Chromosomal Rearrangements and Chromothripsis: The Alternative End Generation Model
Abstract
1. Introduction
2. Chromothripsis: Molecular Characteristics
3. Chromothripsis: Current Models and Their Limitations
3.1. Breakage Fusion Bridge Cycle
3.2. Aborted Apoptosis
3.3. Pulverization of DNA by Exogenous Insults
3.4. Micronuclei as Reaction Chamber of Chromothripsis
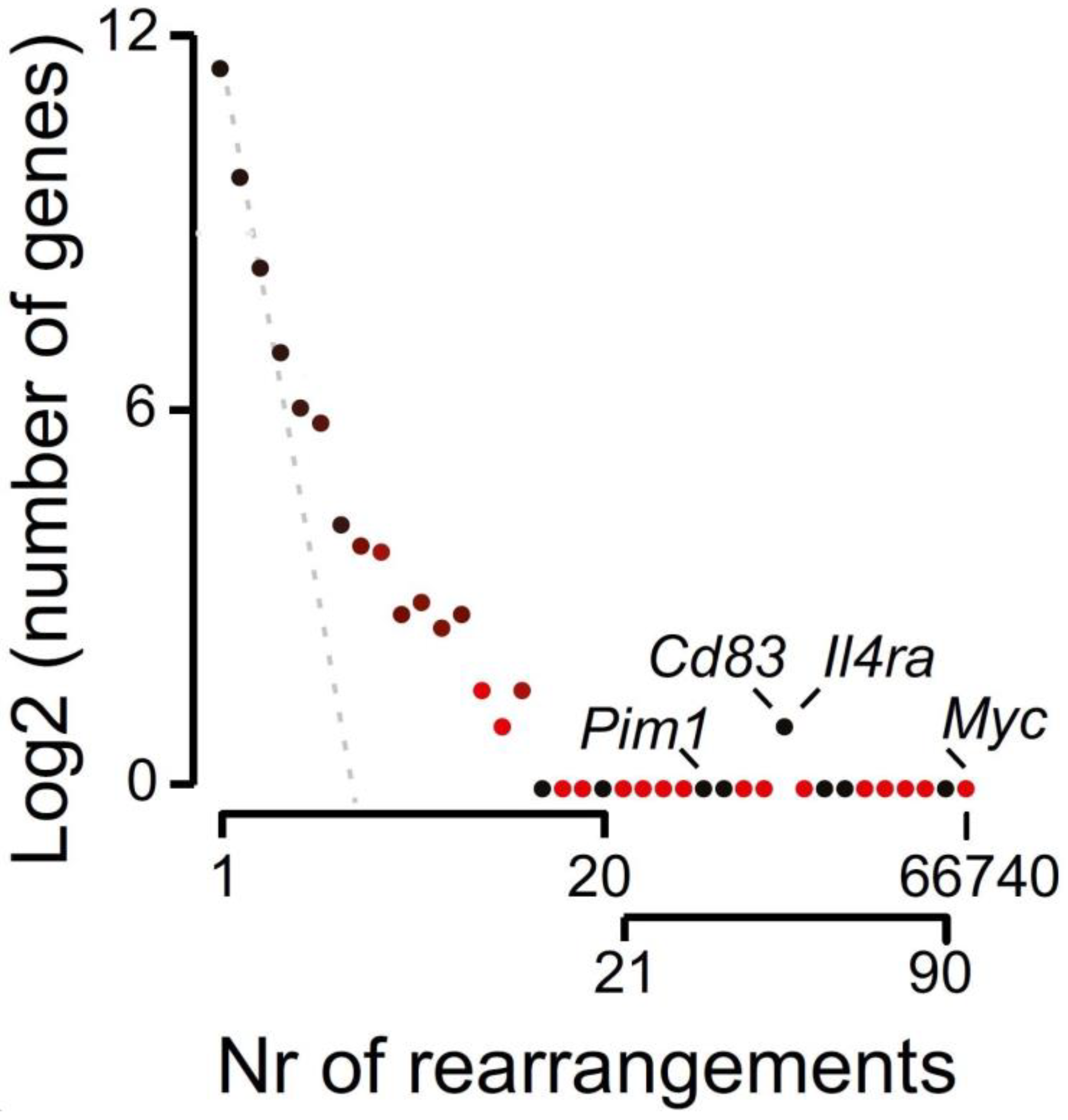
3.5. A Single DSB as Potential Inducer of Chromosomal Rearrangements
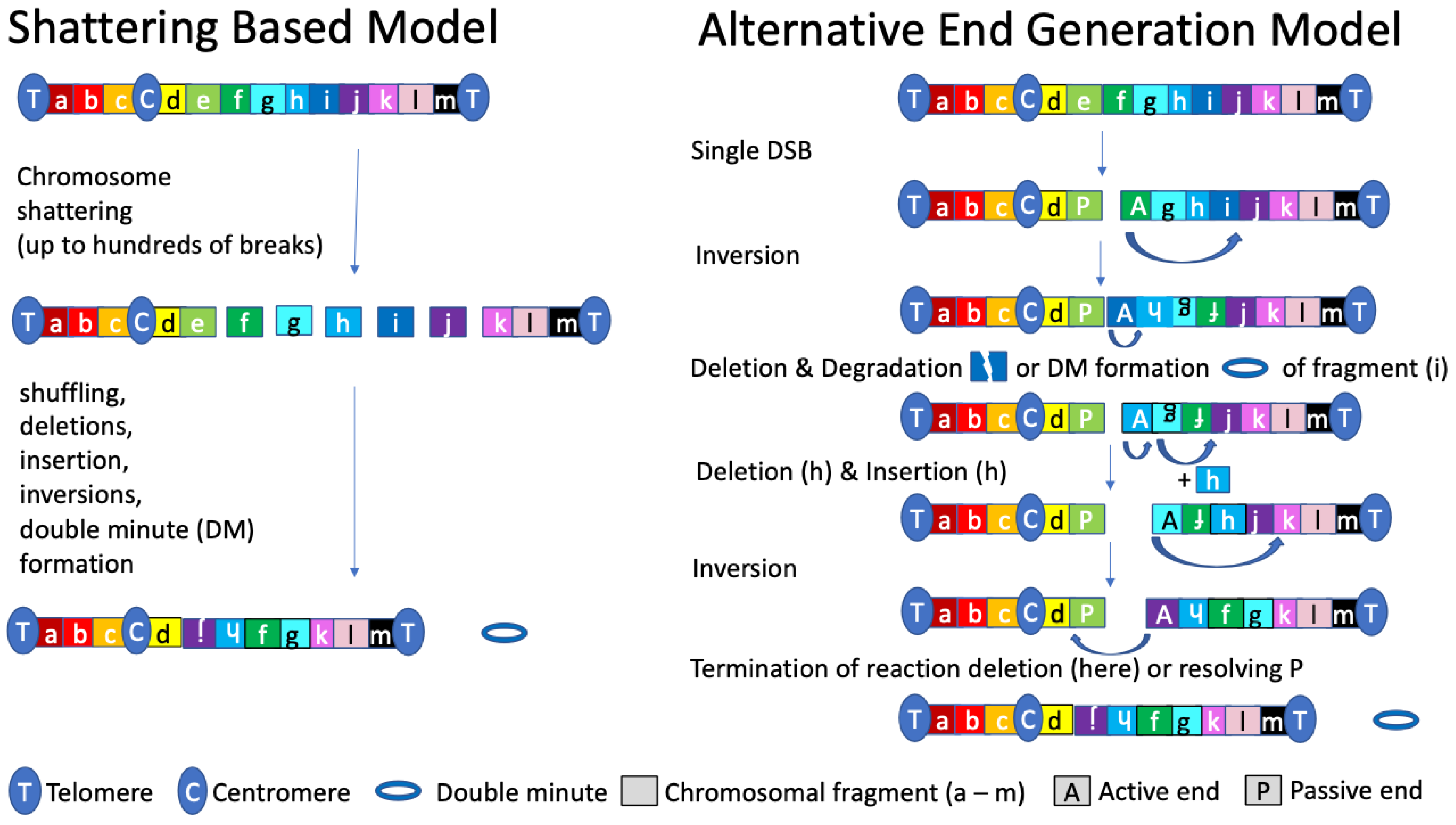
4. The AEG Model of Chromosomal Rearrangements and Chromothripsis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Metzker, M.L. Sequencing technologies the next generation. Nat. Rev. Genet. 2010, 11, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Stephens, P.J.; Greenman, C.D.; Fu, B.; Yang, F.; Bignell, G.R.; Mudie, L.J.; Pleasance, E.D.; Lau, K.W.; Beare, D.; Stebbings, L.A.; et al. Massive Genomic Rearrangement Acquired in a Single Catastrophic Event during Cancer Development. Cell 2011, 144, 27–40. [Google Scholar] [CrossRef] [PubMed]
- Holland, A.J.; Cleveland, D.W. Chromoanagenesis and cancer: Mechanisms and consequences of localized, complex chromosomal rearrangements. Nat. Med. 2012, 18, 1630–1638. [Google Scholar] [CrossRef] [PubMed]
