Associations of miR-181a with Health-Related Quality of Life, Cognitive Functioning, and Clinical Data of Patients with Different Grade Glioma Tumors
Abstract
1. Introduction
2. Results
2.1. Association of miR-181a Expression with Patient Clinicopathological Data
2.2. Associations of miR-181a Expression with IDH1 Status of GBM Tumors
2.3. Associations of mir181a Expression with Functional Status, QoL, and Cognitive Functioning of Patients
3. Discussion
4. Materials and Methods
4.1. Samples and Patient Clinicopathological Data
4.2. Functional Status
4.3. Quality of Life Assessment
4.4. Assessment of Cognitive Functioning
4.5. Gene Expression Analysis
4.6. Statistical Analysis
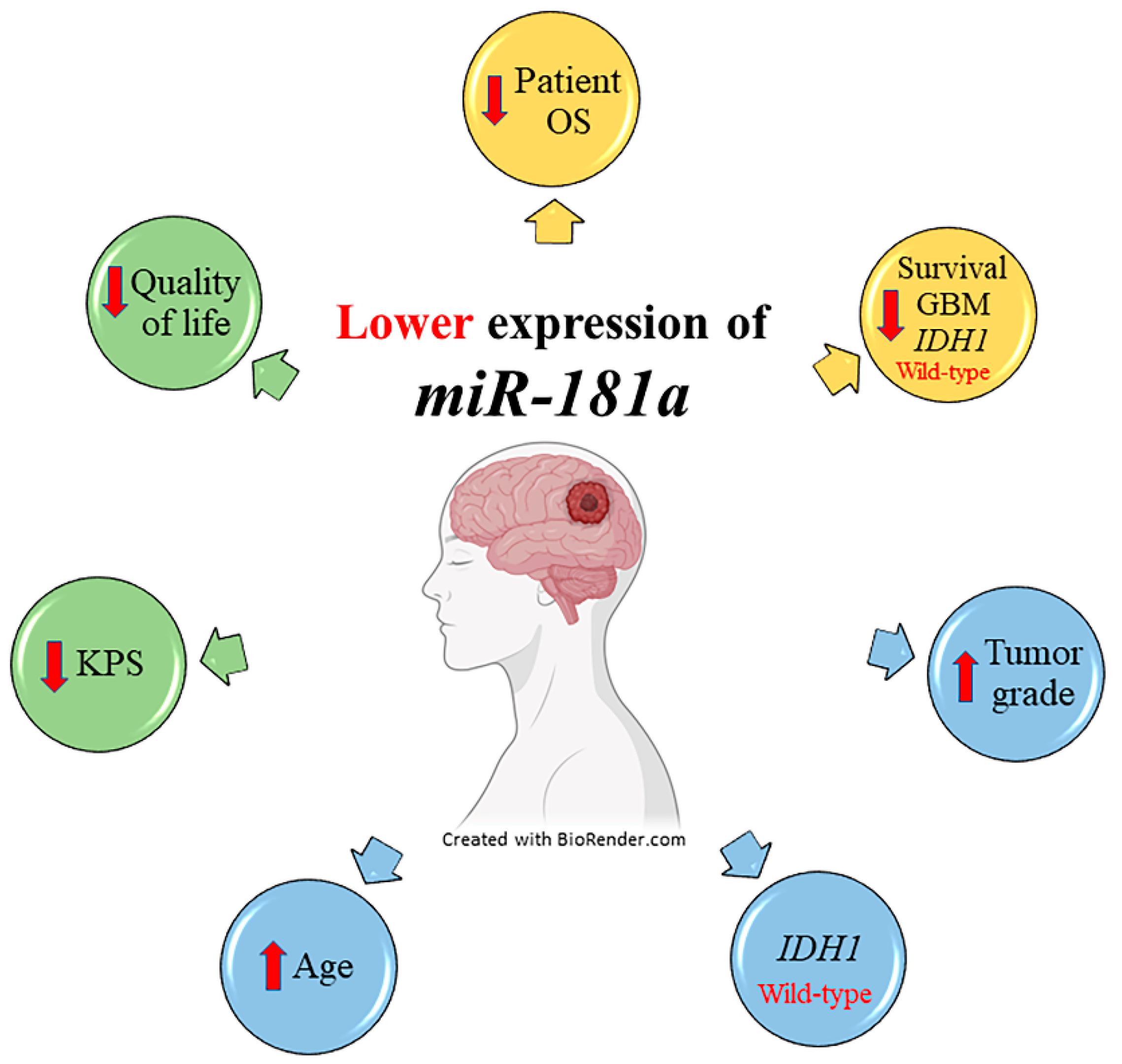
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Louis, D.N.; Perry, A.; Reifenberger, G.; Von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef] [PubMed]
- Weller, M.; dan der Bent, M.; Tonn, J.C.; Stupp, R.; Preusser, M.; Cohen-Jonathan-Moyal, E.; Henriksson, R.; Le Rhun, E.; Balana, C.; Chinot, O.; et al. European Association for Neuro-Oncology (EANO) guideline on the diagnosis and treatment of adult astrocytic and oligodendroglial gliomas. Lancet Oncol. 2017, 18, e315–e329. [Google Scholar] [CrossRef]
- Stupp, R.; Taillibert, S.; Kanner, A.A.; Read, W.; Steinberg, D.M.; Lhermitte, B.; Toms, S.; Idbaih, A.; Ahluwalia, M.S.; Fink, K.; et al. Effect of Tumor-Treating Fields Plus Maintenance Temozolomide vs Maintenance Temozolomide Alone on Survival in Patients With Glioblastoma. JAMA 2017, 318, 2306–2316. [Google Scholar] [CrossRef] [PubMed]
- Minniti, G.; Niyazi, M.; Alongi, F.; Navarria, P.; Belka, C. Current status and recent advances in reirradiation of glioblastoma. Radiat. Oncol. 2021, 16, 36. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro-Oncology 2021, 23, 1231–1251. [Google Scholar] [CrossRef]
- Yang, K.; Wu, Z.; Zhang, H.; Zhang, N.; Wu, W.; Wang, Z.; Dai, Z.; Zhang, X.; Zhang, L.; Peng, Y.; et al. Glioma targeted therapy: Insight into future of molecular approaches. Mol. Cancer 2022, 21, 39. [Google Scholar] [CrossRef]
- Hutóczki, G.; Virga, J.; Birkó, Z.; Klekner, A. Novel Concepts of Glioblastoma Therapy Concerning Its Heterogeneity. Int. J. Mol. Sci. 2021, 22, 10005. [Google Scholar] [CrossRef]
