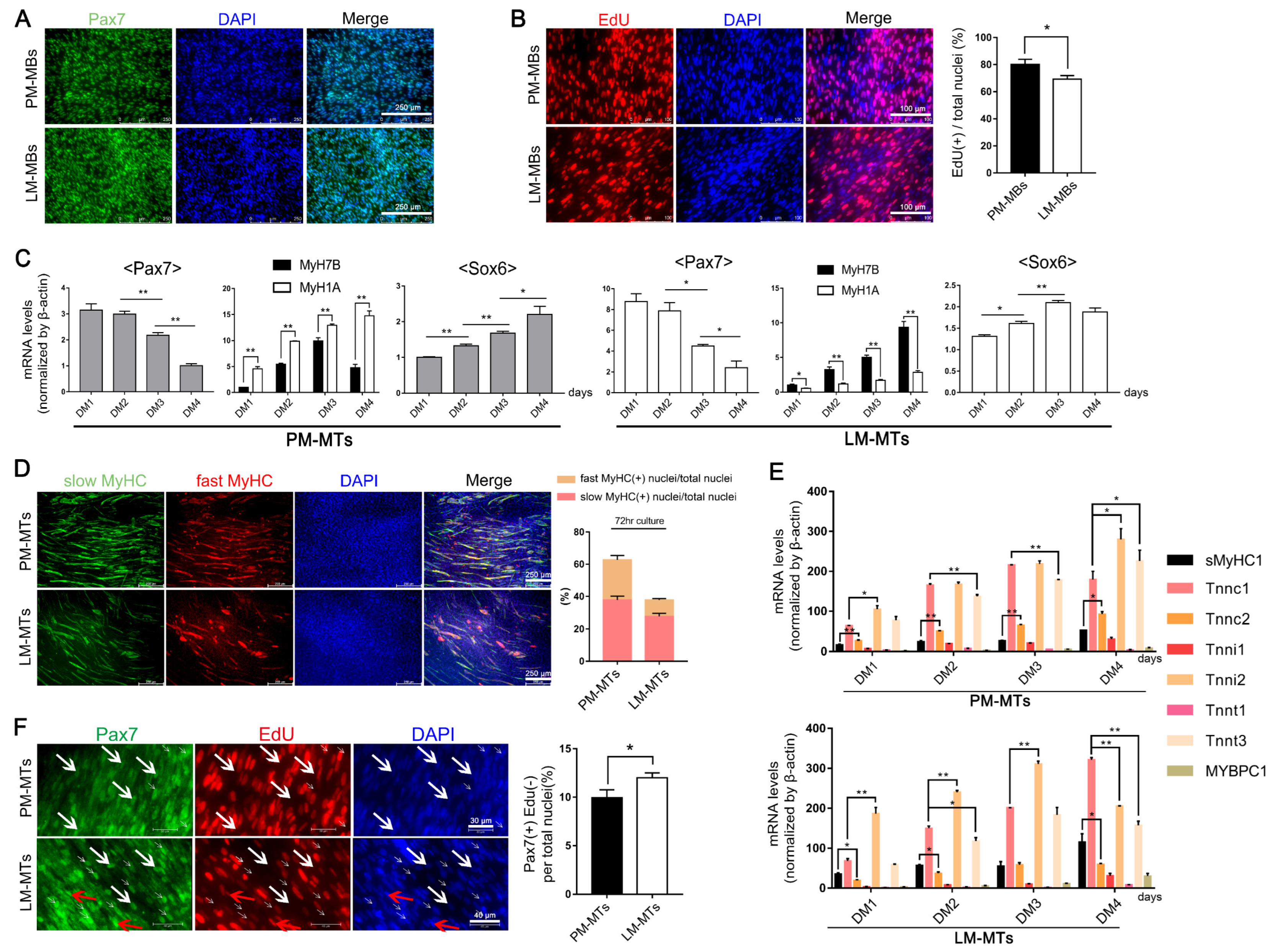
2.1. Different Myogenic Potentials in Satellite Cells of Distinct Muscle Fiber Types
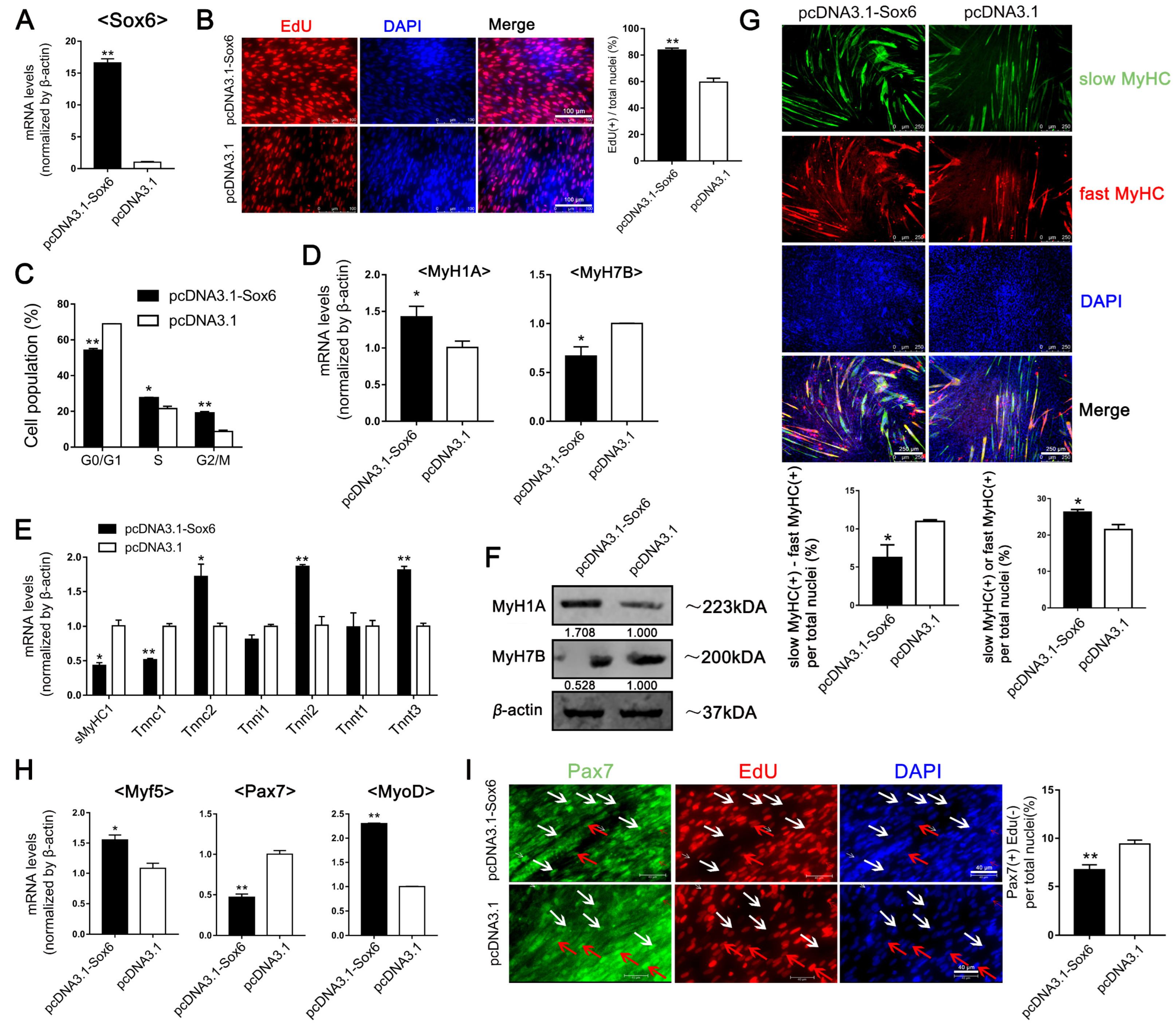
2.2. Sox6 Promotes the Myogenesis and Development of Satellite Cells Derived from Fast Type-Enriched Pectoral Muscles towards Intrinsic Tendency
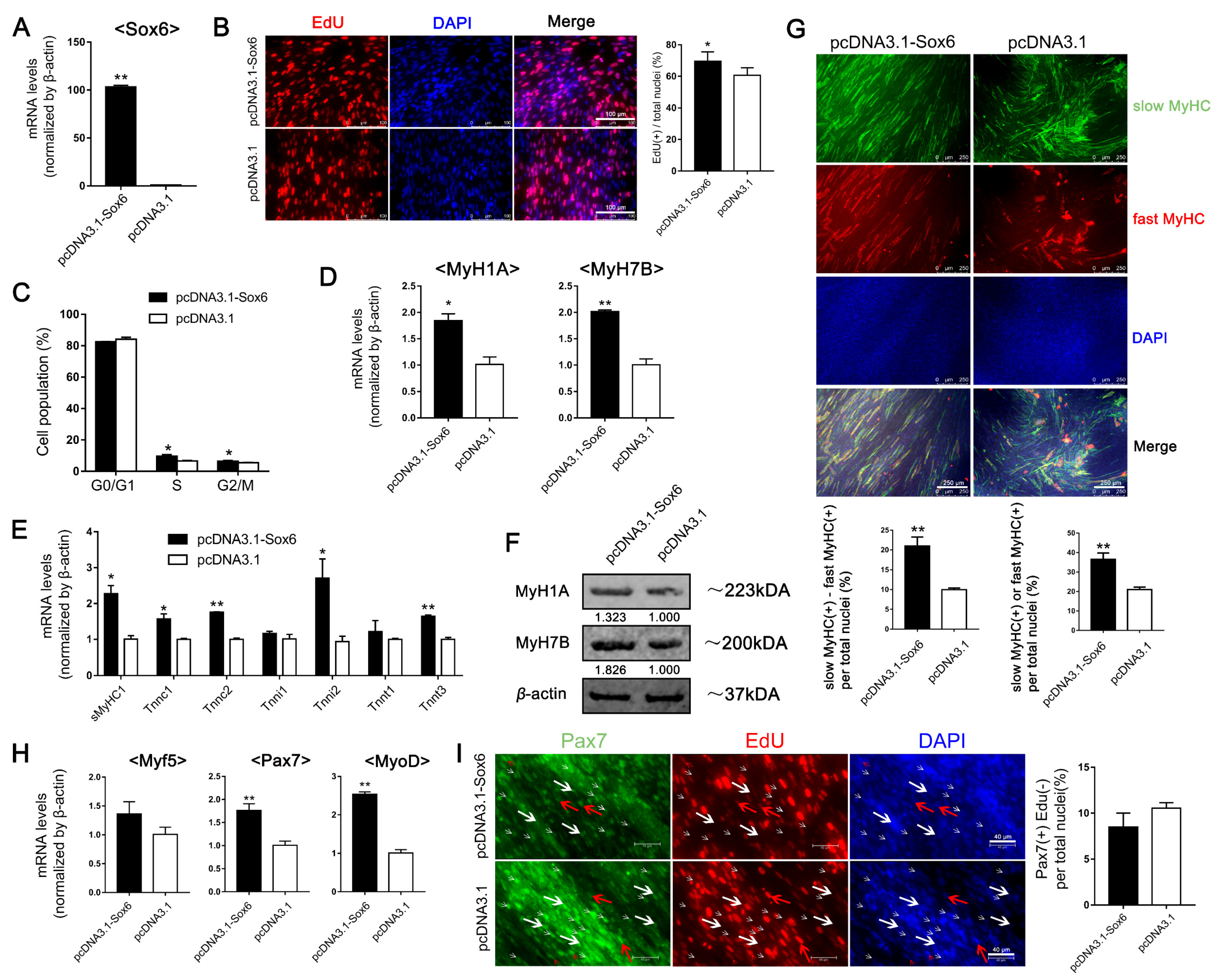
2.3. Different Roles of Sox6 in Satellite Cells Derived from Slow Type-Enriched Leg Muscles
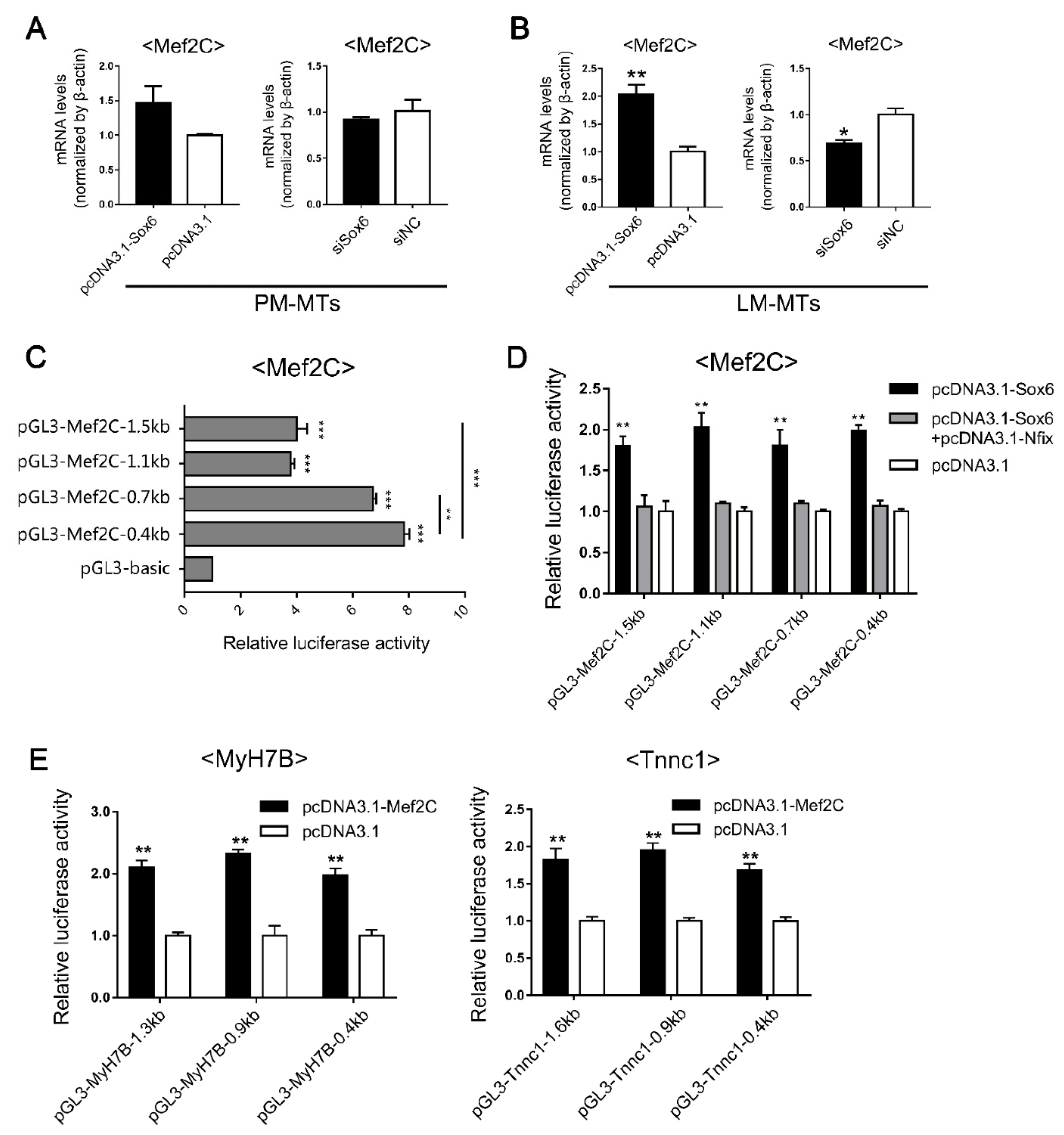
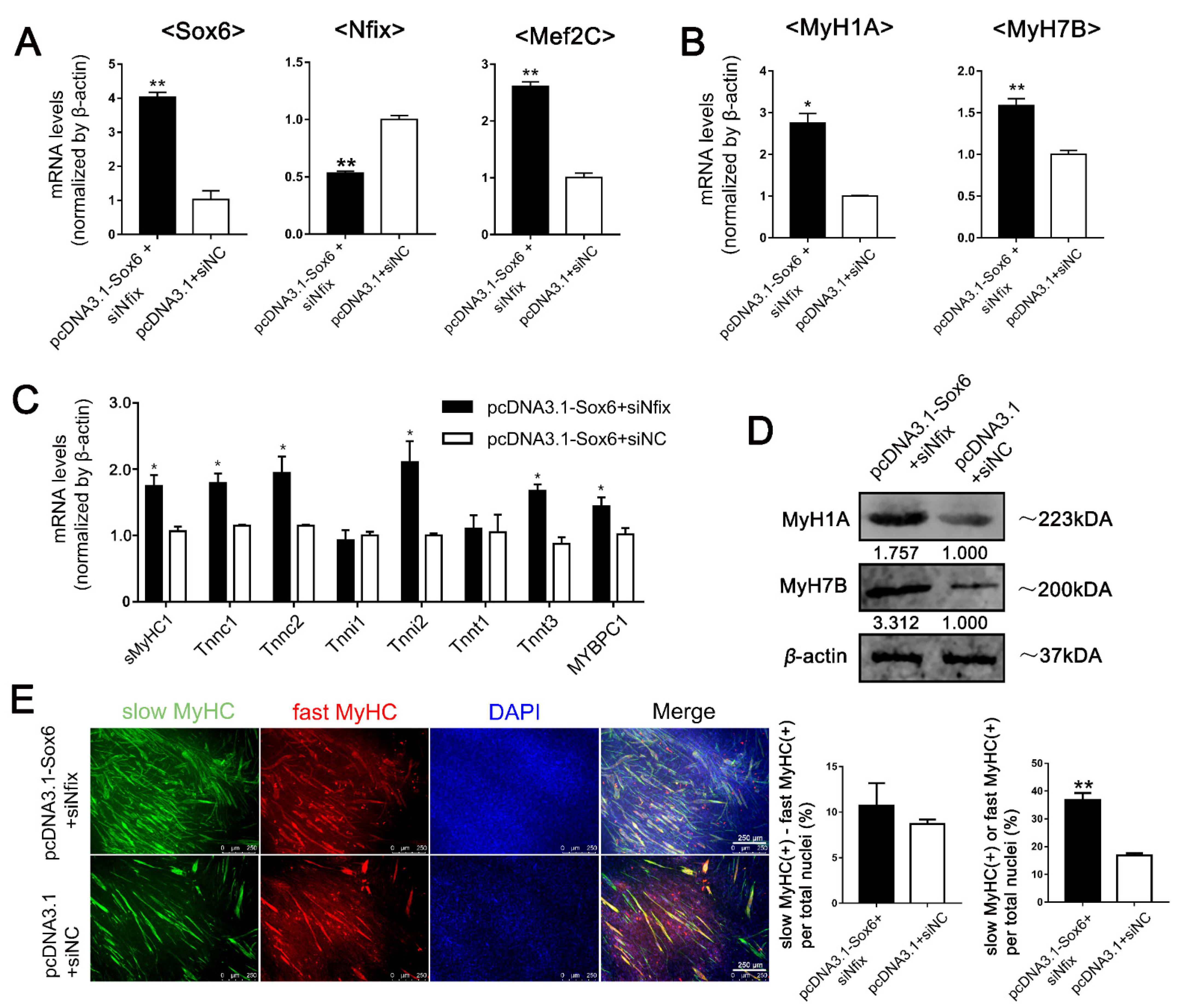
2.4. Differential Expression of Nfix between LM-MBs and PM-MBs at the Differentiation Stage Leads to Different Roles of Sox6 in the Regulation of Slow-Type Fibers
2.5. Sox6 Indirectlyupregulates the Slow-Type Isoforms through the Activation of Mef2C
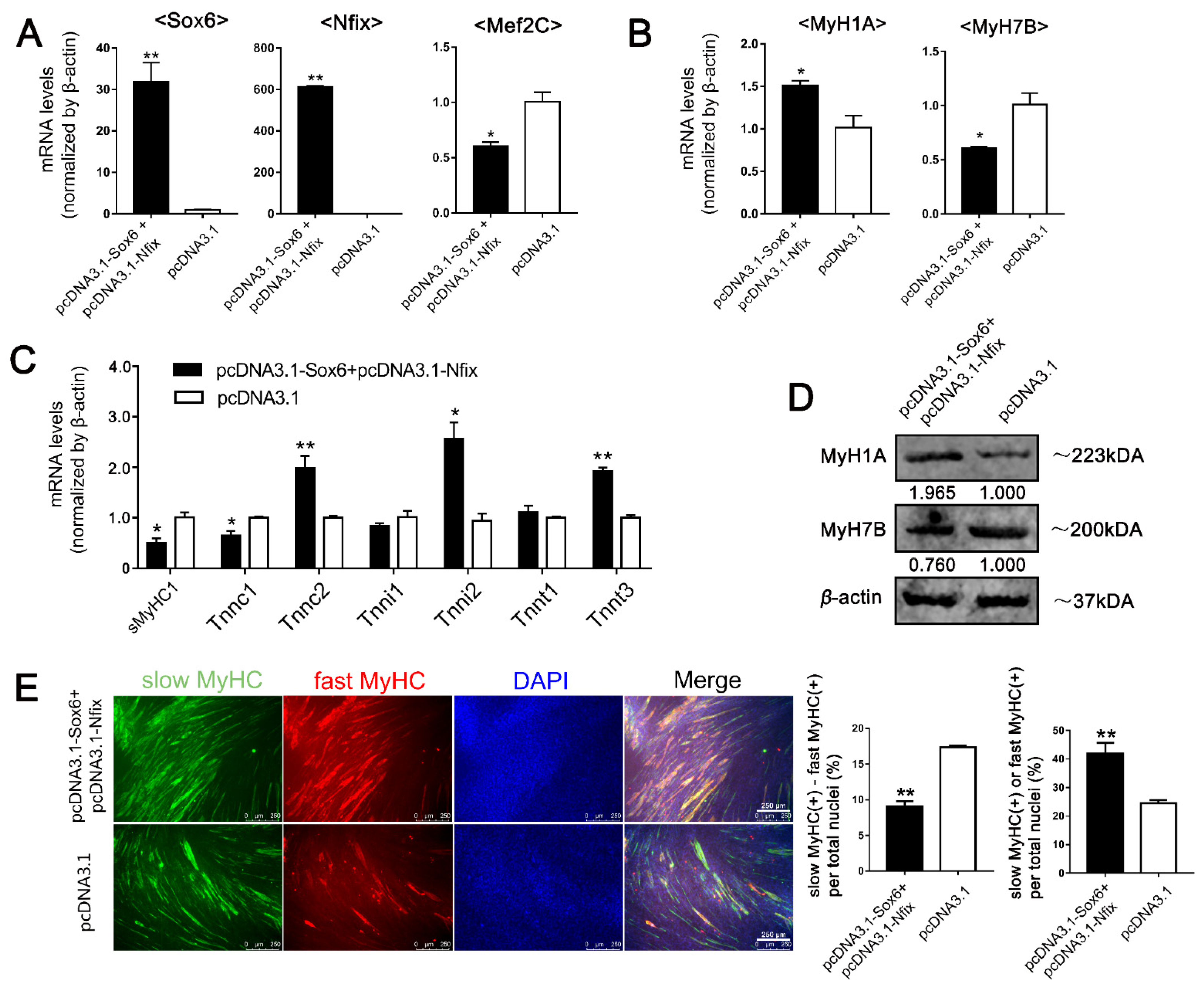
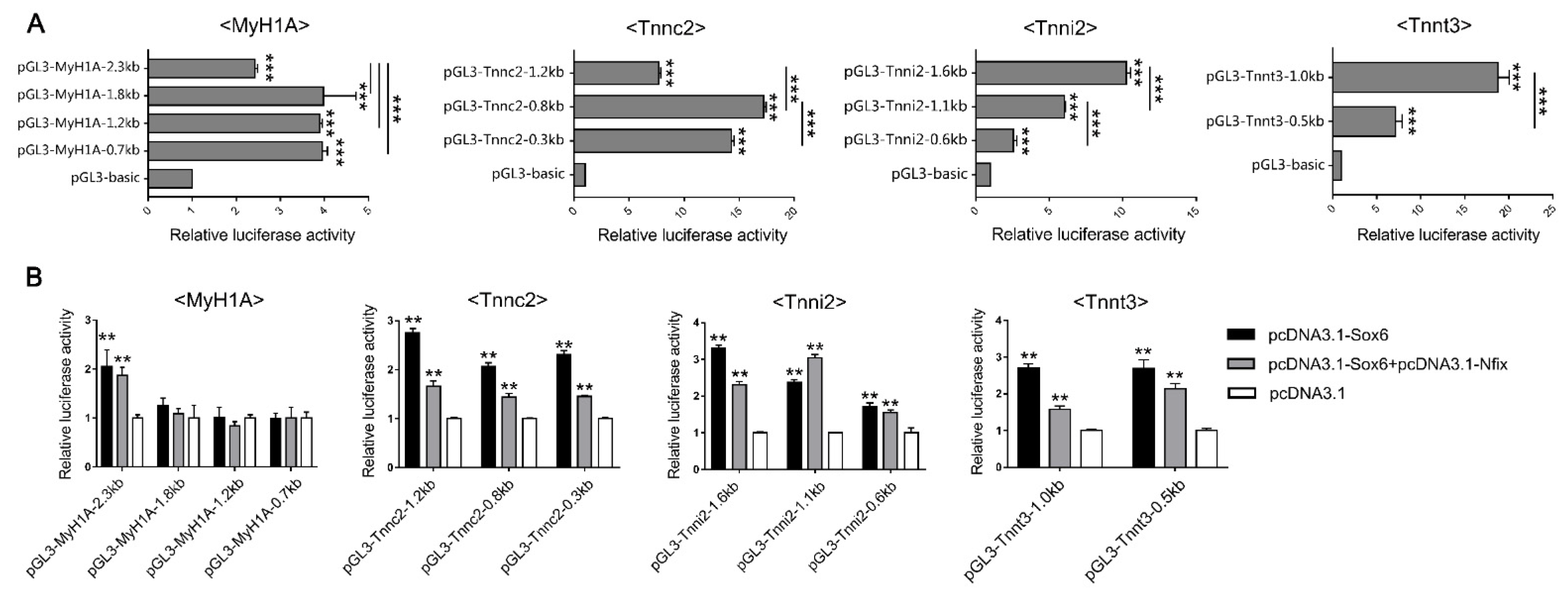
2.6. Sox6 Decreases Slow-Type Isoforms through Binding with Nfix in PM-MTs or Increases Slow-Type Isoforms by Activating Mef2C in LM-MTs
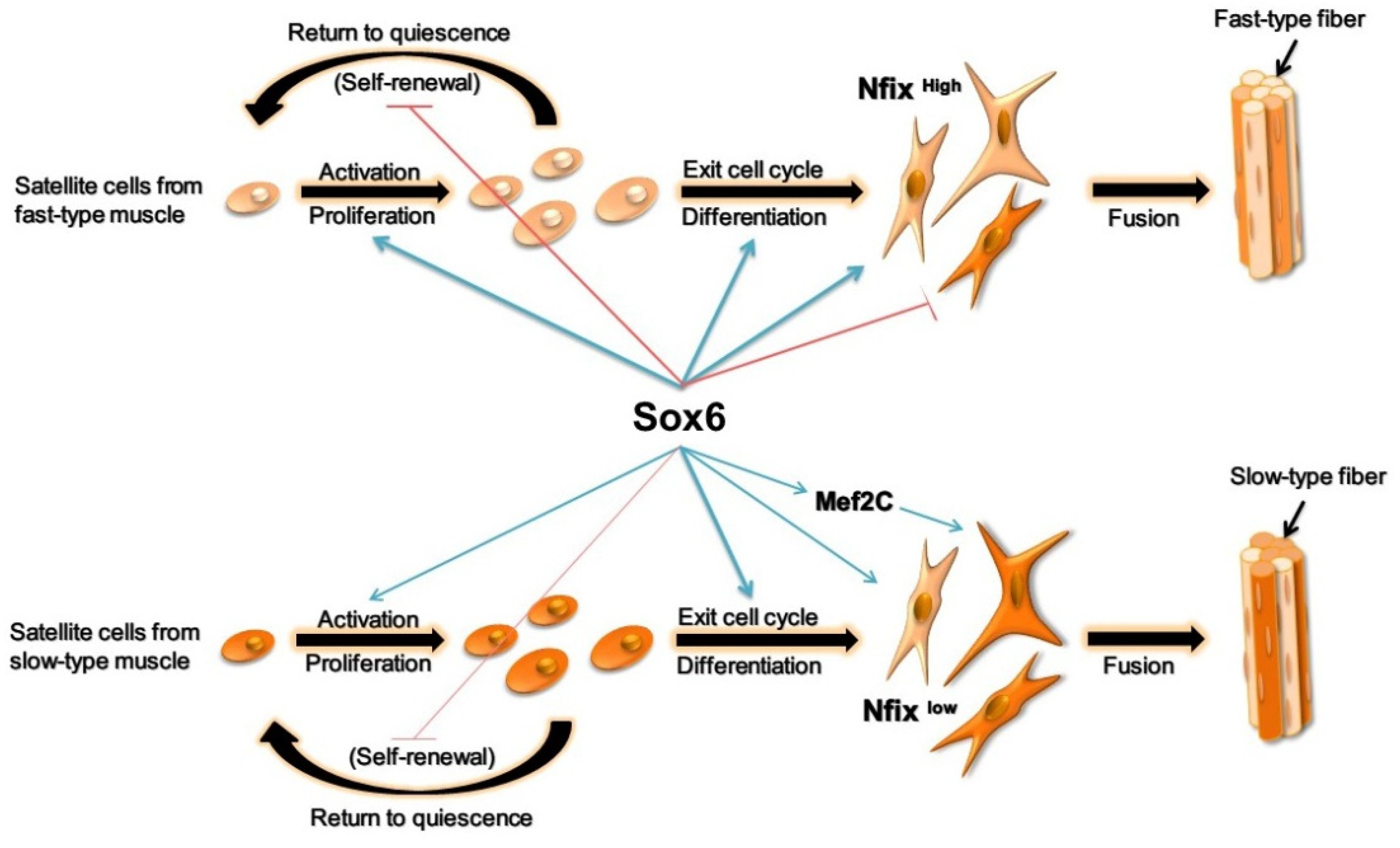
3. Discussion
4. Materials and Methods
4.1. Ethics Standards
4.2. Isolation of Muscle Satellite Cells and Cell Culture
4.3. Immunofluorescence
4.4. Edu and Flow Cytometry Assays
4.5. RNA Extraction, cDNA Synthesis and Quantitative Real-Time PCR
4.6. Plasmid Construction, RNA Oligonucleotides and Cell Transfection
4.7. Western Blot
4.8. Dual-Luciferase Reporter Assay
4.9. Co-Immunoprecipitation Assay
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Schiaffino, S.; Reggiani, C. Fiber Types in Mammalian Skeletal Muscles. Physiol. Rev. 2011, 91, 1447–1531. [Google Scholar] [CrossRef] [PubMed]
- Bassel-Duby, R.; Olson, E.N. Signaling Pathways in Skeletal Muscle Remodeling. Annu. Rev. Biochem. 2006, 75, 19–37. [Google Scholar] [CrossRef] [PubMed]
- Chargé, S.B.P.; Rudnicki, M.A. Cellular and Molecular Regulation of Muscle Regeneration. Physiol. Rev. 2004, 84, 209–238. [Google Scholar] [CrossRef]
