In Planta, In Vitro and In Silico Studies of Chiral N6-Benzyladenine Derivatives: Discovery of Receptor-Specific S-Enantiomers with Cytokinin or Anticytokinin Activities
Abstract
1. Introduction
2. Results and Discussion
2.1. Chemical Synthesis
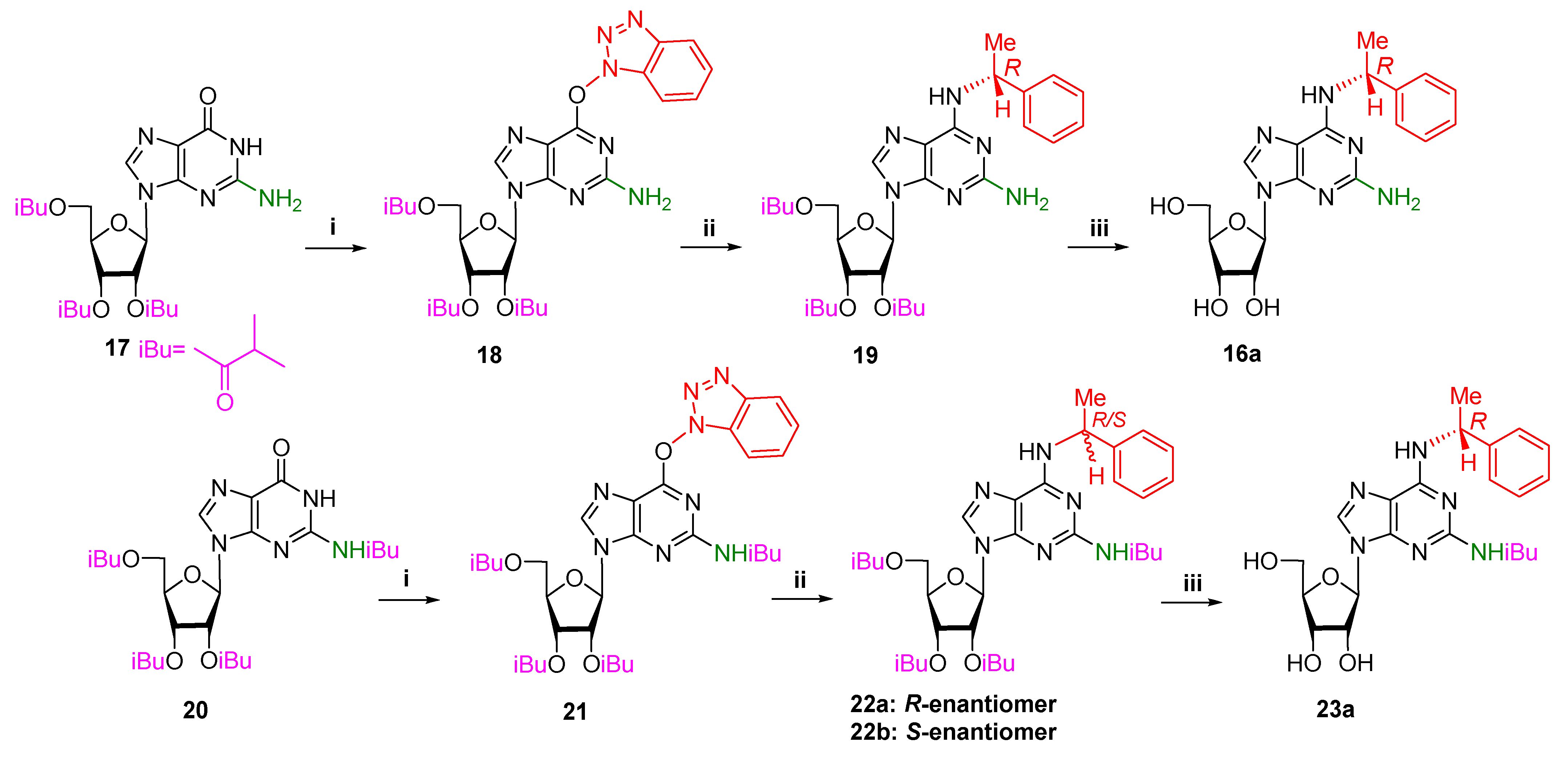
2.1.1. Synthesis of N6-Alkyladenines
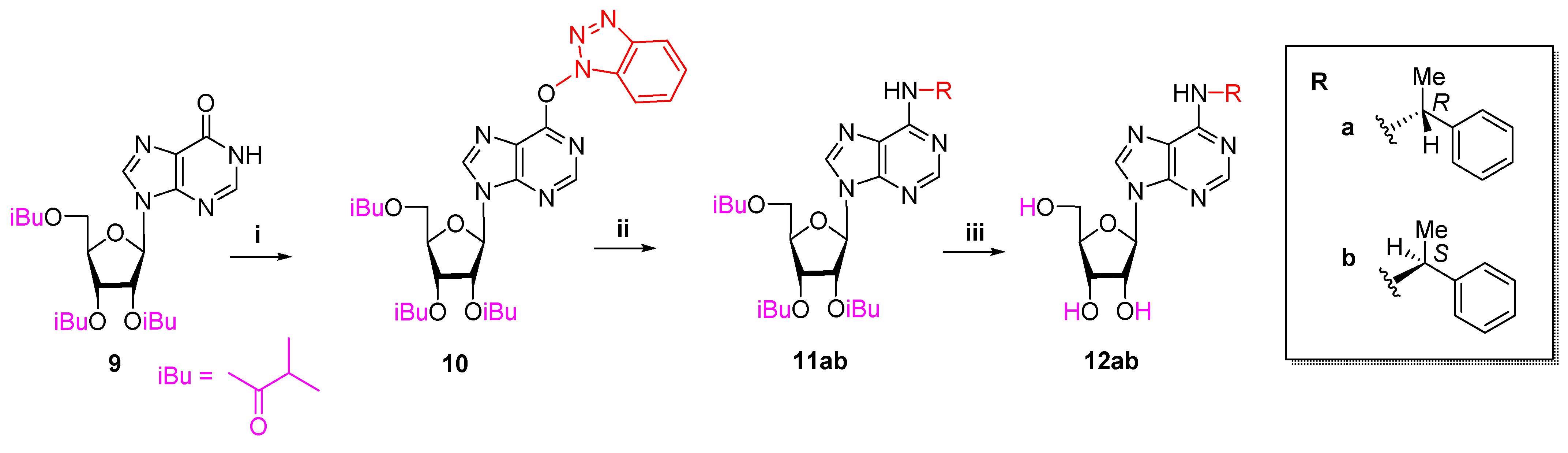
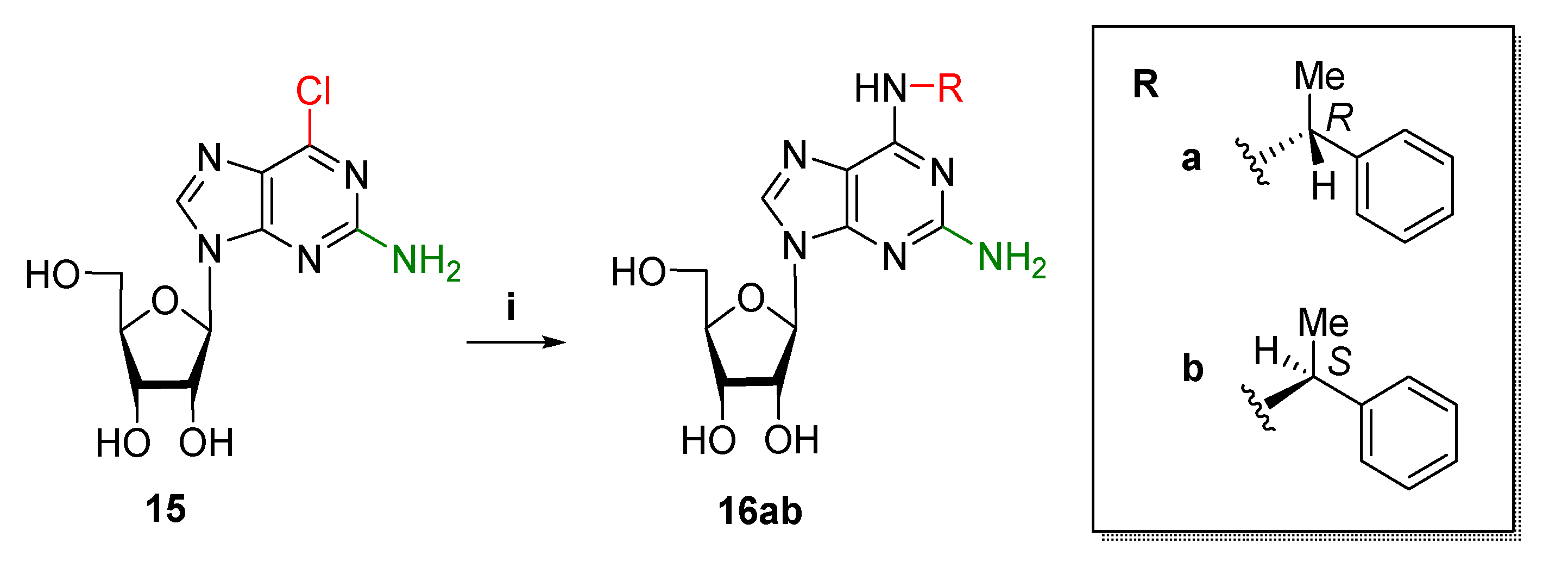
2.1.2. Synthesis of Ribonucleosides
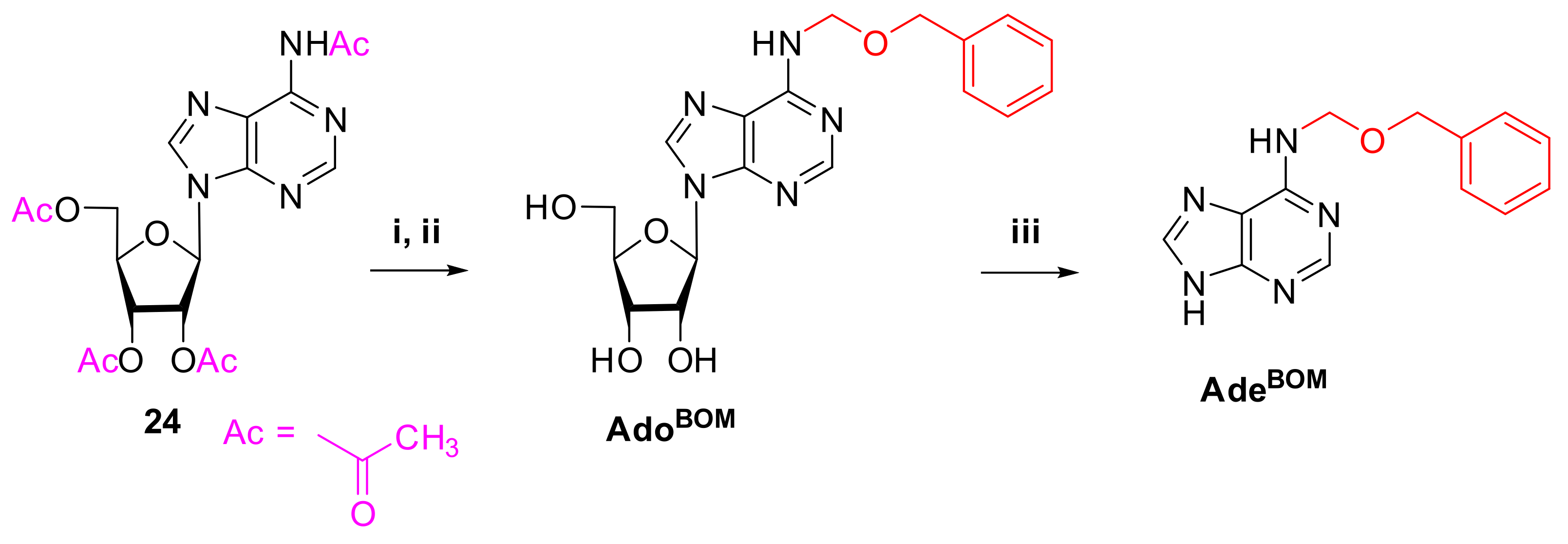
2.1.3. Synthesis of AdoBOM and AdeBOM
2.2. Study of the Biological Activity of the Chiral Compounds
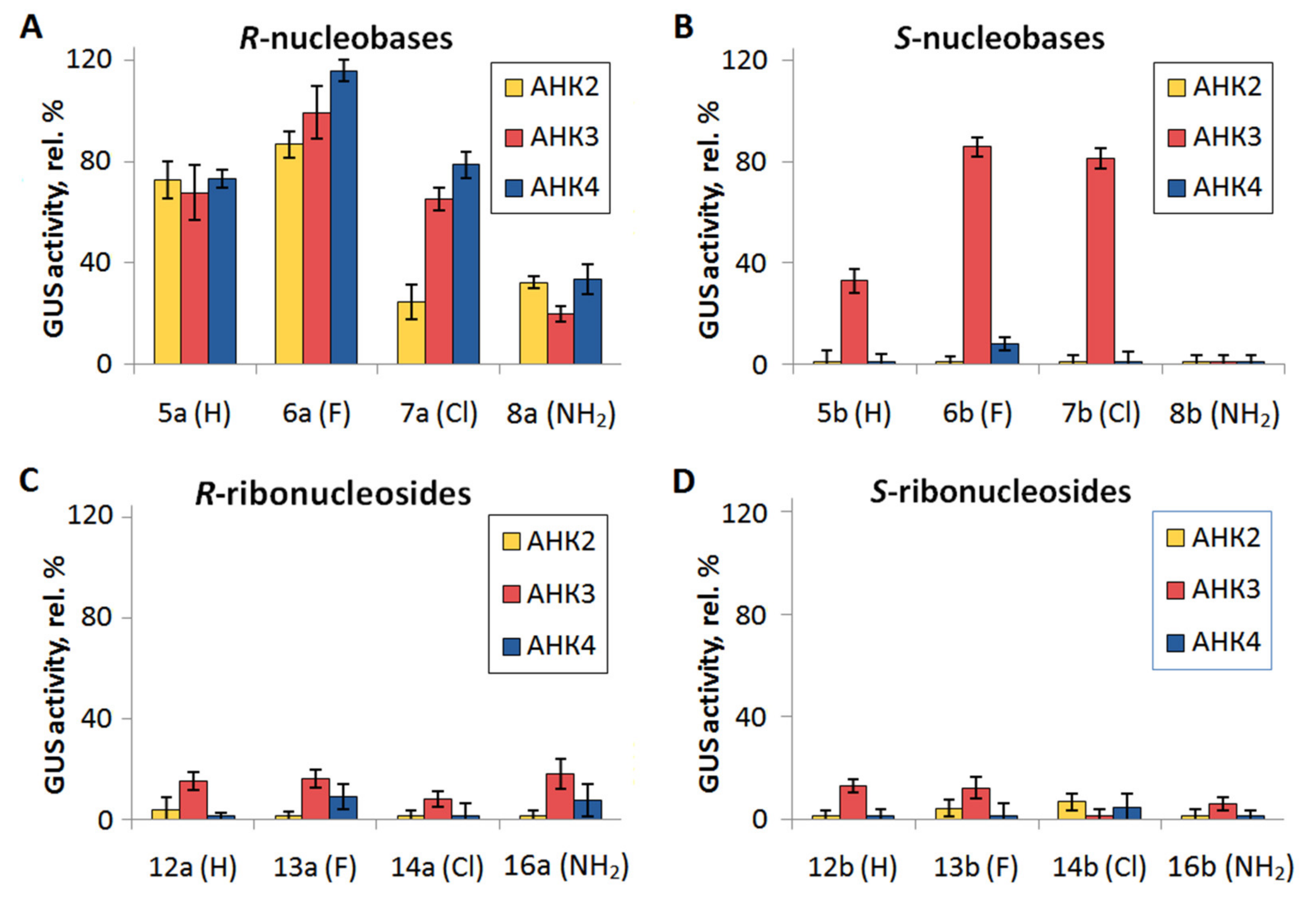
2.2.1. Cytokinin Activity of Compounds in Arabidopsis Bioassays
2.2.2. Anticytokinin Activity of Compounds in Arabidopsis Bioassay
2.2.3. (Anti)Cytokinin Binding Assay
3. Molecular Modeling and Docking
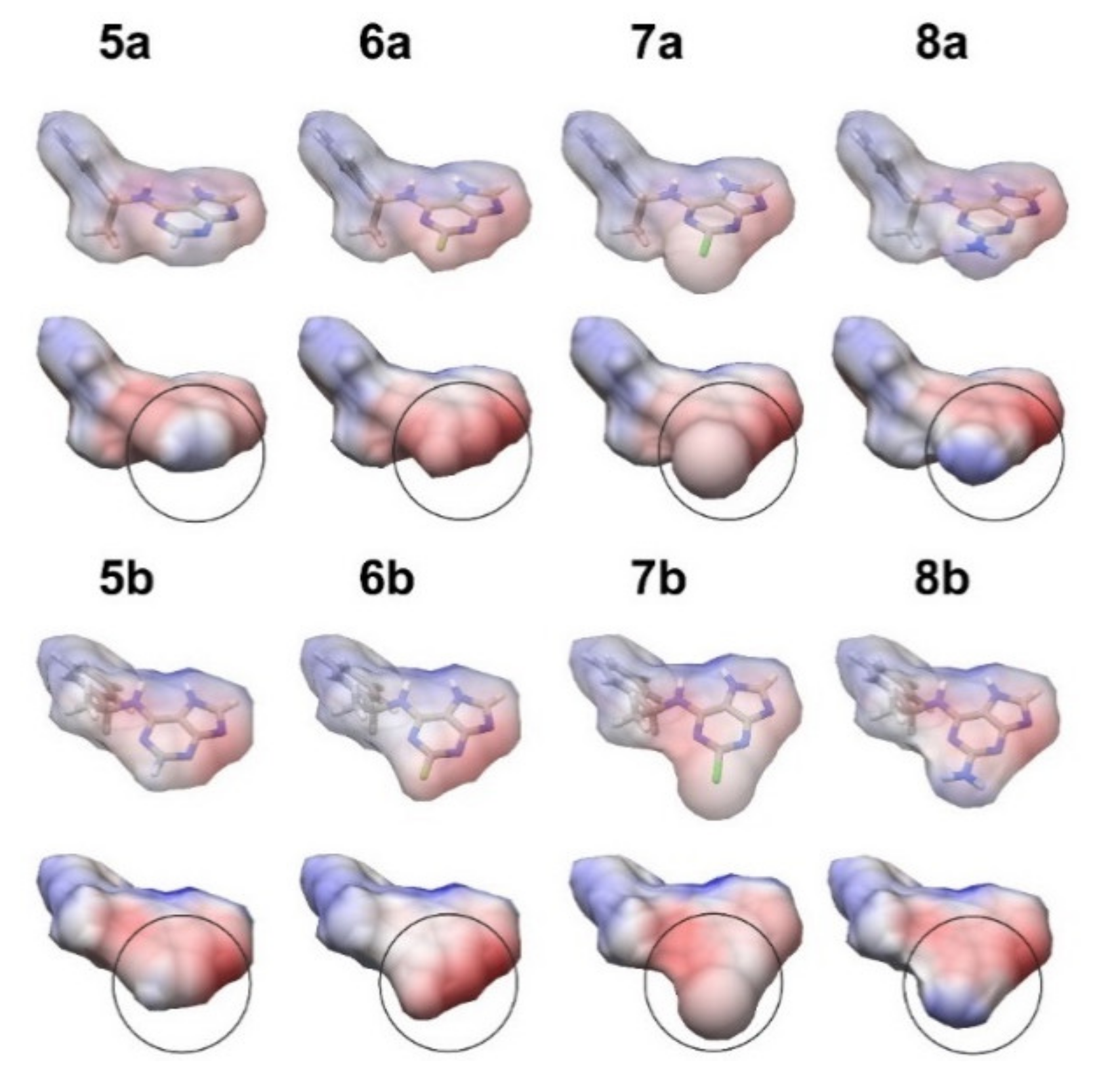
3.1. In Silico Study of the Interaction of Chiral BA Derivatives with Cytokinin Receptors
3.2. Difference between R- and S-Derivative Binding in Molecular Dynamics Simulation
4. Materials and Methods
4.1. Chemical Synthesis
4.1.1. General
4.1.2. Synthesis of N6-Alkyladenines
4.1.3. Synthesis of Ribonucleosides
4.1.4. Synthesis of AdoBOM and AdeBOM
4.2. Plant-Based Methods
4.2.1. Cytokinin and Anticytokinin Activity Assay
4.2.2. Histochemical Determination of GUS Activity
4.2.3. Expression of Cytokinin Receptor Genes and Membrane Fraction Isolation
4.2.4. Cytokinin Binding Assay
4.2.5. Statistical Analysis
4.3. Computational Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sakakibara, H. Cytokinins: Activity, biosynthesis, and translocation. Annu. Rev. Plant Biol. 2006, 57, 431–449. [Google Scholar] [CrossRef] [PubMed]
- Werner, T.; Schmülling, T. Cytokinin action in plant development. Curr. Opin. Plant Biol. 2009, 12, 527–538. [Google Scholar] [CrossRef]
- Spíchal, L. Cytokinins—recent news and views of evolutionally old molecules. Funct. Plant Biol. 2012, 39, 267–284. [Google Scholar] [CrossRef] [PubMed]
- Gruhn, N.; Heyl, A. Updates on the model and the evolution of cytokinin signaling. Curr. Opin. Plant Biol. 2013, 16, 569–574. [Google Scholar] [CrossRef] [PubMed]
- Kieber, J.J.; Schaller, G.E. Cytokinins. Arab. Book 2014, 12, e0168. [Google Scholar] [CrossRef] [PubMed]
- Lomin, S.N.; Savelieva, E.M.; Arkhipov, D.V.; Pashkovskiy, P.P.; Myakushina, Y.A.; Heyl, A.; Romanov, G.A. Cytokinin perception in ancient plants beyond Angiospermae. Int. J. Mol. Sci. 2021, 22, 13077. [Google Scholar] [CrossRef]
- Dello Ioio, R.; Nakamura, K.; Moubayidin, L.; Perilli, S.; Taniguchi, M.; Morita, M.T.; Aoyama, T.; Costantino, P.; Sabatini, S. A genetic framework for the control of cell division and differentiation in the root meristem. Science 2008, 322, 1380–1384. [Google Scholar] [CrossRef]
- Romanov, G.A. How do cytokinins affect the cell? Russ. J. Plant. Physiol. 2009, 56, 268–290. [Google Scholar] [CrossRef]
- Schaller, G.E.; Street, I.H.; Kieber, J.J. Cytokinin and the cell cycle. Curr. Opin. Plant Biol. 2014, 21, 7–15. [Google Scholar] [CrossRef]