- Brennan, C.W.; Verhaak, R.G.W.; McKenna, A.; Campos, B.; Noushmehr, H.; Salama, S.R.; Zheng, S.; Chakravarty, D.; Sanborn, J.Z.; Berman, S.H.; et al. The Somatic Genomic Landscape of Glioblastoma. Cell 2013, 155, 462–477. [Google Scholar] [CrossRef]
- Hou, J.; He, Z.; Liu, T.; Chen, D.; Wang, B.; Wen, Q.; Zheng, X. Evolution of Molecular Targeted Cancer Therapy: Mechanisms of Drug Resistance and Novel Opportunities Identified by CRISPR-Cas9 Screening. Front. Oncol. 2022, 12, 755053. [Google Scholar] [CrossRef]
- Malone, E.R.; Oliva, M.; Sabatini, P.J.B.; Stockley, T.; Siu, L.L. Molecular profiling for precision cancer therapies. Genome Med. 2020, 12, 8. [Google Scholar] [CrossRef]
- Korja, M.; Raj, R.; Seppä, K.; Luostarinen, T.; Malila, N.; Seppälä, M.; Mäenpää, H.; Pitkäniemi, J. Glioblastoma survival is improving despite increasing incidence rates: A nationwide study between 2000 and 2013 in Finland. Neuro-Oncology 2018, 21, 370–379. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Zhao, Z.; Cai, Q.; Zhang, Y.; Zhang, P.; Shi, S.; Xie, H.; Peng, X.; Yin, W.; Tao, Y.; et al. miRNA-based biomarkers, therapies, and resistance in Cancer. Int. J. Biol. Sci. 2020, 16, 2628–2647. [Google Scholar] [CrossRef]
- Si, W.; Shen, J.; Zheng, H.; Fan, W. The role and mechanisms of action of microRNAs in cancer drug resistance. Clin. Epigenet. 2019, 11, 25. [Google Scholar] [CrossRef] [PubMed]
- Giovannetti, E.; Erozenci, A.; Smit, J.; Danesi, R.; Peters, G.J. Molecular mechanisms underlying the role of microRNAs (miRNAs) in anticancer drug resistance and implications for clinical practice. Crit. Rev. Oncol. Hematol. 2011, 81, 103–122. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Shen, X.J.; Zou, Q.; Wang, S.P.; Tang, S.M.; Zhang, G.Z. Biological functions of microRNAs: A review. J. Physiol. Biochem. 2010, 67, 129–139. [Google Scholar] [CrossRef] [PubMed]
- Acunzo, M.; Romano, G.; Wernicke, D.; Croce, C.M. MicroRNA and cancer–A brief overview. Adv. Biol. Regul. 2015, 57, 1–9. [Google Scholar] [CrossRef]