- Relaix, F.; Rocancourt, D.; Mansouri, A.; Buckingham, M. A Pax3/Pax7-dependent population of skeletal muscle progenitor cells. Nature 2005, 435, 948–953. [Google Scholar] [CrossRef] [PubMed]
- Cheung, T.H.; Quach, N.L.; Charville, G.W.; Ling, L.; Lidia, P.; Abdolhossein, E.; Bryan, Y.; Phuong, H.; Rando, T.A. Maintenance of muscle stem-cell quiescence by microRNA-489. Nature 2012, 482, 524–528. [Google Scholar] [CrossRef]
- Sousa-Victor, P.; García-Prat, L.; Muñoz-Cánoves, P. Control of satellite cell function in muscle regeneration and its disruption in ageing. Nat. Rev. Mol. Cell Biol. 2022, 23, 204–226. [Google Scholar] [CrossRef]
- Murach, K.A.; White, S.H.; Wen, Y.; Ho, A.; Dupont-Versteegden, E.E.; McCarthy, J.J.; Peterson, C.A. Differential requirement for satellite cells during overload-induced muscle hypertrophy in growing versus mature mice. Skelet. Muscle 2017, 7, 14. [Google Scholar] [CrossRef]
- Hutcheson, D.A.; Zhao, J.; Merrell, A.; Haldar, M.; Kardon, G. Embryonic and fetal limb myogenic cells are derived from developmentally distinct progenitors and have different requirements for beta-catenin. Genes Dev. 2009, 23, 997–1013. [Google Scholar] [CrossRef]
- Kassar-Duchossoy, L.; Giacone, E.; Gayraud-Morel, B.; Jory, A.; Gomès, D.; Tajbakhsh, S. Pax3/Pax7 mark a novel population of primitive myogenic cells during development. Genes Dev. 2005, 19, 1426–1431. [Google Scholar] [CrossRef]
- Relaix, F.; Montarras, D.; Zaffran, S.; Gayraud-Morel, B.; Rocancourt, D.; Tajbakhsh, S.; Mansouri, A.; Cumano, A.; Buckingham, M. Pax3 and Pax7 have distinct and overlapping functions in adult muscle progenitor cells. J. Cell Biol. 2006, 172, 91–102. [Google Scholar] [CrossRef]
- Seale, P.; Sabourin, L.A.; Girgis-Gabardo, A.; Mansouri, A.; Gruss, P.; Rudnicki, M.A. Pax7 Is Required for the Specification of Myogenic Satellite Cells. Cell 2000, 102, 777–786. [Google Scholar] [CrossRef]
