Dosage Compensation in Drosophila: Its Canonical and Non-Canonical Mechanisms
Abstract
1. Introduction
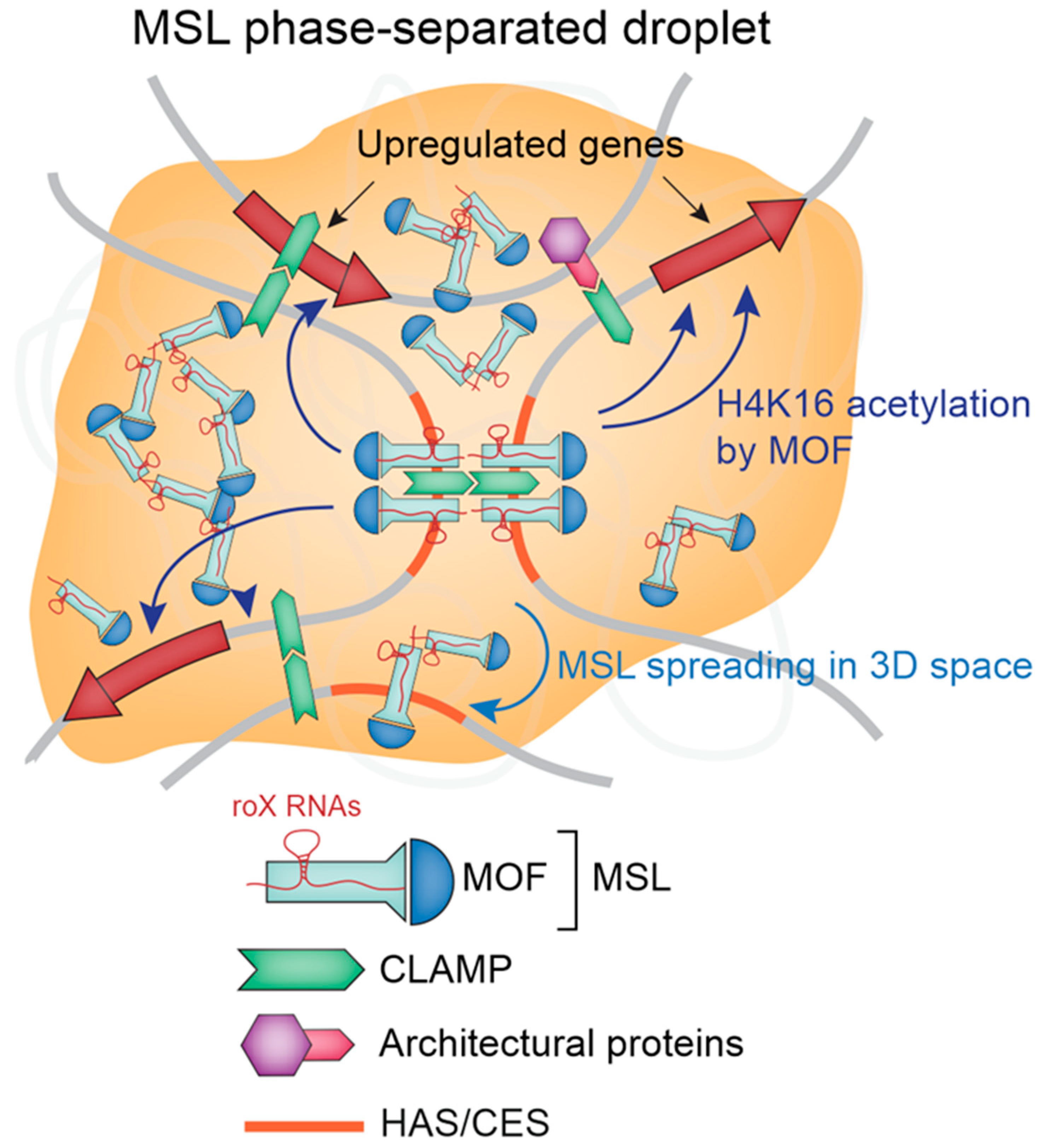
2. MSL Complex Mediates Canonical DC in Drosophila
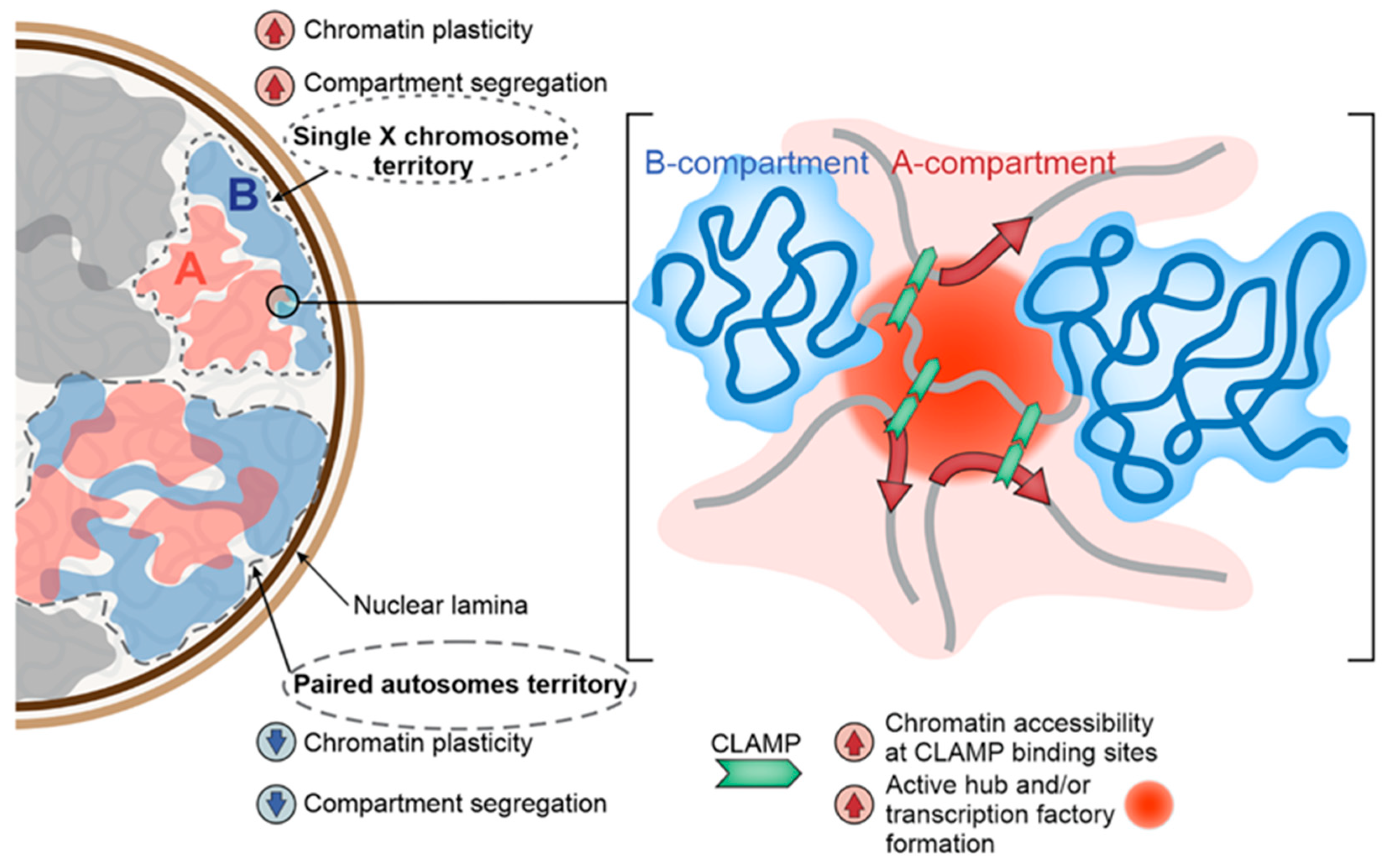
3. Non-Canonical Mechanisms of DC in Drosophila
4. Conclusions and Outlooks
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| 3D | Three-dimensional |
| CES | Chromatin entry sites |
| ChIP | Chromatin immunoprecipitation |
| CLAMP | Chromatin-linked adaptor for MSL proteins |
| DC | Dosage compensation |
| H3K36me3 | Histone H3 tri-methylated at Lys 36 |
| H4K16ac | Histone H4 acetylated at Lys 16 |
| HAS | High-affinity sites |
| LAD | Lamina-associated domain |
| MLE | Maleless |
| MOF | Males absent on the first |
| MRE | MSL recognition element |
| MSL | Male-specific lethal |
| Mtor | Megator |
| NSL | Non-specific lethal |
| PionX | Pioneering on the X |
| Pol II | RNA polymerase II |
| TopoII | Topoisomerase II |
References
- Lucchesi, J.C.; Rawls, J.M., Jr.; Maroni, G. Gene dosage compensation in metafemales (3X;2A) of Drosophila. Nature 1974, 248, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Birchler, J.A.; Hiebert, J.C.; Krietzman, M. Gene expression in adult metafemales of Drosophila melanogaster. Genetics 1989, 122, 869–879. [Google Scholar] [CrossRef] [PubMed]
- Lucchesi, J.C.; Rawls, J.M., Jr. Regulation of gene function: A comparison of enzyme activity levels in relation to gene dosage in diploids and triploids of Drosophila melanogaster. Biochem. Genet. 1973, 9, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Lucchesi, J.C.; Rawls, R.M., Jr. Regulation of gene function: A comparison of X-linked enzyme activity levels in normal and intersexual triploids of Drosophila melanogaster. Genetics 1973, 73, 459–464. [Google Scholar] [CrossRef]
- Maroni, G.; Plaut, W. Dosage compensation in Drosophila melanogaster triploids. II. Glucose-6-phosphate dehydrogenase activity. Genetics 1973, 74, 331–342. [Google Scholar] [CrossRef]
- Lucchesi, J.C. Gene dosage compensation and the evolution of sex chromosomes. Science 1978, 202, 711–716. [Google Scholar] [CrossRef]
- Gupta, V.; Parisi, M.; Sturgill, D.; Nuttall, R.; Doctolero, M.; Dudko, O.K.; Malley, J.D.; Eastman, P.S.; Oliver, B. Global analysis of X-chromosome dosage compensation. J. Biol. 2006, 5, 3. [Google Scholar] [CrossRef]
- Nguyen, D.K.; Disteche, C.M. Dosage compensation of the active X chromosome in mammals. Nat. Genet. 2006, 38, 47–53. [Google Scholar] [CrossRef]
- Deng, X.; Hiatt, J.B.; Nguyen, D.K.; Ercan, S.; Sturgill, D.; Hillier, L.W.; Schlesinger, F.; Davis, C.A.; Reinke, V.J.; Gingeras, T.R.; et al. Evidence for compensatory upregulation of expressed X-linked genes in mammals, Caenorhabditis elegans and Drosophila melanogaster. Nat. Genet. 2011, 43, 1179–1185. [Google Scholar] [CrossRef]
- Kharchenko, P.V.; Xi, R.; Park, P.J. Evidence for dosage compensation between the X chromosome and autosomes in mammals. Nat. Genet. 2011, 43, 1167–1169. [Google Scholar] [CrossRef]
- Sun, L.; Johnson, A.F.; Donohue, R.C.; Li, J.; Cheng, J.; Birchler, J.A. Dosage compensation and inverse effects in triple X metafemales of Drosophila. Proc. Natl. Acad. Sci. USA 2013, 110, 7383–7388. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, F.; Alekseyenko, A.A.; Park, P.J.; Kuroda, M.I. Transcriptional control of a whole chromosome: Emerging models for dosage compensation. Nat. Struct. Mol. Biol. 2014, 21, 118–125. [Google Scholar] [CrossRef] [PubMed]
- Ohno, S.; Kaplan, W.D.; Kinosita, R. Formation of the sex chromatin by a single X-chromosome in liver cells of Rattus norvegicus. Exp. Cell Res. 1959, 18, 415–418. [Google Scholar] [CrossRef]
- Lyon, M.F. Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature 1961, 190, 372–373. [Google Scholar] [CrossRef] [PubMed]
- Lyon, M.F. Sex chromatin and gene action in the mammalian X-chromosome. Am. J. Hum. Genet. 1962, 14, 135–148. [Google Scholar]