- Condrat, C.E.; Thompson, D.C.; Barbu, M.G.; Bugnar, O.L.; Boboc, A.; Cretoiu, D.; Suciu, N.; Cretoiu, S.M.; Voinea, S.C. miRNAs as Biomarkers in Disease: Latest Findings Regarding Their Role in Diagnosis and Prognosis. Cells 2020, 9, 276. [Google Scholar] [CrossRef] [PubMed]
- Indrieri, A.; Carrella, S.; Carotenuto, P.; Banfi, S.; Franco, B. The Pervasive Role of the miR-181 Family in Development, Neurodegeneration, and Cancer. Int. J. Mol. Sci. 2020, 21, 2092. [Google Scholar] [CrossRef]
- Feng, X.; Zhang, C.; Yang, Y.; Hou, D.; Zhu, A. Role of miR-181a in the process of apoptosis of multiple malignant tumors: A literature review. Adv. Clin. Exp. Med. 2018, 27, 263–270. [Google Scholar] [CrossRef] [PubMed]
- Karsy, M.; Arslan, E.; Moy, F. Current Progress on Understanding MicroRNAs in Glioblastoma Multiforme. Genes Cancer 2012, 3, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Saliminejad, K.; Khorram Khorshid, H.R.; Soleymani Fard, S.; Ghaffari, S.H. An overview of microRNAs: Biology, functions, therapeutics, and analysis methods. J. Cell. Physiol. 2019, 234, 5451–5465. [Google Scholar] [CrossRef]
- Zhai, Z.; Mu, T.; Zhao, L.; Li, Y.; Zhu, D.; Pan, Y. MiR-181a-5p facilitates proliferation, invasion, and glycolysis of breast cancer through NDRG2-mediated activation of PTEN/AKT pathway. Bioengineered 2021, 13, 83–95. [Google Scholar] [CrossRef] [PubMed]
- Zuo, L.; Li, X.; Zhu, H.; Li, A.; Wang, Y. Expression of miR-181a in Circulating Tumor Cells of Ovarian Cancer and Its Clinical Application. ACS Omega 2021, 6, 22011–22019. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.; Ma, D.; Zhang, J.; Zhao, J.; Yang, M. MicroRNA-18a induces epithelial-mesenchymal transition like cancer stem cell phenotype via regulating RKIP pathway in pancreatic cancer. Ann. Transl. Med. 2020, 8, 433. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.; Chen, B.; Wang, X.; Wu, K.; Sun, Y. Long non-coding RNA XIST regulates PTEN expression by sponging miR-181a and promotes hepatocellular carcinoma progression. BMC Cancer 2017, 17, 248. [Google Scholar] [CrossRef]
- Yang, C.-C.; Hung, P.-S.; Wang, P.-W.; Liu, C.-J.; Chu, T.-H.; Cheng, H.-W.; Lin, S.-C. miR-181 as a putative biomarker for lymph-node metastasis of oral squamous cell carcinoma. J. Oral Pathol. Med. 2011, 40, 397–404. [Google Scholar] [CrossRef] [PubMed]