- Oshchepkov, M.S.; Kalistratova, A.V.; Savelieva, E.M.; Romanov, G.A.; Bystrova, N.A.; Kochetkov, K.A. Natural and synthetic cytokinins and their applications in biotechnology, agrochemistry and medicine. Russ. Chem. Rev. 2020, 89, 787. [Google Scholar] [CrossRef]
- Werner, T.; Motyka, V.; Strnad, M.; Schmülling, T. Regulation of plant growth by cytokinin. Proc. Natl. Acad. Sci. USA 2001, 98, 10487. [Google Scholar] [CrossRef] [PubMed]
- Riefler, M.; Novak, O.; Strnad, M.; Schmülling, T. Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell 2006, 18, 40–54. [Google Scholar] [CrossRef] [PubMed]
- Schaller, G.E.; Bishopp, A.; Kieber, J.J. The Yin-Yang of hormones: Cytokinin and auxin interactions in plant development. Plant Cell 2015, 27, 44. [Google Scholar] [CrossRef] [PubMed]
- Kakimoto, T. Perception and signal transduction of cytokinins. Annu. Rev. Plant Biol. 2003, 54, 605–627. [Google Scholar] [CrossRef]
- Heyl, A.; Riefler, M.; Romanov, G.A.; Schmülling, T. Properties, functions, and evolution of cytokinin receptors. Eur. J. Cell Biol. 2012, 91, 246–256. [Google Scholar] [CrossRef]
- Lomin, S.N.; Krivosheev, D.M.; Steklov, M.Y.; Osolodkin, D.I.; Romanov, G.A. Receptor properties and features of cytokinin signaling. Acta Nat. 2012, 4, 31–45. [Google Scholar] [CrossRef]
- Steklov, M.Y.; Lomin, S.N.; Osolodkin, D.I.; Romanov, G.A. Structural basis for cytokinin receptor signaling: An evolutionary approach. Plant Cell Rep. 2013, 32, 781–793. [Google Scholar] [CrossRef]
- Higuchi, M.; Pischke, M.S.; Mähönen, A.P.; Miyawaki, K.; Hashimoto, Y.; Seki, M.; Kobayashi, M.; Shinozaki, K.; Kato, T.; Tabata, S.; et al. In planta functions of the Arabidopsis cytokinin receptor family. Proc. Natl. Acad. Sci. USA 2004, 101, 8821–8826. [Google Scholar] [CrossRef]
- Doležal, K.; Popa, I.; Krystof, V.; Spíchal, L.; Fojtíkova, M.; Holub, J.; Lenobel, R.; Schmülling, T.; Strnad, M. Preparation and biological activity of 6-benzylaminopurine derivatives in plants and human cancer cells. Bioorg. Med. Chem. 2006, 14, 875–884. [Google Scholar] [CrossRef]
- Doležal, K.; Popa, I.; Hauserova, E.; Spíchal, L.; Chakrabarty, K.; Novak, O.; Krystof, V.; Voller, J.; Holub, J.; Strnad, M. Preparation, biological activity and endogenous occurrence of N6-benzyladenosines. Bioorg. Med. Chem. 2007, 15, 3737–3747. [Google Scholar] [CrossRef]
- Szücova, L.; Spíchal, L.; Doležal, K.; Zatloukal, M.; Greplova, J.; Galuszka, P.; Krystof, V.; Voller, J.; Popa, I.; Massino, F.J.; et al. Synthesis, characterization and biological activity of ring-substituted 6-benzylamino-9-tetrahydropyran-2-yl and 9-tetrahydrofuran-2-ylpurine derivatives. Bioorg. Med. Chem. 2009, 17, 1938–1947. [Google Scholar] [CrossRef] [PubMed]
- Podlesakova, K.; Zalabak, D.; Cudejkova, M.; Plíhal, O.; Szücova, L.; Doležal, K.; Spíchal, L.; Strnad, M.; Galuszka, P. Novel cytokinin derivatives do not show negative effects on root growth and proliferation in submicromolar range. PLoS ONE 2012, 7, e39293. [Google Scholar] [CrossRef] [PubMed]
- Plíhal, O.; Szücova, L.; Galuszka, P. N9-substituted aromatic cytokinins with negligible side effects on root development are an emerging tool for in vitro culturing. Plant Signal. Behav. 2013, 8, e24392. [Google Scholar] [CrossRef] [PubMed]