- Mukherjee, A.S.; Beermann, W. Synthesis of ribonucleic acid by the X-chromosomes of Drosophila melanogaster and the problem of dosage compensation. Nature 1965, 207, 785–786. [Google Scholar] [CrossRef] [PubMed]
- Hilfiker, A.; Yang, Y.; Hayes, D.H.; Beard, C.A.; Manning, J.E.; Lucchesi, J.C. Dosage compensation in Drosophila: The X-chromosomal binding of MSL-1 and MLE is dependent on Sxl activity. EMBO J. 1994, 13, 3542–3550. [Google Scholar] [CrossRef]
- Hilfiker, A.; Hilfiker-Kleiner, D.; Pannuti, A.; Lucchesi, J.C. mof, a putative acetyl transferase gene related to the Tip60 and MOZ human genes and to the SAS genes of yeast, is required for dosage compensation in Drosophila. EMBO J. 1997, 16, 2054–2060. [Google Scholar] [CrossRef]
- Akhtar, A.; Becker, P.B. Activation of transcription through histone H4 acetylation by MOF, an acetyltransferase essential for dosage compensation in Drosophila. Mol. Cell 2000, 5, 367–375. [Google Scholar] [CrossRef]
- Meller, V.H.; Gordadze, P.R.; Park, Y.; Chu, X.; Stuckenholz, C.; Kelley, R.L.; Kuroda, M.I. Ordered assembly of roX RNAs into MSL complexes on the dosage-compensated X chromosome in Drosophila. Curr. Biol. 2000, 10, 136–143. [Google Scholar] [CrossRef]
- Smith, E.R.; Pannuti, A.; Gu, W.; Steurnagel, A.; Cook, R.G.; Allis, C.D.; Lucchesi, J.C. The drosophila MSL complex acetylates histone H4 at lysine 16, a chromatin modification linked to dosage compensation. Mol. Cell. Biol. 2000, 20, 312–318. [Google Scholar] [CrossRef] [PubMed]
- Larschan, E.; Bishop, E.P.; Kharchenko, P.V.; Core, L.J.; Lis, J.T.; Park, P.J.; Kuroda, M.I. X chromosome dosage compensation via enhanced transcriptional elongation in Drosophila. Nature 2011, 471, 115–118. [Google Scholar] [CrossRef] [PubMed]
- Conrad, T.; Cavalli, F.M.; Holz, H.; Hallacli, E.; Kind, J.; Ilik, I.; Vaquerizas, J.M.; Luscombe, N.M.; Akhtar, A. The MOF chromobarrel domain controls genome-wide H4K16 acetylation and spreading of the MSL complex. Dev. Cell 2012, 22, 610–624. [Google Scholar] [CrossRef]
- Hamada, F.N.; Park, P.J.; Gordadze, P.R.; Kuroda, M.I. Global regulation of X chromosomal genes by the MSL complex in Drosophila melanogaster. Genes Dev. 2005, 19, 2289–2294. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Malone, J.H.; Powell, S.K.; Periwal, V.; Spana, E.; Macalpine, D.M.; Oliver, B. Expression in aneuploid Drosophila S2 cells. PLoS Biol. 2010, 8, e1000320. [Google Scholar] [CrossRef] [PubMed]
- Rastelli, L.; Kuroda, M.I. An analysis of maleless and histone H4 acetylation in Drosophila melanogaster spermatogenesis. Mech. Dev. 1998, 71, 107–117. [Google Scholar] [CrossRef]
- Meiklejohn, C.D.; Landeen, E.L.; Cook, J.M.; Kingan, S.B.; Presgraves, D.C. Sex chromosome-specific regulation in the Drosophila male germline but little evidence for chromosomal dosage compensation or meiotic inactivation. PLoS Biol. 2011, 9, e1001126. [Google Scholar] [CrossRef]
- Mahadevaraju, S.; Fear, J.M.; Akeju, M.; Galletta, B.J.; Pinheiro, M.M.L.S.; Avelino, C.C.; Cabral-de-Mello, D.C.; Conlon, K.; Dell’Orso, S.; Demere, Z.; et al. Dynamic sex chromosome expression in Drosophila male germ cells. Nat. Commun. 2021, 12, 892. [Google Scholar] [CrossRef]
- Witt, E.; Shao, Z.; Hu, C.; Krause, H.M.; Zhao, L. Single-cell RNA-sequencing reveals pre-meiotic X-chromosome dosage compensation in Drosophila testis. PLoS Genet. 2021, 17, e1009728. [Google Scholar] [CrossRef]
- Ilyin, A.A.; Kononkova, A.D.; Golova, A.V.; Shloma, V.V.; Olenkina, O.M.; Nenasheva, V.V.; Abramov, Y.A.; Kotov, A.A.; Maksimov, D.A.; Laktionov, P.P.; et al. Comparison of genome architecture at two stages of male germline cell differentiation in Drosophila. Nucleic Acids Res. 2022, 50, 3203–3225. [Google Scholar] [CrossRef]