- Xue, W.-X.; Zhang, M.-Y.; Li, R.; Liu, X.; Yin, Y.-H.; Qu, Y.-Q. Serum miR-1228-3p and miR-181a-5p as Noninvasive Biomarkers for Non-Small Cell Lung Cancer Diagnosis and Prognosis. BioMed Res. Int. 2020, 2020, 9601876. [Google Scholar] [CrossRef]
- Shen, H.; Weng, X.D.; Liu, X.H.; Yang, D.; Wang, L.; Guo, J.; Wang, M.; Wang, X.; Diao, C.H. miR-181a-5p is downregulated and inhibits proliferation and the cell cycle in prostate cancer. Int. J. Clin. Exp. Pathol. 2018, 11, 3969–3976. [Google Scholar]
- Yang, L.; Ma, Y.; Xin, Y.; Han, R.; Li, R.; Hao, X. Role of the microRNA 181 family in glioma development. Mol. Med. Rep. 2017, 17, 322–329. [Google Scholar] [CrossRef]
- Shi, L.; Cheng, Z.; Zhang, J.; Li, R.; Zhao, P.; Fu, Z.; You, Y. hsa-mir-181a and hsa-mir-181b function as tumor suppressors in human glioma cells. Brain Res. 2008, 1236, 185–193. [Google Scholar] [CrossRef]
- Huang, S.-X.; Zhao, Z.-Y.; Weng, G.-H.; He, X.-Y.; Wu, C.-J.; Fu, C.-Y.; Sui, Z.-Y.; Ma, Y.-S.; Liu, T. Upregulation of miR-181a suppresses the formation of glioblastoma stem cells by targeting the Notch2 oncogene and correlates with good prognosis in patients with glioblastoma multiforme. Biochem. Biophys. Res. Commun. 2017, 486, 1129–1136. [Google Scholar] [CrossRef] [PubMed]
- Macharia, L.W.; Muriithi, W.; Heming, C.P.; Nyaga, D.K.; Aran, V.; Mureithi, M.W.; Ferrer, V.P.; Pane, A.; Filho, P.N.; Moura-Neto, V. The genotypic and phenotypic impact of hypoxia microenvironment on glioblastoma cell lines. BMC Cancer 2021, 21, 1248. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Ma, J.; Wang, X.; Peng, F.; Chen, X.; Zheng, B.; Wang, C.; Dai, Z.; Ai, J.; Zhao, S. Kaiso (ZBTB33) Downregulation by miRNA-181a Inhibits Cell Proliferation, Invasion, and the Epithelial–Mesenchymal Transition in Glioma Cells. Cell. Physiol. Biochem. 2018, 48, 947–958. [Google Scholar] [CrossRef] [PubMed]
- Wen, X.; Li, S.; Guo, M.; Liao, H.; Chen, Y.; Kuang, X.; Liao, X.; Ma, L.; Li, Q. miR-181a-5p inhibits the proliferation and invasion of drug-resistant glioblastoma cells by targeting F-box protein 11 expression. Oncol. Lett. 2020, 20, 235. [Google Scholar] [CrossRef]