- Vylícilova, H.; Husickova, A.; Spíchal, L.; Srovnal, J.; Doležal, K.; Plíhal, O.; Plíhalova, L. C-2-substituted aromatic cytokinin sugar conjugates delay the onset of senescence by maintaining the activity of the photosynthetic apparatus. Phytochemistry 2016, 122, 22–33. [Google Scholar] [CrossRef]
- Zahajska, L.; Nisler, J.; Voller, J.; Gucký, T.; Pospísil, T.; Spíchal, L.; Strnad, M. Preparation, characterization and biological activity of C8-substituted cytokinins. Phytochemistry 2017, 135, 115–127. [Google Scholar] [CrossRef]
- Savelieva, E.M.; Oslovsky, V.E.; Karlov, D.S.; Kurochkin, N.N.; Getman, I.A.; Lomin, S.N.; Sidorov, G.V.; Mikhailov, S.N.; Osolodkin, D.I.; Romanov, G.A. Cytokinin activity of N6-benzyladenine derivatives assayed by interaction with the receptors in planta, in vitro, and in silico. Phytochemistry 2018, 149, 161–177. [Google Scholar] [CrossRef]
- Koprna, R.; De Diego, N.; Dundálková, L.; Spíchal, L. Use of cytokinins as agrochemicals. Bioorg. Med. Chem. 2016, 24, 484–492. [Google Scholar] [CrossRef]
- Spíchal, L.; Werner, T.; Popa, I.; Riefler, M.; Schmülling, T.; Strnad, M. The purine derivative PI-55 blocks cytokinin action via receptor inhibition. FEBS J. 2009, 276, 244–253. [Google Scholar] [CrossRef]
- Nisler, J.; Zatloukal, M.; Popa, I.; Doležal, K.; Strnad, M.; Spíchal, L. Cytokinin receptor antagonists derived from 6-benzylaminopurine. Phytochemistry 2010, 71, 823–830. [Google Scholar] [CrossRef]
- Arata, Y.; Nagasawa-Iida, A.; Uneme, H.; Nakajima, H.; Kakimoto, T.; Sato, R. The phenylquinazoline compound S-4893 is a non-competitive cytokinin antagonist that targets Arabidopsis cytokinin receptor CRE1 and promotes root growth in Arabidopsis and rice. Plant Cell Physiol. 2010, 51, 2047. [Google Scholar] [CrossRef]
- Krivosheev, D.M.; Kolyachkina, S.V.; Mikhailov, S.N.; Tararov, V.I.; Vanyushin, B.F.; Romanov, G.A. N6(Benzyloxymethyl)adenosine is a novel anticytokinin, an antagonist of cytokinin receptor CRE1/AHK4 of Arabidopsis. Dokl. Biochem. Biophys. 2012, 444, 178–181. [Google Scholar] [CrossRef] [PubMed]
- Romanov, G.A.; Spíchal, L.; Lomin, S.N.; Strnad, M.; Schmülling, T. A live cell hormone-binding assay on transgenic bacteria expressing a eukaryotic receptor protein. Anal. Biochem. 2005, 347, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Romanov, G.A.; Lomin, S.N. Hormone-binding assay using living bacteria expressing eukaryotic receptors. In Plant Hormones: Methods and Protocols, 2nd ed.; Cutler, S., Bonetta, D., Eds.; Methods in Molecular Biology; Humana Press: Humana Totowa, NJ, USA, 2009; Volume 495, pp. 111–120. [Google Scholar]
- Lomin, S.A.; Krivosheev, D.M.; Steklov, M.Y.; Arkhipov, D.A.; Osolodkin, D.I.; Schmülling, T.; Romanov, G.A. Plant membrane assays with cytokinin receptors underpin the unique role of free cytokinin bases as biologically active ligands. J. Exp. Bot. 2015, 66, 1851. [Google Scholar] [CrossRef] [PubMed]
- Savelieva, E.M.; Lomin, S.N.; Romanov, G.A. A modified method for quantification of cytokinin-receptor binding using isolated plant microsomes enriched with cognate transmembrane receptors. Russ. J. Plant Physiol. 2022, 69, 6. [Google Scholar]