- Lott, S.E.; Villalta, J.E.; Schroth, G.P.; Luo, S.; Tonkin, L.A.; Eisen, M.B. Noncanonical compensation of zygotic X transcription in early Drosophila melanogaster development revealed through single-embryo RNA-seq. PLoS Biol. 2011, 9, e1000590. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Birchler, J.A. Interaction study of the male specific lethal (MSL) complex and trans-acting dosage effects in metafemales of Drosophila melanogaster. Cytogenet. Genome Res. 2009, 124, 298–311. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Oliver, B. Dosage compensation goes global. Curr. Opin. Genet. Dev. 2007, 17, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Stenberg, P.; Larsson, J. Buffering and the evolution of chromosome-wide gene regulation. Chromosoma 2011, 120, 213–225. [Google Scholar] [CrossRef][Green Version]
- Birchler, J.A. Parallel universes for models of X chromosome dosage compensation in Drosophila: A review. Cytogenet. Genome Res. 2016, 148, 52–67. [Google Scholar] [CrossRef]
- Kuroda, M.I.; Hilfiker, A.; Lucchesi, J.C. Dosage compensation in Drosophila—A model for the coordinate regulation of transcription. Genetics 2016, 204, 435–450. [Google Scholar] [CrossRef]
- Samata, M.; Akhtar, A. Dosage compensation of the X chromosome: A complex epigenetic assignment involving chromatin regulators and long noncoding RNAs. Annu. Rev. Biochem. 2018, 87, 323–350. [Google Scholar] [CrossRef]
- Makki, R.; Meller, V.H. When down is up: Heterochromatin, nuclear organization and X upregulation. Cells 2021, 10, 3416. [Google Scholar] [CrossRef]
- Belote, J.M.; Lucchesi, J.C. Male-specific lethal mutations of Drosophila melanogaster. Genetics 1980, 96, 165–186. [Google Scholar] [CrossRef]
- Kuroda, M.I.; Kernan, M.J.; Kreber, R.; Ganetzky, B.; Baker, B.S. The maleless protein associates with the X chromosome to regulate dosage compensation in Drosophila. Cell 1991, 66, 935–947. [Google Scholar] [CrossRef]
- Gorman, M.; Kuroda, M.I.; Baker, B.S. Regulation of the sex-specific binding of the maleless dosage compensation protein to the male X chromosome in Drosophila. Cell 1993, 72, 39–49. [Google Scholar] [CrossRef]
- Palmer, M.J.; Mergner, V.A.; Richman, R.; Manning, J.E.; Kuroda, M.I.; Lucchesi, J.C. The male-specific lethal-one (msl-1) gene of Drosophila melanogaster encodes a novel protein that associates with the X chromosome in males. Genetics 1993, 134, 545–557. [Google Scholar] [CrossRef] [PubMed]
- Bashaw, G.J.; Baker, B.S. The msl-2 dosage compensation gene of Drosophila encodes a putative DNA-binding protein whose expression is sex specifically regulated by Sex-lethal. Development 1995, 121, 3245–3258. [Google Scholar] [CrossRef] [PubMed]
- Gorman, M.; Franke, A.; Baker, B.S. Molecular characterization of the male-specific lethal-3 gene and investigations of the regulation of dosage compensation in Drosophila. Development 1995, 121, 463–475. [Google Scholar] [CrossRef]
- Kelley, R.L.; Solovyeva, I.; Lyman, L.M.; Richman, R.; Solovyev, V.; Kuroda, M.I. Expression of msl-2 causes assembly of dosage compensation regulators on the X chromosomes and female lethality in Drosophila. Cell 1995, 81, 867–877. [Google Scholar] [CrossRef]
- Zhou, S.; Yang, Y.; Scott, M.J.; Pannuti, A.; Fehr, K.C.; Eisen, A.; Koonin, E.V.; Fouts, D.L.; Wrightsman, R.; Manning, J.E.; et al. Male-specific lethal 2, a dosage compensation gene of Drosophila, undergoes sex-specific regulation and encodes a protein with a RING finger and a metallothionein-like cysteine cluster. EMBO J. 1995, 14, 2884–2895. [Google Scholar] [CrossRef]
- Amrein, H.; Axel, R. Genes expressed in neurons of adult male Drosophila. Cell 1997, 88, 459–469. [Google Scholar] [CrossRef]
- Meller, V.H.; Wu, K.H.; Roman, G.; Kuroda, M.I.; Davis, R.L. roX1 RNA paints the X chromosome of male Drosophila and is regulated by the dosage compensation system. Cell 1997, 88, 445–457. [Google Scholar] [CrossRef]
- Meller, V.H.; Rattner, B.P. The roX genes encode redundant male-specific lethal transcripts required for targeting of the MSL complex. EMBO J. 2002, 21, 1084–1091. [Google Scholar] [CrossRef]
- Mendjan, S.; Taipale, M.; Kind, J.; Holz, H.; Gebhardt, P.; Schelder, M.; Vermeulen, M.; Buscaino, A.; Duncan, K.; Mueller, J.; et al. Nuclear pore components are involved in the transcriptional regulation of dosage compensation in Drosophila. Mol. Cell 2006, 21, 811–823. [Google Scholar] [CrossRef]