- Schäfer, A.; Evers, L.; Meier, L.; Schlomann, U.; Bopp, M.H.A.; Dreizner, G.-L.; Lassmann, O.; Ben Bacha, A.; Benescu, A.-C.; Pojskic, M.; et al. The Metalloprotease-Disintegrin ADAM8 Alters the Tumor Suppressor miR-181a-5p Expression Profile in Glioblastoma Thereby Contributing to Its Aggressiveness. Front. Oncol. 2022, 12, 826273. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Chen, G.; Zhu, W.; Shi, D.; Lv, L.; Zhang, C.; Liu, P.; Hu, W. MicroRNA-181a sensitizes human malignant glioma U87MG cells to radiation by targeting Bcl-2. Oncol. Rep. 2010, 23, 997–1003. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Al-Zahrani, A.; Beylerli, O.; Sufianov, R.; Talybov, R.; Meshcheryakova, S.; Sufianova, G.; Gareev, I.; Sufianov, A. Circulating miRNAs as Diagnostic and Prognostic Biomarkers in High-Grade Gliomas. Front. Oncol. 2022, 12, 898537. [Google Scholar] [CrossRef]
- Han, S.; Liu, Y.; Cai, S.J.; Qian, M.; Ding, J.; Larion, M.; Gilbert, M.R.; Yang, C. IDH mutation in glioma: Molecular mechanisms and potential therapeutic targets. Br. J. Cancer 2020, 122, 1580–1589. [Google Scholar] [CrossRef]
- Sippl, C.; Schoeneberger, L.; Teping, F.; Schulz-Schaeffer, W.; Urbschat, S.; Ketter, R.; Oertel, J. Impact of miRNA-181a2 on the Clinical Course of IDH1 Wild Type Glioblastoma. Processes 2021, 9, 728. [Google Scholar] [CrossRef]
- Presnell, S.R.; Al-Attar, A.; Cichocki, F.; Miller, J.S.; Lutz, C.T. Human natural killer cell microRNA: Differential expression of MIR181A1B1 and MIR181A2B2 genes encoding identical mature microRNAs. Genes Immun. 2015, 16, 89–98. [Google Scholar] [CrossRef]
- Vaitkiene, P.; Pranckeviciene, A.; Stakaitis, R.; Steponaitis, G.; Tamasauskas, A.; Bunevicius, A. Association of miR-34a Expression with Quality of Life of Glioblastoma Patients: A Prospective Study. Cancers 2019, 11, 300. [Google Scholar] [CrossRef] [PubMed]
- Stakaitis, R.; Pranckeviciene, A.; Steponaitis, G.; Tamasauskas, A.; Bunevicius, A.; Vaitkiene, P. Unique Interplay Between Molecular miR-181b/d Biomarkers and Health Related Quality of Life Score in the Predictive Glioma Models. Int. J. Mol. Sci. 2020, 21, 7450. [Google Scholar] [CrossRef] [PubMed]