- Ono, Y.; Boldrin, L.; Knopp, P.; Morgan, J.E.; Zammit, P.S. Muscle satellite cells are a functionally heterogeneous population in both somite-derived and branchiomeric muscles. Dev. Biol. 2010, 337, 29–41. [Google Scholar] [CrossRef] [PubMed]
- Collins, C.A.; Olsen, I.; Zammit, P.S.; Heslop, L.; Petrie, A.; Partridge, T.A.; Morgan, J.E. Stem Cell Function, Self-Renewal, and Behavioral Heterogeneity of Cells from the Adult Muscle Satellite Cell Niche. Cell 2005, 122, 289–301. [Google Scholar] [CrossRef] [PubMed]
- Almada, A.E.; Wagers, A.J. Molecular circuitry of stem cell fate in skeletal muscle regeneration, ageing and disease. Nat. Rev. Mol. Cell Biol. 2016, 17, 267–279. [Google Scholar] [CrossRef]
- Turner, D.L.; Butler, P.J. The aerobic capacity of locomotory muscles in the tufted duck, Aythya fuligula. J. Exp. Biol. 1988, 135, 445–460. [Google Scholar] [CrossRef]
- Li, L.; Liu, H.-H.; Xu, F.; Si, J.-M.; Jia, J.; Wang, J.-W. MyoD expression profile and developmental differences of leg and breast muscle in Peking duck (Anas platyrhynchos Domestica) during embryonic to neonatal stages. Micron 2010, 41, 847–852. [Google Scholar] [CrossRef]
- Ma, M.; Cai, B.; Jiang, L.; Abdalla, B.A.; Li, Z.; Nie, Q.; Zhang, X. lncRNA-Six1 Is a Target of miR-1611 that Functions as a ceRNA to Regulate Six1 Protein Expression and Fiber Type Switching in Chicken Myogenesis. Cells 2018, 7, 243. [Google Scholar] [CrossRef]
- Huo, W.; Weng, K.; Li, Y.; Zhang, Y.; Zhang, Y.; Xu, Q.; Chen, G. Comparison of muscle fiber characteristics and glycolytic potential between slow-and fast-growing broilers. Poult. Sci. 2022, 101, 101649. [Google Scholar] [CrossRef]
- Lin, S.; Lin, X.; Zhang, Z.; Jiang, M.; Rao, Y.; Nie, Q.; Zhang, X. Copy Number Variation in SOX6 Contributes to Chicken Muscle Development. Genes 2018, 9, 42. [Google Scholar] [CrossRef]