- Jin, Y.; Wang, Y.; Walker, D.L.; Dong, H.; Conley, C.; Johansen, J.; Johansen, K.M. JIL-1: A novel chromosomal tandem kinase implicated in transcriptional regulation in Drosophila. Mol. Cell 1999, 4, 129–135. [Google Scholar] [CrossRef]
- Jin, Y.; Wang, Y.; Johansen, J.; Johansen, K.M. JIL-1, a chromosomal kinase implicated in regulation of chromatin structure, associates with the male specific lethal (MSL) dosage compensation complex. J. Cell Biol. 2000, 149, 1005–1010. [Google Scholar] [CrossRef] [PubMed]
- Cugusi, S.; Ramos, E.; Ling, H.; Yokoyama, R.; Luk, K.M.; Lucchesi, J.C. Topoisomerase II plays a role in dosage compensation in Drosophila. Transcription 2013, 4, 238–250. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Vaquerizas, J.M.; Suyama, R.; Kind, J.; Miura, K.; Luscombe, N.M.; Akhtar, A. Nuclear pore proteins nup153 and megator define transcriptionally active regions in the Drosophila genome. PLoS Genet. 2010, 6, e1000846. [Google Scholar] [CrossRef] [PubMed]
- Albig, C.; Tikhonova, E.; Krause, S.; Maksimenko, O.; Regnard, C.; Becker, P.B. Factor cooperation for chromosome discrimination in Drosophila. Nucleic Acids Res. 2019, 47, 1706–1724. [Google Scholar] [CrossRef] [PubMed]
- Aleman, J.R.; Kuhn, T.M.; Pascual-Garcia, P.; Gospocic, J.; Lan, Y.; Bonasio, R.; Little, S.C.; Capelson, M. Correct dosage of X chromosome transcription is controlled by a nuclear pore component. Cell Rep. 2021, 35, 109236. [Google Scholar] [CrossRef] [PubMed]
- Lindehell, H.; Kim, M.; Larsson, J. Proximity ligation assays of protein and RNA interactions in the male-specific lethal complex on Drosophila melanogaster polytene chromosomes. Chromosoma 2015, 124, 385–395. [Google Scholar] [CrossRef]
- Raja, S.J.; Charapitsa, I.; Conrad, T.; Vaquerizas, J.M.; Gebhardt, P.; Holz, H.; Kadlec, J.; Fraterman, S.; Luscombe, N.M.; Akhtar, A. The nonspecific lethal complex is a transcriptional regulator in Drosophila. Mol. Cell 2010, 38, 827–841. [Google Scholar] [CrossRef]
- Lam, K.C.; Mühlpfordt, F.; Vaquerizas, J.M.; Raja, S.J.; Holz, H.; Luscombe, N.M.; Manke, T.; Akhtar, A. The NSL complex regulates housekeeping genes in Drosophila. PLoS Genet. 2012, 8, e1002736. [Google Scholar] [CrossRef]
- Franke, A.; Baker, B.S. The rox1 and rox2 RNAs are essential components of the compensasome, which mediates dosage compensation in Drosophila. Mol. Cell 1999, 4, 117–122. [Google Scholar] [CrossRef]
- Kim, M.; Faucillion, M.L.; Larsson, J. RNA-on-X 1 and 2 in Drosophila melanogaster fulfill separate functions in dosage compensation. PLoS Genet. 2018, 14, e1007842. [Google Scholar] [CrossRef] [PubMed]
- Copps, K.; Richman, R.; Lyman, L.M.; Chang, K.A.; Rampersad-Ammons, J.; Kuroda, M.I. Complex formation by the Drosophila MSL proteins: Role of the MSL2 RING finger in protein complex assembly. EMBO J. 1998, 17, 5409–5417. [Google Scholar] [CrossRef] [PubMed]
- Scott, M.J.; Pan, L.L.; Cleland, S.B.; Knox, A.L.; Heinrich, J. MSL1 plays a central role in assembly of the MSL complex, essential for dosage compensation in Drosophila. EMBO J. 2000, 19, 144–155. [Google Scholar] [CrossRef]
- Kadlec, J.; Hallacli, E.; Lipp, M.; Holz, H.; Sanchez-Weatherby, J.; Cusack, S.; Akhtar, A. Structural basis for MOF and MSL3 recruitment into the dosage compensation complex by MSL1. Nat. Struct. Mol. Biol. 2011, 18, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Larschan, E.; Alekseyenko, A.A.; Gortchakov, A.A.; Peng, S.; Li, B.; Yang, P.; Workman, J.L.; Park, P.J.; Kuroda, M.I. MSL complex is attracted to genes marked by H3K36 trimethylation using a sequence-independent mechanism. Mol. Cell 2007, 28, 121–133. [Google Scholar] [CrossRef]
- Bell, O.; Conrad, T.; Kind, J.; Wirbelauer, C.; Akhtar, A.; Schübeler, D. Transcription-coupled methylation of histone H3 at lysine 36 regulates dosage compensation by enhancing recruitment of the MSL complex in Drosophila melanogaster. Mol. Cell. Biol. 2008, 28, 3401–3409. [Google Scholar] [CrossRef]