- Karnofsky, D.A.; Abelmann, W.H.; Craver, L.F.; Burchenal, J.H. The use of the nitrogen mustards in the palliative treatment of carcinoma. With particular reference to bronchogenic carcinoma. Cancer 1948, 1, 534–656. [Google Scholar] [CrossRef]
- Fayers, P.; Aaronson, N.K.; Bjordal, K.; Groenvold, M.; Curran, D.; Bottomley, A. EORTC QLQ-C30 Scoring Manual, 3rd ed.; European Organisation for Research and Treatment of Cancer: Brussels, Belgium, 2001. [Google Scholar]
- Osoba, D.; Aaronson, N.K.; Muller, M.; Sneeuw, K.; Hsu, M.-A.; Yung, W.K.A.; Brada, M.; Newlands, E. The development and psychometric validation of a brain cancer quality-of-life questionnaire for use in combination with general cancer-specific questionnaires. Qual. Life Res. 1996, 5, 139–150. [Google Scholar] [CrossRef] [PubMed]
- Kroenke, K.; Spitzer, R.L.; Williams, J.B. The PHQ-9: Validity of a brief depression severity measure. J. Gen. Intern. Med. 2001, 16, 606–613. [Google Scholar] [CrossRef] [PubMed]
- Pranckeviciene, A.; Bunevicius, A. Depression screening in patients with brain tumors: A review. CNS Oncol. 2015, 4, 71–78. [Google Scholar] [CrossRef]
- Rooney, A.G.; McNamara, S.; MacKinnon, M.; Fraser, M.; Rampling, R.; Carson, A.; Grant, R. Screening for major depressive disorder in adults with cerebral glioma: An initial validation of 3 self-report instruments. Neuro-Oncology 2012, 15, 122–129. [Google Scholar] [CrossRef]
- Bent, M.V.D.; Wefel, J.; Schiff, D.; Taphoorn, M.; Jaeckle, K.; Junck, L.; Armstrong, T.; Choucair, A.; Waldman, A.; Gorlia, T.; et al. Response assessment in neuro-oncology (a report of the RANO group): Assessment of outcome in trials of diffuse low-grade gliomas. Lancet Oncol. 2011, 12, 583–593. [Google Scholar] [CrossRef]
- Benedict, R.H.; Schretlen, D.; Groninger, L.; Brandt, J. Hopkins Verbal Learning Test—Revised: Normative Data and Analysis of Inter-Form and Test-Retest Reliability. Clin. Neuropsychol. 1998, 12, 43–55. [Google Scholar] [CrossRef]
- Reitan, R.M. Validity of the Trail Making Test as an Indicator of Organic Brain Damage. Percept. Mot. Ski. 1958, 8, 271–276. [Google Scholar] [CrossRef]