- Quiat, D.; Voelker, K.A.; Pei, J.; Grishin, N.V.; Grange, R.W.; Bassel-Duby, R.; Olson, E.N. Concerted regulation of myofiber-specific gene expression and muscle performance by the transcriptional repressor Sox6. Proc. Natl. Acad. Sci. USA 2011, 108, 10196–10201. [Google Scholar] [CrossRef]
- Zammit, P.; Relaix, F.; Nagata, Y.; Ruiz, A.; Collins, C.; Partridge, T., Jr. Pax7 and myogenic progression in skeletal muscle satellite cells. J. Cell Sci. 2006, 119, 1824–1832. [Google Scholar] [CrossRef] [PubMed]
- Zammit, P.S.; Golding, J.P.; Yosuke, N.; Valérie, H.; Partridge, T.A.; Beauchamp, J.R. Muscle satellite cells adopt divergent fates: A mechanism for self-renewal? J. Cell Biol. 2004, 166, 347–357. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Price, F.; Rudnicki, M.A. Satellite Cells and the Muscle Stem Cell Niche. Physiol. Rev. 2013, 93, 23–67. [Google Scholar] [CrossRef] [PubMed]
- Rossi, G.; Antonini, S.; Bonfanti, C.; Monteverde, S.; Vezzali, C.; Tajbakhsh, S.; Cossu, G.; Messina, G. Nfix Regulates Temporal Progression of Muscle Regeneration through Modulation of Myostatin Expression. Cell Rep. 2016, 14, 2238–2249. [Google Scholar] [CrossRef]
- Taglietti, V.; Maroli, G.; Cermenati, S.; Monteverde, S.; Ferrante, A.; Rossi, G.; Cossu, G.; Beltrame, M.; Messina, G. Nfix Induces a Switch in Sox6 Transcriptional Activity to Regulate MyHC-I Expression in Fetal Muscle. Cell Rep. 2016, 17, 2354–2366. [Google Scholar] [CrossRef]
- Anderson, C.M.; Hu, J.; Barnes, R.M.; Heidt, A.B.; Cornelissen, I.; Black, B.L. Myocyte enhancer factor 2C function in skeletal muscle is required for normal growth and glucose metabolism in mice. Skelet. Muscle 2015, 5, 7. [Google Scholar] [CrossRef]
- Potthoff, M.J.; Arnold, M.A.; McAnally, J.; Richardson, J.A.; Bassel-Duby, R.; Olson, E.N. Regulation of Skeletal Muscle Sarcomere Integrity and Postnatal Muscle Function by Mef2c. Mol. Cell. Biol. 2007, 27, 8143–8151. [Google Scholar] [CrossRef]