- Sural, T.H.; Peng, S.; Li, B.; Workman, J.L.; Park, P.J.; Kuroda, M.I. The MSL3 chromodomain directs a key targeting step for dosage compensation of the Drosophila melanogaster X chromosome. Nat. Struct. Mol. Biol. 2008, 15, 1318–1325. [Google Scholar] [CrossRef]
- Alekseyenko, A.A.; Larschan, E.; Lai, W.R.; Park, P.J.; Kuroda, M.I. High-resolution ChIP-chip analysis reveals that the Drosophila MSL complex selectively identifies active genes on the male X chromosome. Genes Dev. 2006, 20, 848–857. [Google Scholar] [CrossRef]
- Gilfillan, G.D.; Straub, T.; de Wit, E.; Greil, F.; Lamm, R.; van Steensel, B.; Becker, P.B. Chromosome-wide gene-specific targeting of the Drosophila dosage compensation complex. Genes Dev. 2006, 20, 858–870. [Google Scholar] [CrossRef]
- Lee, C.G.; Chang, K.A.; Kuroda, M.I.; Hurwitz, J. The NTPase/helicase activities of Drosophila maleless, an essential factor in dosage compensation. EMBO J. 1997, 16, 2671–2681. [Google Scholar] [CrossRef]
- Gu, W.; Wei, X.; Pannuti, A.; Lucchesi, J.C. Targeting the chromatin-remodeling MSL complex of Drosophila to its sites of action on the X chromosome requires both acetyl transferase and ATPase activities. EMBO J. 2000, 19, 5202–5211. [Google Scholar] [CrossRef] [PubMed]
- Fauth, T.; Müller-Planitz, F.; König, C.; Straub, T.; Becker, P.B. The DNA binding CXC domain of MSL2 is required for faithful targeting the Dosage Compensation Complex to the X chromosome. Nucleic Acids Res. 2010, 38, 3209–3221. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Villa, R.; Wang, J.; Feng, Y.; Wang, J.; Becker, P.B.; Ye, K. Structural basis of X chromosome DNA recognition by the MSL2 CXC domain during Drosophila dosage compensation. Genes Dev. 2014, 28, 2652–2662. [Google Scholar] [CrossRef] [PubMed]
- Villa, R.; Forné, I.; Müller, M.; Imhof, A.; Straub, T.; Becker, P.B. MSL2 combines sensor and effector functions in homeostatic control of the Drosophila dosage compensation machinery. Mol. Cell 2012, 48, 647–654. [Google Scholar] [CrossRef]
- Kelley, R.L.; Meller, V.H.; Gordadze, P.R.; Roman, G.; Davis, R.L.; Kuroda, M.I. Epigenetic spreading of the Drosophila dosage compensation complex from roX RNA genes into flanking chromatin. Cell 1999, 98, 513–522. [Google Scholar] [CrossRef]
- Alekseyenko, A.A.; Peng, S.; Larschan, E.; Gorchakov, A.A.; Lee, O.K.; Kharchenko, P.; McGrath, S.D.; Wang, C.I.; Mardis, E.R.; Park, P.J.; et al. A sequence motif within chromatin entry sites directs MSL establishment on the Drosophila X chromosome. Cell 2008, 134, 599–609. [Google Scholar] [CrossRef]
- Straub, T.; Grimaud, C.; Gilfillan, G.D.; Mitterweger, A.; Becker, P.B. The chromosomal high-affinity binding sites for the Drosophila dosage compensation complex. PLoS Genet. 2008, 4, e1000302. [Google Scholar] [CrossRef]
- Lindehell, H.; Glotov, A.; Dorafshan, E.; Schwartz, Y.B.; Larsson, J. The role of H3K36 methylation and associated methyltransferases in chromosome-specific gene regulation. Sci. Adv. 2021, 7, eabh4390. [Google Scholar] [CrossRef]
- Lyman, L.M.; Copps, K.; Rastelli, L.; Kelley, R.L.; Kuroda, M.I. Drosophila male-specific lethal-2 protein: Structure/function analysis and dependence on MSL-1 for chromosome association. Genetics 1997, 147, 1743–1753. [Google Scholar] [CrossRef]
- Li, F.; Parry, D.A.; Scott, M.J. The amino-terminal region of Drosophila MSL1 contains basic, glycine-rich, and leucine zipper-like motifs that promote X chromosome binding, self-association, and MSL2 binding, respectively. Mol. Cell. Biol. 2005, 25, 8913–8924. [Google Scholar] [CrossRef]
- Hallacli, E.; Lipp, M.; Georgiev, P.; Spielman, C.; Cusack, S.; Akhtar, A.; Kadlec, J. Msl1-mediated dimerization of the dosage compensation complex is essential for male X-chromosome regulation in Drosophila. Mol. Cell 2012, 48, 587–600. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo, M.L.; Kim, M.; Philip, P.; Allgardsson, A.; Stenberg, P.; Larsson, J. Non-coding roX RNAs prevent the binding of the MSL-complex to heterochromatic regions. PLoS Genet. 2014, 10, e1004865. [Google Scholar] [CrossRef] [PubMed]