- Devinsky, F. Chirality and the origin of life. Symmetry 2021, 13, 2277. [Google Scholar] [CrossRef]
- Dyakin, V.V.; Uversky, V.N. Arrow of time, entropy, and protein folding: Holistic view on biochirality. Int. J. Mol. Sci. 2022, 23, 3687. [Google Scholar] [CrossRef]
- Chen, Y.; Ma, W. The origin of biological homochirality along with the origin of life. PLoS Comput. Biol. 2020, 16, e1007592. [Google Scholar] [CrossRef]
- Skolnick, J.; Zhou, H.; Gao, M. On the possible origin of protein homochirality, structure, and biochemical function. Proc. Natl. Acad. Sci. USA 2019, 116, 26571–26579. [Google Scholar] [CrossRef]
- Oda, A.; Nakayoshi, T.; Kato, K.; Fukuyoshi, S.; Kurimoto, E. Three dimensional structures of putative, primitive proteins to investigate the origin of homochirality. Sci. Rep. 2019, 9, 11594. [Google Scholar] [CrossRef]
- Metzler, D.E. Biochemistry. The Chemical Reactions of Living Cells, 2nd ed.; Metzler, D.E., Ed.; Academic Press: San Diego, CA, USA, 2001; Volume 1, pp. 677–712. [Google Scholar]
- Kamada-Nobusada, T.; Sakakibara, H. Molecular basis for cytokinin biosynthesis. Phytochemistry 2009, 70, 444–449. [Google Scholar] [CrossRef]
- Stolz, A.; Riefler, M.; Lomin, S.N.; Achazi, K.; Romanov, G.A.; Schmülling, T. The specificity of cytokinin signalling in Arabidopsis thaliana is mediated by differing ligand affinities and expression profiles of the receptors. Plant J. 2011, 67, 157–168. [Google Scholar] [CrossRef] [PubMed]
- Romanov, G.A.; Schmulling, T. On the biological activity of cytokinin free bases and their ribosides. Planta 2022, 255, 27. [Google Scholar] [CrossRef] [PubMed]
- Oslovsky, V.E.; Savelieva, E.M.; Drenichev, M.S.; Romanov, G.A.; Mikhailov, S.N. Distinct peculiarities of in planta synthesis of isoprenoid and aromatic cytokinins. Biomolecules 2020, 10, 86. [Google Scholar] [CrossRef] [PubMed]
- Brunoni, F.; Rolli, E.; Polverini, E.; Spíchal, L.; Ricci, A. The adjuvant activity of two urea derivatives on cytokinins: An example of serendipitous dual effect. Plant Growth Regul. 2021, 95, 169–190. [Google Scholar] [CrossRef]
- Heyl, A.; Wulfetange, K.; Pils, B.; Nielsen, N.; Romanov, G.A.; Schmülling, T. Evolutionary proteomics identifies amino acids essential for ligand binding of the cytokinin receptor CHASE domain. BMC Evol. Biol. 2007, 7, 62. [Google Scholar] [CrossRef]
- Hothorn, M.; Dabi, T.; Chory, J. Structural basis for cytokinin recognition by Arabidopsis thaliana histidine kinase 4. Nat. Chem. Biol. 2011, 7, 766–768. [Google Scholar] [CrossRef]
- Belyakov, P.A.; Kadentsev, V.I.; Chizhov, A.O.; Kolotyrkina, N.G.; Shashkov, A.S.; Ananikov, V.P. Mechanistic insight into organic and catalytic reactions by joint studies using mass spectrometry and NMR spectroscopy. Mendeleev Commun. 2010, 20, 125–131. [Google Scholar] [CrossRef]
- Steklov, M.Y.; Tararov, V.I.; Romanov, G.A.; Mikhailov, S.N. Facile synthesis of 8-azido-6-benzylaminopurine. Nucleosides Nucleotides Nucleic Acids 2011, 30, 503–511. [Google Scholar] [CrossRef]
- Hu, Y.L.; Liu, X.; Lu, M. Synthesis and biological activity of novel 6-substituted purine derivatives. J. Mex. Chem. Soc. 2010, 54, 74–78. [Google Scholar] [CrossRef]