- Strauss, E.; Sherman, E.M.S.; Spreen, O. A Compendium of Neuropsychological Tests: Administration, Norms, and Commentary, 3rd ed.; Oxford University Press: New York, NY, USA, 2006. [Google Scholar]
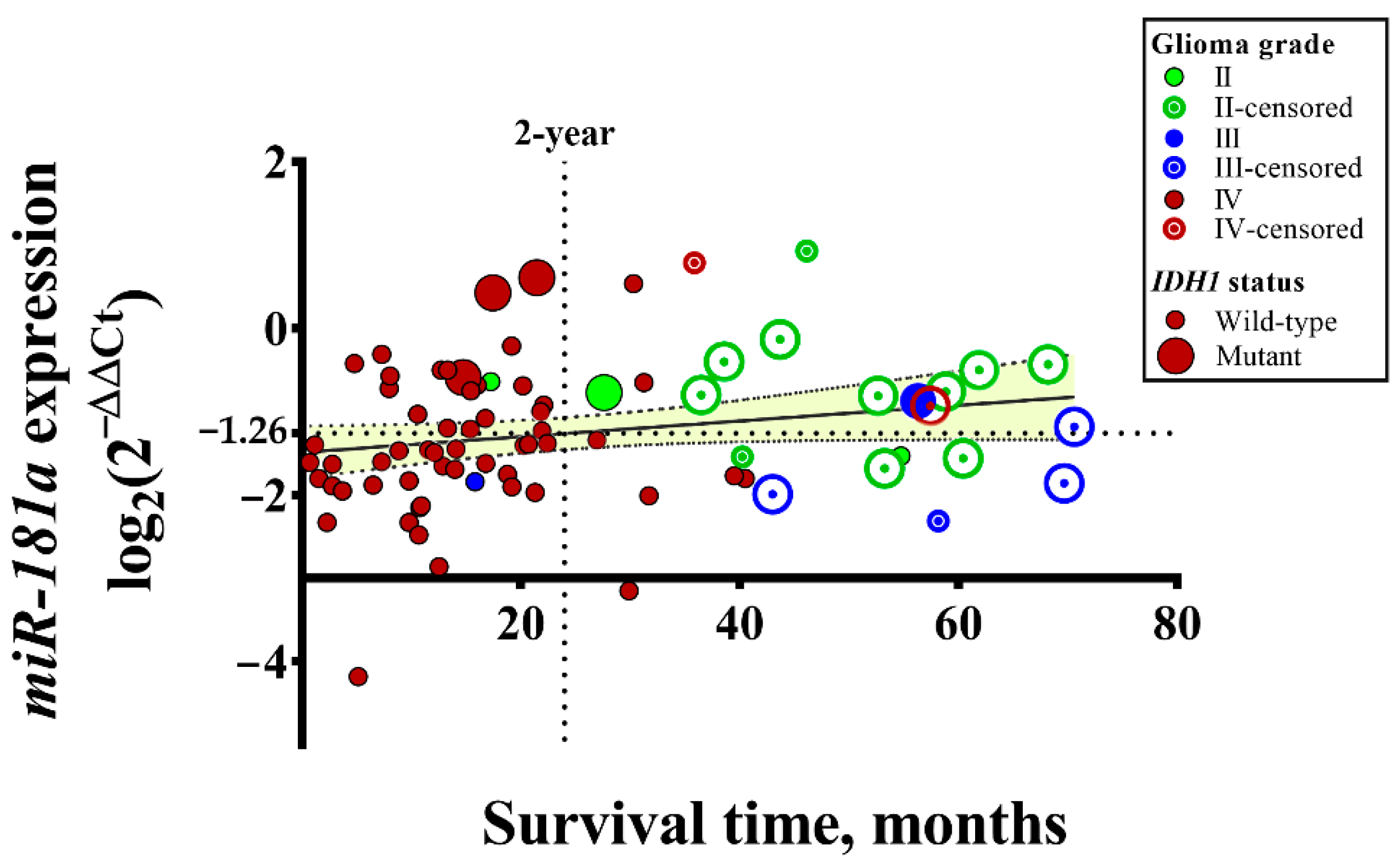


| Variables | Total No. | Expression of miR-181a | ||
|---|---|---|---|---|
| Low (%) | High (%) | p-Value | ||
| Gender | ||||
| Male | 43 | 20 (46.5) | 23 (53.5) | 0.15 |
| Female | 35 | 22 (62.9) | 13 (37.1) | |
| Age, year | ||||
| <54 | 39 | 15 (38.5) | 24 (61.5) | 0.006 |
| ≥54 | 39 | 27 (69.2) | 12 (30.8) | |
| Grade | ||||
| II | 14 | 4 (28.6) | 10 (71.4) | 0.036 |
| III–IV | 64 | 38 (59.4) | 26 (40.6) | |
| MGMT | ||||
| Unmet | 34 | 19 (55.9) | 15 (44.1) | 0.978 |
| Met | 36 | 20 (55.6) | 16 (44.4) | |
| IDH1 | ||||
| Wt | 57 | 36 (63.2) | 21 (36.8) | 0.002 |
| Mut | 18 | 4 (22.2) | 14 (77.8) | |
| Tumor location | ||||
| Right hemisphere | 37 | 23 (62.2) | 14 (37.8) | 0.25 |
| Left hemisphere | 37 | 18 (48.6) | 19 (54.4) | |
| Bilateral | 4 | 1 (25.0) | 3 (75.0) | |
| Variables | Univariate | Multivariate | ||
|---|---|---|---|---|
| HR (95% CI) | p-Value | HR (95% CI) | p-Value | |
| Gender | 0.732 (0.441–1.214) | 0.226 | N/A | |
| Female vs. Male | ||||
| Age, year | 3.856 (2.205–6.741) | <0.001 | 1.417 (0.778–2.581) | 0.254 |
| <54 vs. ≥54 | ||||
| Grade | 9.748 (3.029–31.373) | <0.001 | 2.921 (1.459–5.846) | 0.002 |
| II vs. III–IV | ||||
| MGMT | 0.712 (0.417–1.213) | 0.211 | N/A | |
| Unmeth vs. Meth | ||||
| IDH1 | 0.105 (0.041–0.272) | <0.001 | 0.314 (0.100–0.989) | 0.048 |
| Wt vs. Mut | ||||
| Expression of miR-181a | 0.584 (0.350–0.974) | 0.039 | 0.909 (0.521–1.585) | 0.737 |
| Low vs. High | ||||
| Variables | Total | Female | Male | GBM | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | r | p | N | r | p | N | r | p | N | r | p | |
| EORTC QLQ-C30 Quality of Life | 68 | 0.310 | 0.010 | 31 | 0.135 | 0.469 | 37 | 0.423 | 0.009 | 50 | 0.290 | 0.041 |
| EORTC QLQ-BN20 Tumor Related Symptoms | 68 | −0.209 | 0.088 | 31 | −0.005 | 0.978 | 37 | −0.311 | 0.061 | 50 | −0.223 | 0.120 |
| PHQ-9 Depression | 68 | −0.128 | 0.299 | 31 | 0.060 | 0.750 | 37 | −0.211 | 0.210 | 50 | −0.126 | 0.385 |