- Alekseyenko, A.A.; Ho, J.W.; Peng, S.; Gelbart, M.; Tolstorukov, M.Y.; Plachetka, A.; Kharchenko, P.V.; Jung, Y.L.; Gorchakov, A.A.; Larschan, E.; et al. Sequence-specific targeting of dosage compensation in Drosophila favors an active chromatin context. PLoS Genet. 2012, 8, e1002646. [Google Scholar] [CrossRef] [PubMed]
- Villa, R.; Schauer, T.; Smialowski, P.; Straub, T.; Becker, P.B. PionX sites mark the X chromosome for dosage compensation. Nature 2016, 537, 244–248. [Google Scholar] [CrossRef] [PubMed]
- Larschan, E.; Soruco, M.M.; Lee, O.K.; Peng, S.; Bishop, E.; Chery, J.; Goebel, K.; Feng, J.; Park, P.J.; Kuroda, M.I. Identification of chromatin-associated regulators of MSL complex targeting in Drosophila dosage compensation. PLoS Genet. 2012, 8, e1002830. [Google Scholar] [CrossRef]
- Soruco, M.M.; Chery, J.; Bishop, E.P.; Siggers, T.; Tolstorukov, M.Y.; Leydon, A.R.; Sugden, A.U.; Goebel, K.; Feng, J.; Xia, P.; et al. The CLAMP protein links the MSL complex to the X chromosome during Drosophila dosage compensation. Genes Dev. 2013, 27, 1551–1556. [Google Scholar] [CrossRef]
- Kuzu, G.; Kaye, E.G.; Chery, J.; Siggers, T.; Yang, L.; Dobson, J.R.; Boor, S.; Bliss, J.; Liu, W.; Jogl, G.; et al. Expansion of GA dinucleotide repeats increases the density of CLAMP binding sites on the X-chromosome to promote Drosophila dosage compensation. PLoS Genet. 2016, 12, e1006120. [Google Scholar] [CrossRef]
- Tikhonova, E.; Fedotova, A.; Bonchuk, A.; Mogila, V.; Larschan, E.N.; Georgiev, P.; Maksimenko, O. The simultaneous interaction of MSL2 with CLAMP and DNA provides redundancy in the initiation of dosage compensation in Drosophila males. Development 2019, 146, dev179663. [Google Scholar] [CrossRef]
- Urban, J.; Kuzu, G.; Bowman, S.; Scruggs, B.; Henriques, T.; Kingston, R.; Adelman, K.; Tolstorukov, M.; Larschan, E. Enhanced chromatin accessibility of the dosage compensated Drosophila male X-chromosome requires the CLAMP zinc finger protein. PLoS ONE 2017, 12, e0186855. [Google Scholar] [CrossRef]
- Park, Y.; Kelley, R.L.; Oh, H.; Kuroda, M.I.; Meller, V.H. Extent of chromatin spreading determined by roX RNA recruitment of MSL proteins. Science 2002, 298, 1620–1623. [Google Scholar] [CrossRef]
- Ilik, I.A.; Quinn, J.J.; Georgiev, P.; Tavares-Cadete, F.; Maticzka, D.; Toscano, S.; Wan, Y.; Spitale, R.C.; Luscombe, N.; Backofen, R.; et al. Tandem stem-loops in roX RNAs act together to mediate X chromosome dosage compensation in Drosophila. Mol. Cell 2013, 51, 156–173. [Google Scholar] [CrossRef] [PubMed]
- Maenner, S.; Müller, M.; Fröhlich, J.; Langer, D.; Becker, P.B. ATP-dependent roX RNA remodeling by the helicase maleless enables specific association of MSL proteins. Mol. Cell 2013, 51, 174–184. [Google Scholar] [CrossRef] [PubMed]
- Ilik, I.A.; Maticzka, D.; Georgiev, P.; Gutierrez, N.M.; Backofen, R.; Akhtar, A. A mutually exclusive stem-loop arrangement in roX2 RNA is essential for X-chromosome regulation in Drosophila. Genes Dev. 2017, 31, 1973–1987. [Google Scholar] [CrossRef] [PubMed]
- Lv, M.; Yao, Y.; Li, F.; Xu, L.; Yang, L.; Gong, Q.; Xu, Y.Z.; Shi, Y.; Fan, Y.J.; Tang, Y. Structural insights reveal the specific recognition of roX RNA by the dsRNA-binding domains of the RNA helicase MLE and its indispensable role in dosage compensation in Drosophila. Nucleic Acids Res. 2019, 47, 3142–3157. [Google Scholar] [CrossRef] [PubMed]
- Müller, M.; Schauer, T.; Krause, S.; Villa, R.; Thomae, A.W.; Becker, P.B. Two-step mechanism for selective incorporation of lncRNA into a chromatin modifier. Nucleic Acids Res. 2020, 48, 7483–7501. [Google Scholar] [CrossRef] [PubMed]
- Valsecchi, C.I.K.; Basilicata, M.F.; Georgiev, P.; Gaub, A.; Seyfferth, J.; Kulkarni, T.; Panhale, A.; Semplicio, G.; Manjunath, V.; Holz, H.; et al. RNA nucleation by MSL2 induces selective X chromosome compartmentalization. Nature 2021, 589, 137–142. [Google Scholar] [CrossRef]