- Gerster, J.F.; Jones, J.W.; Robins, R.K. Purine Nucleosides. IV. The Synthesis of 6-halogenated 9-β-D-ribofuranosylpurines from inosine and guanosine. J. Org. Chem. 1963, 28, 945–948. [Google Scholar] [CrossRef]
- Bae, S.; Lakshman, M.K. O6-(Benzotriazol-1-yl) inosine derivatives: Easily synthesized, reactive nucleosides. J. Am. Chem. Soc. 2007, 129, 782–789. [Google Scholar] [CrossRef] [PubMed]
- Wan, Z.K.; Binnun, E.; Wilson, D.P.; Lee, J. A highly facile and efficient one-step synthesis of N6-adenosine and N6-2’-deoxyadenosine derivatives. Org. Lett. 2005, 7, 5877–5880. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.H.; Patel, D.V. “BOP” as a reagent for mild and efficient preparation of esters. Tetrahedron Lett. 1994, 35, 5603–5606. [Google Scholar] [CrossRef]
- Wan, Z.K.; Wacharasindhu, S.; Binnun, E.; Mansour, T. An efficient direct amination of cyclic amides and cyclic ureas. Org. Lett. 2006, 8, 2425–2428. [Google Scholar] [CrossRef]
- Wan, Z.K.; Wacharasindhu, S.; Levins, C.G.; Lin, M.; Tabei, K.; Mansour, T.S. The scope and mechanism of phosphonium-mediated snarreactions in heterocyclic amides and ureas. J. Org. Chem. 2007, 72, 10194–10210. [Google Scholar] [CrossRef]
- Kore, A.R.; Yang, B.; Srinivasan, B. Recent developments in the synthesis of substituted purine nucleosides and nucleotides. Curr. Org. Chem. 2014, 18, 2072–2107. [Google Scholar] [CrossRef]
- Devine, S.M.; Scammells, P.J. Synthesis and utility of 2-halo-O6-(benzotriazol-1-yl)-functionalized purine nucleosides. Eur. J. Org. Chem. 2011, 6, 1092–1098. [Google Scholar] [CrossRef]
- Oslovsky, V.E.; Drenichev, M.S.; Sun, L.; Kurochkin, N.N.; Kunetsky, V.E.; Mirabelli, C.; Neyts, J.; Leyssen, P.; Mikhailov, S.N. Fluorination of naturally occurring N6-benzyladenosine remarkably increased its antiviral activity and selectivity. Molecules 2017, 7, 1219. [Google Scholar] [CrossRef]
- Zhong, M.; Nowak, I.; Robins, M.J. 6-(2-alkylimidazol-1-yl)purines undergo regiospecific glycosylation at N9. Org. Lett. 2005, 7, 4601–4603. [Google Scholar] [CrossRef]
- Sniady, A.; Bedore, M.W.; Jamison, T.F. One-flow multistep synthesis of nucleosides by Brønsted acid-catalyzed glycosylation. Angew. Chem. 2011, 123, 2203–2206. [Google Scholar] [CrossRef]
- Dumbre, S.G.; Jang, M.Y.; Herdewijn, P. Synthesis of α-L-threose nucleoside phosphonates via regioselective sugar protection. J. Org. Chem. 2013, 78, 7137–7144. [Google Scholar] [CrossRef] [PubMed]
- Framski, G.; Gdaniec, Z.; Gdaniec, M.; Boryski, J. A reinvestigated mechanism of ribosylation of adenine under silylating conditions. Tetrahedron 2006, 62, 10123–10129. [Google Scholar] [CrossRef]
- Drenichev, M.S.; Oslovsky, V.E.; Sun, L.; Tijsma, A.; Kurochkin, N.N.; Tararov, V.I.; Chizhov, A.O.; Neyts, J.; Pannecouque, C.; Leyssen, P.; et al. Modification of the length and structure of the linker of N6-benzyladenosine modulates its selective antiviral activity against enterovirus 71. Eur. J. Med. Chem. 2016, 111, 84–94. [Google Scholar] [CrossRef]