- Aare, S.; Spendiff, S.; Vuda, M. Failed reinnervation in aging skeletal muscle. Skelet. Muscle 2016, 6, 29. [Google Scholar] [CrossRef]
- Lee, S.-H.; Kim, B.-J.; Park, D.-R.; Kim, U.-H. Exercise induces muscle fiber type switching via transient receptor potential melastatin 2 (TRPM2)-dependent Ca 2+ signaling. J. Appl. Physiol. 2018, 124, 364–373. [Google Scholar] [CrossRef]
- Alastair, K.; Keith, B. Contractile and metabolic properties of engineered skeletal muscle derived from slow and fast phenotype mouse muscle. J. Cell Physiol. 2015, 230, 1750–1757. [Google Scholar]
- Luo, W.; Chen, J.; Li, L.; Ren, X.; Cheng, T.; Lu, S.; Lawal, R.A.; Nie, Q.; Zhang, X.; Hanotte, O. c-Myc inhibits myoblast differentiation and promotes myoblast proliferation and muscle fibre hypertrophy by regulating the expression of its target genes, miRNAs and lincRNAs. Cell Death Differ. 2019, 26, 426–442. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Lin, Z.; Chen, J.; Chen, G.; Zhang, S.; Liu, M.; Li, H.; He, D.; Liang, S.; Luo, Q.; et al. TMEM182 interacts with integrin beta 1 and regulates myoblast differentiation and muscle regeneration. J. Cachexia Sarcopenia Muscle 2021, 12, 1704–1723. [Google Scholar] [CrossRef] [PubMed]
- Talbot, J.; Maves, L. Skeletal muscle fiber type: Using insights from muscle developmental biology to dissect targets for susceptibility and resistance to muscle disease. Wiley Interdiscip. Rev. Dev. Biol. 2016, 5, 518–534. [Google Scholar] [CrossRef] [PubMed]
- Agbulut, O.; Noirez, P.; Beaumont, F.; Butler-Browne, G. Myosin heavy chain isoforms in postnatal muscle development of mice. Biol. Cell 2003, 95, 399–406. [Google Scholar] [CrossRef]
- Bjornson, C.R.R.; Cheung, T.H.; Liu, L.; Tripathi, P.V.; Steeper, K.M.; Rando, T.A. Notch Signaling Is Necessary to Maintain Quiescence in Adult Muscle Stem Cells. Stem Cells 2012, 30, 232–242. [Google Scholar] [CrossRef]