- Tikhonova, E.; Mariasina, S.; Efimov, S.; Polshakov, V.; Maksimenko, O.; Georgiev, P.; Bonchuk, A. Structural basis for interaction between CLAMP and MSL2 proteins involved in the specific recruitment of the dosage compensation complex in Drosophila. Nucleic Acids Res. 2022, 50, 6521–6531. [Google Scholar] [CrossRef]
- Villa, R.; Jagtap, P.K.A.; Thomae, A.W.; Campos Sparr, A.; Forné, I.; Hennig, J.; Straub, T.; Becker, P.B. Divergent evolution toward sex chromosome-specific gene regulation in Drosophila. Genes Dev. 2021, 35, 1055–1070. [Google Scholar] [CrossRef]
- Ramírez, F.; Lingg, T.; Toscano, S.; Lam, K.C.; Georgiev, P.; Chung, H.R.; Lajoie, B.R.; de Wit, E.; Zhan, Y.; de Laat, W.; et al. High-affinity sites form an interaction network to facilitate spreading of the MSL complex across the X chromosome in Drosophila. Mol. Cell 2015, 60, 146–162. [Google Scholar] [CrossRef]
- Schauer, T.; Ghavi-Helm, Y.; Sexton, T.; Albig, C.; Regnard, C.; Cavalli, G.; Furlong, E.E.; Becker, P.B. Chromosome topology guides the Drosophila Dosage Compensation Complex for target gene activation. EMBO Rep. 2017, 18, 1854–1868. [Google Scholar] [CrossRef]
- Pal, K.; Forcato, M.; Jost, D.; Sexton, T.; Vaillant, C.; Salviato, E.; Mazza, E.M.C.; Lugli, E.; Cavalli, G.; Ferrari, F. Global chromatin conformation differences in the Drosophila dosage compensated chromosome X. Nat. Commun. 2019, 10, 5355. [Google Scholar] [CrossRef] [PubMed]
- Jordan, W., 3rd; Larschan, E. The zinc finger protein CLAMP promotes long-range chromatin interactions that mediate dosage compensation of the Drosophila male X-chromosome. Epigenetics Chromatin 2021, 14, 29. [Google Scholar] [CrossRef] [PubMed]
- Tikhonova, E.; Mariasina, S.; Arkova, O.; Maksimenko, O.; Georgiev, P.; Bonchuk, A. Dimerization activity of a disordered N-terminal domain from Drosophila CLAMP protein. Int. J. Mol. Sci. 2022, 23, 3862. [Google Scholar] [CrossRef] [PubMed]
- Bag, I.; Dale, R.K.; Palmer, C.; Lei, E.P. The zinc-finger protein CLAMP promotes gypsy chromatin insulator function in Drosophila. J. Cell Sci. 2019, 132, jcs226092. [Google Scholar] [CrossRef] [PubMed]
- Turner, B.M.; Birley, A.J.; Lavender, J. Histone H4 isoforms acetylated at specific lysine residues define individual chromosomes and chromatin domains in Drosophila polytene nuclei. Cell 1992, 69, 375–384. [Google Scholar] [CrossRef]
- Bone, J.R.; Lavender, J.; Richman, R.; Palmer, M.J.; Turner, B.M.; Kuroda, M.I. Acetylated histone H4 on the male X chromosome is associated with dosage compensation in Drosophila. Genes Dev. 1994, 8, 96–104. [Google Scholar] [CrossRef]
- Kind, J.; Vaquerizas, J.M.; Gebhardt, P.; Gentzel, M.; Luscombe, N.M.; Bertone, P.; Akhtar, A. Genome-wide analysis reveals MOF as a key regulator of dosage compensation and gene expression in Drosophila. Cell 2008, 133, 813–828. [Google Scholar] [CrossRef]
- Gelbart, M.E.; Larschan, E.; Peng, S.; Park, P.J.; Kuroda, M.I. Drosophila MSL complex globally acetylates H4K16 on the male X chromosome for dosage compensation. Nat. Struct. Mol. Biol. 2009, 16, 825–832. [Google Scholar] [CrossRef]
- Barnes, C.E.; English, D.M.; Cowley, S.M. Acetylation & Co: An expanding repertoire of histone acylations regulates chromatin and transcription. Essays Biochem. 2019, 63, 97–107. [Google Scholar]
- Shogren-Knaak, M.; Ishii, H.; Sun, J.M.; Pazin, M.J.; Davie, J.R.; Peterson, C.L. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 2006, 311, 844–847. [Google Scholar] [CrossRef]
- Allahverdi, A.; Yang, R.; Korolev, N.; Fan, Y.; Davey, C.A.; Liu, C.F.; Nordenskiöld, L. The effects of histone H4 tail acetylations on cation-induced chromatin folding and self-association. Nucleic Acids Res. 2011, 39, 1680–1691. [Google Scholar] [CrossRef] [PubMed]
- Smith, E.R.; Allis, C.D.; Lucchesi, J.C. Linking global histone acetylation to the transcription enhancement of X-chromosomal genes in Drosophila males. J. Biol. Chem. 2001, 276, 31483–31486. [Google Scholar] [CrossRef] [PubMed]