- Drenichev, M.S.; Oslovsky, V.E.; Zenchenko, A.A.; Danilova, C.V.; Varga, M.A.; Esipov, R.S.; Lykoshin, D.D.; Alexeev, C.S. Comparative analysis of enzymatic transglycosylation using E. coli nucleoside phosphorylases: A synthetic concept for the preparation of purine modified 2′-deoxyribonucleosides from ribonucleosides. Int. J. Mol. Sci. 2022, 23, 2795. [Google Scholar] [CrossRef] [PubMed]
- Romanov, G.A.; Kieber, J.J.; Schmülling, T. A rapid cytokinin response assay in Arabidopsis indicates a role for phospholipase D in cytokinin signaling. FEBS Lett. 2002, 515, 39–43. [Google Scholar] [CrossRef]
- Zvereva, S.D.; Romanov, G.A. Reporter genes for plant genetic engineering: Characteristics and detection. Russ. J. Plant Physiol. 2000, 47, 424–432. [Google Scholar]
- Sparkes, I.A.; Runions, J.; Kearns, A.; Hawes, C. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 2006, 1, 2019–2025. [Google Scholar] [CrossRef]
- Voinnet, O.; Rivas, S.; Mestre, P.; Baulcombe, D. An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 2003, 33, 949–956. [Google Scholar] [CrossRef]
- Sidorov, G.V.; Myasoedov, N.F.; Lomin, S.N.; Romanov, G.A. Synthesis of tritium- and deuterium-labeled isopentenyladenine. Radiochemistry 2015, 57, 108–110. [Google Scholar] [CrossRef]
- Arkhipov, D.V.; Lomin, S.N.; Myakushina, Y.A.; Savelieva, E.M.; Osolodkin, D.I.; Romanov, G.A. Modeling of protein–protein interactions in cytokinin signal transduction. Int. J. Mol. Sci. 2019, 20, 2096. [Google Scholar] [CrossRef]
- Smith, T.J. MOLView: A program for analyzing and displaying atomic structures on the Macintosh personal computer. J. Mol. Graph. 1995, 13, 122–125. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Krieger, E.; Vriend, G. YASARA View—molecular graphics for all devices - from smartphones to workstations. Bioinformatics 2014, 30, 2981–2982. [Google Scholar] [CrossRef] [PubMed]
- Maglic, J.B.; Lavendomme, R. MoloVol: An easy-to-use program for analyzing cavities, volumes and surface areas of chemical structures. J. Appl. Crystallogr. 2022, 55. [Google Scholar] [CrossRef]
- Krieger, E.; Dunbrack, R.L., Jr.; Hooft, R.W.; Krieger, B. Assignment of protonation states in proteins and ligands: Combining pKa prediction with hydrogen bonding network optimization. Methods Mol. Biol. 2012, 819, 405–421. [Google Scholar] [CrossRef]
- Krieger, E.; Nielsen, J.E.; Spronk, C.A.; Vriend, G. Fast empirical pKa prediction by Ewald summation. J. Mol. Graph. Modell. 2006, 25, 481–486. [Google Scholar] [CrossRef]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef]
- Jakalian, A.; Jack, D.B.; Bayly, C.I. Fast, efficient generation of high-quality atomic charges. AM1-BCC model: II. Parameterization and validation. J. Comput. Chem. 2002, 23, 1623–1641. [Google Scholar] [CrossRef]
- Hornak, V.; Abel, R.; Okur, A.; Strockbine, B.; Roitberg, A.; Simmerling, C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins 2006, 65, 712–725. [Google Scholar] [CrossRef]
- Essman, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef]
- Krieger, E.; Vriend, G. New ways to boost molecular dynamics simulations. J. Comp. Chem. 2015, 36, 996–1007. [Google Scholar] [CrossRef] [PubMed]