| KPS Functional status evaluated by a clinician | 70 | 0.237 | 0.049 | 33 | 0.041 | 0.822 | 37 | 0.291 | 0.080 | 54 | 0.260 | 0.057 |
| HVLT-R Cumulative learning | 68 | −0.175 | 0.155 | 32 | −0.102 | 0.580 | 36 | −0.194 | 0.256 | 50 | −0.245 | 0.087 |
| HVLT-R Delayed recall | 68 | −0.226 | 0.064 | 32 | −0.094 | 0.607 | 36 | −0.282 | 0.096 | 50 | −0.291 | 0.040 |
| TMT-A Psychomotor speed | 63 | 0.010 | 0.941 | 28 | −0.218 | 0.265 | 35 | 0.112 | 0.523 | 45 | 0.026 | 0.866 |
| TMT-B Executive functions | 63 | 0.079 | 0.538 | 28 | −0.240 | 0.218 | 35 | 0.224 | 0.197 | 45 | 0.031 | 0.838 |
| Verbal fluency | 68 | −0.134 | 0.276 | 32 | −0.213 | 0.241 | 36 | −0.078 | 0.652 | 50 | −0.167 | 0.245 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Valiulyte, I.; Pranckeviciene, A.; Bunevicius, A.; Tamasauskas, A.; Svitina, H.; Skrypkina, I.; Vaitkiene, P. Associations of miR-181a with Health-Related Quality of Life, Cognitive Functioning, and Clinical Data of Patients with Different Grade Glioma Tumors. Int. J. Mol. Sci. 2022, 23, 11149. https://doi.org/10.3390/ijms231911149
Valiulyte I, Pranckeviciene A, Bunevicius A, Tamasauskas A, Svitina H, Skrypkina I, Vaitkiene P. Associations of miR-181a with Health-Related Quality of Life, Cognitive Functioning, and Clinical Data of Patients with Different Grade Glioma Tumors. International Journal of Molecular Sciences. 2022; 23(19):11149. https://doi.org/10.3390/ijms231911149
Chicago/Turabian StyleValiulyte, Indre, Aiste Pranckeviciene, Adomas Bunevicius, Arimantas Tamasauskas, Hanna Svitina, Inessa Skrypkina, and Paulina Vaitkiene. 2022. "Associations of miR-181a with Health-Related Quality of Life, Cognitive Functioning, and Clinical Data of Patients with Different Grade Glioma Tumors" International Journal of Molecular Sciences 23, no. 19: 11149. https://doi.org/10.3390/ijms231911149
APA StyleValiulyte, I., Pranckeviciene, A., Bunevicius, A., Tamasauskas, A., Svitina, H., Skrypkina, I., & Vaitkiene, P. (2022). Associations of miR-181a with Health-Related Quality of Life, Cognitive Functioning, and Clinical Data of Patients with Different Grade Glioma Tumors. International Journal of Molecular Sciences, 23(19), 11149. https://doi.org/10.3390/ijms231911149