- Gu, W.; Szauter, P.; Lucchesi, J.C. Targeting of MOF, a putative histone acetyl transferase, to the X chromosome of Drosophila melanogaster. Dev. Genet. 1998, 22, 56–64. [Google Scholar] [CrossRef]
- Copur, Ö.; Gorchakov, A.; Finkl, K.; Kuroda, M.I.; Müller, J. Sex-specific phenotypes of histone H4 point mutants establish dosage compensation as the critical function of H4K16 acetylation in Drosophila. Proc. Natl. Acad. Sci. USA 2018, 115, 13336–13341. [Google Scholar] [CrossRef] [PubMed]
- Samata, M.; Alexiadis, A.; Richard, G.; Georgiev, P.; Nuebler, J.; Kulkarni, T.; Renschler, G.; Basilicata, M.F.; Zenk, F.L.; Shvedunova, M.; et al. Intergenerationally maintained histone H4 lysine 16 acetylation is instructive for future gene activation. Cell 2020, 182, 127–144.e23. [Google Scholar] [CrossRef]
- Conrad, T.; Cavalli, F.M.; Vaquerizas, J.M.; Luscombe, N.M.; Akhtar, A. Drosophila dosage compensation involves enhanced Pol II recruitment to male X-linked promoters. Science 2012, 337, 742–746. [Google Scholar] [CrossRef]
- Vaquerizas, J.M.; Cavalli, F.M.; Conrad, T.; Akhtar, A.; Luscombe, N.M. Response to comments on "Drosophila dosage compensation involves enhanced Pol II recruitment to male X-linked promoters". Science 2013, 340, 273. [Google Scholar] [CrossRef]
- Ferrari, F.; Plachetka, A.; Alekseyenko, A.A.; Jung, Y.L.; Ozsolak, F.; Kharchenko, P.V.; Park, P.J.; Kuroda, M.I. “Jump start and gain” model for dosage compensation in Drosophila based on direct sequencing of nascent transcripts. Cell Rep. 2013, 5, 629–636. [Google Scholar] [CrossRef]
- Shi, Z.; Lim, C.; Tran, V.; Cui, K.; Zhao, K.; Chen, X. Single-cyst transcriptome analysis of Drosophila male germline stem cell lineage. Development 2020, 147, dev184259. [Google Scholar] [CrossRef]
- Stenberg, P.; Lundberg, L.E.; Johansson, A.M.; Rydén, P.; Svensson, M.J.; Larsson, J. Buffering of segmental and chromosomal aneuploidies in Drosophila melanogaster. PLoS Genet. 2009, 5, e1000465. [Google Scholar] [CrossRef]
- Kind, J.; Pagie, L.; Ortabozkoyun, H.; Boyle, S.; de Vries, S.S.; Janssen, H.; Amendola, M.; Nolen, L.D.; Bickmore, W.A.; van Steensel, B. Single-cell dynamics of genome-nuclear lamina interactions. Cell 2013, 153, 178–192. [Google Scholar] [CrossRef] [PubMed]
- Kind, J.; Pagie, L.; de Vries, S.S.; Nahidiazar, L.; Dey, S.S.; Bienko, M.; Zhan, Y.; Lajoie, B.; de Graaf, C.A.; Amendola, M.; et al. Genome-wide maps of nuclear lamina interactions in single human cells. Cell 2015, 163, 134–147. [Google Scholar] [CrossRef] [PubMed]
- Ulianov, S.V.; Doronin, S.A.; Khrameeva, E.E.; Kos, P.I.; Luzhin, A.V.; Starikov, S.S.; Galitsyna, A.A.; Nenasheva, V.V.; Ilyin, A.A.; Flyamer, I.M.; et al. Nuclear lamina integrity is required for proper spatial organization of chromatin in Drosophila. Nat. Commun. 2019, 10, 1176. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shevelyov, Y.Y.; Ulianov, S.V.; Gelfand, M.S.; Belyakin, S.N.; Razin, S.V. Dosage Compensation in Drosophila: Its Canonical and Non-Canonical Mechanisms. Int. J. Mol. Sci. 2022, 23, 10976. https://doi.org/10.3390/ijms231810976
Shevelyov YY, Ulianov SV, Gelfand MS, Belyakin SN, Razin SV. Dosage Compensation in Drosophila: Its Canonical and Non-Canonical Mechanisms. International Journal of Molecular Sciences. 2022; 23(18):10976. https://doi.org/10.3390/ijms231810976
Chicago/Turabian StyleShevelyov, Yuri Y., Sergey V. Ulianov, Mikhail S. Gelfand, Stepan N. Belyakin, and Sergey V. Razin. 2022. "Dosage Compensation in Drosophila: Its Canonical and Non-Canonical Mechanisms" International Journal of Molecular Sciences 23, no. 18: 10976. https://doi.org/10.3390/ijms231810976
APA StyleShevelyov, Y. Y., Ulianov, S. V., Gelfand, M. S., Belyakin, S. N., & Razin, S. V. (2022). Dosage Compensation in Drosophila: Its Canonical and Non-Canonical Mechanisms. International Journal of Molecular Sciences, 23(18), 10976. https://doi.org/10.3390/ijms231810976