- Shihuan, K.; Chargé, S.B.; Patrick, S.; Michael, H.; Rudnicki, M.A. Distinct roles for Pax7 and Pax3 in adult regenerative myogenesis. J. Cell Biol. 2006, 172, 103–113. [Google Scholar]
- Fry, C.S.; Noehren, B.; Mula, J.; Ubele, M.F.; Westgate, P.M.; Kern, P.A.; Peterson, C.A. Fibre type-specific satellite cell response to aerobic training in sedentary adults. J. Physiol. 2014, 592, 2625–2635. [Google Scholar] [CrossRef]
- Darius, E.F.; Bianca, M.; Christian, H.; Tim, N.; Katrin, H.; Assmann, B.E.; Heiko, R. Disruption of SOX6 is associated with a rapid-onset dopa-responsive movement disorder, delayed development, and dysmorphic features. Pediatr. Neurol. 2015, 52, 115–118. [Google Scholar]
- Hagiwara, N.; Klewer, S.E.; Samson, R.A.; Erickson, D.T.; Lyon, M.F.; Brilliant, M.H. Sox6 is a candidate gene for p100H myopathy, heart block, and sudden neonatal death. Proc. Natl. Acad. Sci. USA 2000, 97, 4180–4185. [Google Scholar] [CrossRef]
- Hagiwara, N.; Yeh, M.; Liu, A. Sox6 is required for normal fiber type differentiation of fetal skeletal muscle in mice. Dev. Dyn. 2007, 236, 2062–2076. [Google Scholar] [CrossRef]
- Saclier, M.; Lapi, M.; Bonfanti, C.; Rossi, G.; Antonini, S.; Messina, G. The Transcription Factor Nfix Requires RhoA-ROCK1 Dependent Phagocytosis to Mediate Macrophage Skewing during Skeletal Muscle Regeneration. Cells 2020, 9, 708. [Google Scholar] [CrossRef]
- Hagiwara, N. Sox6, jack of all trades: A versatile regulatory protein in vertebrate development. Dev. Dyn. 2011, 240, 1311–1321. [Google Scholar] [CrossRef]
- Rossi, G.; Bonfanti, C.; Antonini, S.; Bastoni, M.; Monteverde, S.; Innocenzi, A.; Saclier, M.; Taglietti, V.; Messina, G. Silencing Nfix rescues muscular dystrophy by delaying muscle regeneration. Nat. Commun. 2017, 8, 1055. [Google Scholar] [CrossRef]
- Messina, G.; Biressi, S.; Monteverde, S.; Magli, A.; Cassano, M.; Perani, L.; Roncaglia, E.; Tagliafico, E.; Starnes, L.; Campbell, C.E.; et al. Nfix Regulates Fetal-Specific Transcription in Developing Skeletal Muscle. Cell 2010, 140, 554–566. [Google Scholar] [CrossRef] [PubMed]