- Wallace, A.C.; Laskowski, R.A.; Thornton, J.M. LIGPLOT: A program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995, 8, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A. PDBsum new things. Nucleic Acids Res. 2009, 37, D355–D359. [Google Scholar] [CrossRef] [PubMed]
- BIOVIA, Dassault Systèmes. Discovery Studio Visualizer. 2020, v. 21.1.0.20298. San Diego. Available online: https://discover.3ds.com/discovery-studio-visualizer-download (accessed on 8 January 2022).
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera-a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]





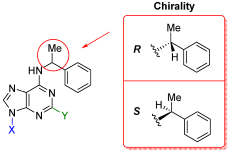 | ||||
|---|---|---|---|---|
| Compound | Chemical Name | Chirality | N9-Substituent (X) | C2-Substituent (Y) |
| Nucleobases | ||||
| 5a | N6-((R)-α-methylbenzyl) adenine | R | H | H |
| 6a | 2-fluoro,N6-((R)-α-methylbenzyl) adenine | R | H | F |
| 7a | 2-chloro,N6-((R)-α-methylbenzyl) adenine | R | H | Cl |
| 8a | 2-amino,N6-((R)-α-methylbenzyl) adenine | R | H | NH2 |
| 5b | N6-((S)-α-methylbenzyl) adenine | S | H | H |
| 6b | 2-fluoro,N6-((S)-α-methylbenzyl) adenine | S | H | F |
| 7b | 2-chloro,N6-((S)-α-methylbenzyl) adenine | S | H | Cl |
| 8b | 2-amino,N6-((S)-α-methylbenzyl) adenine | S | H | NH2 |
| Ribonucleosides | ||||
| 12a | N6-((R)-α-methylbenzyl) adenosine | R | Rib | H |
| 13a | 2-fluoro,N6-((R)-α-methylbenzyl) adenosine | R | Rib | F |
| 14a | 2-chloro,N6-((R)-α-methylbenzyl) adenosine | R | Rib | Cl |
| 16a | 2-amino, N6-((R)-α-methylbenzyl) adenosine | R | Rib | NH2 |
| 12b | N6-((S)-α-methylbenzyl) adenosine | S | Rib | H |
| 13b | 2-fluoro,N6-((S)-α-methylbenzyl) adenosine | S | Rib | F |
| 14b | 2-chloro,N6-((S)-α-methylbenzyl) adenosine | S | Rib | Cl |
| 16b | 2-amino,N6-((S)-α-methylbenzyl) adenosine | S | Rib | NH2 |
 | ||||
|---|---|---|---|---|
| Derivative Number | АНК2 | АНК3 | CRE1/АНК4 | |
| S-nucleobases | 5b | 71.3 ± 5.1 | 107.6 ± 11.5 | 43.0 ±5,5 |
| 6b | 116.5 ± 10.5 | 122.3 ± 2.8 | 93.7 ± 4.2 | |
| 7b | 105.0 ± 6.0 | 111.5 ± 4.8 | 100.3 ± 2.5 | |
| 8b | 9.8 ± 4.8 | 79.5 ± 9.0 | 38.5 ± 7.7 | |
| 12a | 89.1 ± 12.6 | 74.4 ± 7.7 | 142.5 ± 8.1 | |
| R-ribonucleosides | 13a | 86.1 ± 2.6 | 95.2 ± 7.6 | 101.2 ± 11.1 |
| 14a | 105.3 ± 2.6 | 79.8 ± 3.5 | 109.2 ± 13.2 | |
| 16a | 47.3 ± 5.4 | 26.6 ± 2.8 | 48.6 ± 2.6 | |
| S-ribonucleosides | 12b | 9.5 ± 2.6 | 71.9 ± 8.3 | 0.0 ± 2.6 |
| 13b | 0.4 ± 3.6 | 61.0 ± 3.5 | 0.0 ± 2.7 | |
| 14b | 1.3 ± 3.7 | 84.3 ± 10.1 | 0.0 ± 2.5 | |
| 16b | 8.5 ± 2.2 | 77.8 ± 12.9 | 0.0 ± 5.4 | |
 | ||||
|---|---|---|---|---|
| Derivative Number | АНК2 | АНК3 | CRE1/АНК4 | |
| S-nucleobases | 5b | 124.5 ± 3.9 | 129.3 ± 4.8 | 162.5 ± 10.1 |
| 8b | 136.80 ± 4.4 | 140.5 ± 9.0 | 136.7 ± 19.0 | |
| R-ribonucleosides | 12a | 197.5 ± 5.2 | 240.6 ± 2.9 | 212.7 ± 6.1 |
| 16a | 126.8 ± 10.4 | 108.4 ± 11.4 | 112.5 ± 6.7 | |
| S-ribonucleosides | 12b | 64.6 ± 2.5 | 152.1 ± 6.1 | 60.9 ± 8.6 |
| 13b | 102.9 ± 3.8 | n/d * | 110.6 ± 3.4 | |
| 14b | 116.7 ± 15.7 | n/d * | 126.4 ± 11.5 | |
| 16b | 109.8 ± 5.3 | 124.6 ± 11.0 | 64.8 ± 4.5 | |
 | |||||||
|---|---|---|---|---|---|---|---|
| Derivative Number | АНК2 | АНК3 | CRE1/AHK4 | ||||
| 1 µM | 50 µM | 1 µM | 50 µM | 1 µM | 50 µM | ||