- Perroud, J.; Bernheim, L.; Frieden, M.; Koenig, S. Distinct roles of NFATc1 and NFATc4 in human primary myoblast differentiation and in the maintenance of reserve cells. J. Cell Sci. 2017, 130, 3083–3093. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Du, J.; Shen, L.; Tan, Z.; Jiang, D.; Jiang, A.; Li, Q.; Tang, G.; Jiang, Y.; Wang, J.; et al. MiR-204-5p regulates C2C12 myoblast differentiation by targeting MEF2C and ERRγ. Biomed. Pharmacother. 2018, 101, 528–535. [Google Scholar] [CrossRef] [PubMed]
- Jama, A.; Huang, D.; Alshudukhi, A.A.; Chrast, R.; Ren, H. Lipin1 is required for skeletal muscle development by regulating MEF2c and MyoD expression. J. Physiol. 2019, 597, 889–901. [Google Scholar] [CrossRef]
- Sakuma, K.; Akiho, M.; Nakashima, H.; Nakao, R.; Hirata, M.; Inashima, S.; Yamaguchi, A.; Yasuhara, M. Cyclosporin A modulates cellular localization of MEF2C protein and blocks fiber hypertrophy in the overloaded soleus muscle of mice. Acta Neuropathol. 2008, 115, 663–674. [Google Scholar] [CrossRef]
- Zhang, C.-Y.; Yang, C.-Q.; Chen, Q.; Liu, J.; Zhang, G.; Dong, C.; Liu, X.-L.; Farooq, H.M.U.; Zhao, S.-Q.; Luo, L.-H.; et al. miR-194-Loaded Gelatin Nanospheres Target MEF2C to Suppress Muscle Atrophy in a Mechanical Unloading Model. Mol. Pharm. 2021, 18, 2959–2973. [Google Scholar] [CrossRef]
- Liu, N.; Nelson, B.R.; Bezprozvannaya, S.; Shelton, J.M.; Richardson, J.A.; Bassel-Duby, R.; Olson, E.N. Requirement of MEF2A, C, and D for skeletal muscle regeneration. Proc. Natl. Acad. Sci. USA 2014, 111, 4109–4114. [Google Scholar] [CrossRef]
- Song, M.-Y.; Han, C.Y.; Moon, Y.J.; Lee, J.H.; Bae, E.J.; Park, B.-H. Sirt6 reprograms myofibers to oxidative type through CREB-dependent Sox6 suppression. Nat. Commun. 2022, 13, 1808. [Google Scholar] [CrossRef] [PubMed]
- Bai, C.; Hou, L.; Li, F.; He, X.; Zhang, M.; Guan, W. Isolation and Biological Characteristics of Beijing Fatty Chicken Skeletal Muscle Satellite Cells. Cell Commun. Adhes. 2012, 19, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhong, H.; Lin, S.; Liang, L.; Ye, S.; Xu, Z.; Ji, C.; Zhang, Z.; Zhang, D.; Zhang, X. Polymorphisms of AMY1A gene and their association with growth, carcass traits and feed intake efficiency in chickens. Genomics 2021, 113, 583–594. [Google Scholar] [CrossRef] [PubMed]
- Kenneth, J.L.; Thomas, D.S. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−△△Ct Method. Methods 2001, 25, 402–408. [Google Scholar]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Z.; Lin, S.; Luo, W.; Ren, T.; Huang, X.; Li, W.; Zhang, X. Sox6 Differentially Regulates Inherited Myogenic Abilities and Muscle Fiber Types of Satellite Cells Derived from Fast- and Slow-Type Muscles. Int. J. Mol. Sci. 2022, 23, 11327. https://doi.org/10.3390/ijms231911327
Zhang Z, Lin S, Luo W, Ren T, Huang X, Li W, Zhang X. Sox6 Differentially Regulates Inherited Myogenic Abilities and Muscle Fiber Types of Satellite Cells Derived from Fast- and Slow-Type Muscles. International Journal of Molecular Sciences. 2022; 23(19):11327. https://doi.org/10.3390/ijms231911327
Chicago/Turabian StyleZhang, Zihao, Shudai Lin, Wen Luo, Tuanhui Ren, Xing Huang, Wangyu Li, and Xiquan Zhang. 2022. "Sox6 Differentially Regulates Inherited Myogenic Abilities and Muscle Fiber Types of Satellite Cells Derived from Fast- and Slow-Type Muscles" International Journal of Molecular Sciences 23, no. 19: 11327. https://doi.org/10.3390/ijms231911327
APA StyleZhang, Z., Lin, S., Luo, W., Ren, T., Huang, X., Li, W., & Zhang, X. (2022). Sox6 Differentially Regulates Inherited Myogenic Abilities and Muscle Fiber Types of Satellite Cells Derived from Fast- and Slow-Type Muscles. International Journal of Molecular Sciences, 23(19), 11327. https://doi.org/10.3390/ijms231911327