| R-nucleobases | 5a | 88.9 ± 0.5 | n/d * | 97.7 ± 1.0 | n/d * | 87.7 ± 1.0 | n/d * |
| 6a | 92.1 ± 1.0 | n/d * | 99.8 ± 1.5 | n/d * | 97.2 ± 0.9 | n/d * | |
| 7a | 42.6 ± 0.4 | n/d * | 88.9 ± 0.7 | n/d * | 74.1 ± 1.6 | n/d * | |
| 8a | 58.8 ± 1.3 | n/d * | 54.7 ± 1.1 | n/d * | 30.4 ± 1.0 | n/d * | |
| S-nucleobases | 5b | 8.9 ± 4.9 | 48.2 ± 1.0 | 46.5 ± 2.2 | 97.0 ± 0.6 | 11.1 ± 1.4 | 48.6 ± 3.3 |
| 6b | 8.7 ± 0.9 | 84.7 ± 0.6 | 74.7 ± 1.7 | 99.7 ± 5.3 | 45.8 ± 0.3 | 98.4 ± 0.2 | |
| 7b | 3.1 ± 0.4 | 82.5 ± 0.7 | 80.6 ± 1.5 | 97.4 ± 2.6 | 37.1 ± 1.2 | 94.9 ± 0.1 | |
| 8b | 2.3 ± 3.7 | 28.0 ± 0.1 | 3.7 ± 0.1 | 85.4 ± 1.5 | 4.8 ± 0.5 | 70.4 ± 1.7 | |
| R-ribonucleosides | 12a | 9.3 ± 0.6 | 16.3 ± 0.5 | 4.3 ±1.4 | 35.7 ± 0.9 | 10.7 ± 1.6 | 10.6 ± 1.8 |
| 13a | 2.6 ± 5.1 | 15.2 ± 2.0 | 26.9 ± 2.3 | 81.3 ± 6.1 | 0.5 ± 1.9 | 39.5 ±1.0 | |
| 14a | 1.4 ± 0.1 | 23.7 ± 3.2 | 3.5 ± 0.1 | 45.0 ± 7.7 | 3.2 ± 0.1 | 64.3 ± 1.2 | |
| 16a | 1.7 ± 2.9 | 11.2 ± 0.2 | 3.6 ± 2.9 | 38.8 ± 0.4 | 2.3 ± 0.9 | 19.6 ± 2.8 | |
| S-ribonucleosides | 12b | 9.9 ± 2.1 | 12.2 ± 1.2 | 0.6 ± 0.1 | 8.3 ± 1.2 | 4.1 ± 1.5 | 4.0 ± 0.2 |
| 13b | 0.5 ± 1.7 | 0.2 ± 0.01 | 3.1 ± 5.8 | 40.9 ± 2.8 | 4.1 ± 2.6 | 2.9 ± 0.6 | |
| 14b | 0.6 ± 0.2 | 1.2 ± 0.3 | 3.8 ± 0.6 | 12.1 ± 8.4 | 1.2 ± 0.4 | 4.3 ± 1.6 | |
| 16b | 3.4 ± 1.0 | 6.2 ± 0.1 | 8.6 ± 1.2 | 22.9 ± 1.8 | 2.9 ± 2.1 | 6.9 ± 3.5 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Savelieva, E.M.; Zenchenko, A.A.; Drenichev, M.S.; Kozlova, A.A.; Kurochkin, N.N.; Arkhipov, D.V.; Chizhov, A.O.; Oslovsky, V.E.; Romanov, G.A. In Planta, In Vitro and In Silico Studies of Chiral N6-Benzyladenine Derivatives: Discovery of Receptor-Specific S-Enantiomers with Cytokinin or Anticytokinin Activities. Int. J. Mol. Sci. 2022, 23, 11334. https://doi.org/10.3390/ijms231911334
Savelieva EM, Zenchenko AA, Drenichev MS, Kozlova AA, Kurochkin NN, Arkhipov DV, Chizhov AO, Oslovsky VE, Romanov GA. In Planta, In Vitro and In Silico Studies of Chiral N6-Benzyladenine Derivatives: Discovery of Receptor-Specific S-Enantiomers with Cytokinin or Anticytokinin Activities. International Journal of Molecular Sciences. 2022; 23(19):11334. https://doi.org/10.3390/ijms231911334
Chicago/Turabian StyleSavelieva, Ekaterina M., Anastasia A. Zenchenko, Mikhail S. Drenichev, Anna A. Kozlova, Nikolay N. Kurochkin, Dmitry V. Arkhipov, Alexander O. Chizhov, Vladimir E. Oslovsky, and Georgy A. Romanov. 2022. "In Planta, In Vitro and In Silico Studies of Chiral N6-Benzyladenine Derivatives: Discovery of Receptor-Specific S-Enantiomers with Cytokinin or Anticytokinin Activities" International Journal of Molecular Sciences 23, no. 19: 11334. https://doi.org/10.3390/ijms231911334
APA StyleSavelieva, E. M., Zenchenko, A. A., Drenichev, M. S., Kozlova, A. A., Kurochkin, N. N., Arkhipov, D. V., Chizhov, A. O., Oslovsky, V. E., & Romanov, G. A. (2022). In Planta, In Vitro and In Silico Studies of Chiral N6-Benzyladenine Derivatives: Discovery of Receptor-Specific S-Enantiomers with Cytokinin or Anticytokinin Activities. International Journal of Molecular Sciences, 23(19), 11334. https://doi.org/10.3390/ijms231911334