- Korbel, J.O.; Campbell, P.J. Criteria for inference of chromothripsis in cancer genomes. Cell 2013, 152, 1226–1236. [Google Scholar] [CrossRef] [PubMed]
- Pellestor, F.; Gaillard, J.; Schneider, A.; Puechberty, J.; Gatinois, V. Chromoanagenesis, the mechanisms of a genomic chaos. Semin Cell Dev. Biol. 2022, 123, 90–99. [Google Scholar] [CrossRef] [PubMed]
- Brás., A.; Rodrigues, A.S.; Rueff, J. Copy number variations and constitutional chromothripsis. Biomed. Rep. 2020, 13, 11. [Google Scholar] [CrossRef] [PubMed]
- Leibowitz, M.L.; Zhang, C.Z.; Pellman, D. Chromothripsis: A New Mechanism for Rapid Karyotype Evolution. Ann. Rev. Genet. 2015, 49, 183–211. [Google Scholar] [CrossRef]
- Shoshani, O.; Brunner, S.F.; Yaeger, R.; Ly, P.; Nechemia-Arbely, Y.; Kim, D.H.; Fang, R.; Castillon, G.A.; Yu, M.; Li, J.S.Z.; et al. Chromothripsis drives the evolution of gene amplification in cancer. Nature 2021, 591, 137–141. [Google Scholar] [CrossRef]
- Munisha, M.; Schimenti, J.C. Genome maintenance during embryogenesis. DNA Repair 2021, 106, 103195. [Google Scholar] [CrossRef]
- Liu, P.; Erez, A.; Nagamani, S.C.; Dhar, S.U.; Kołodziejska, K.E.; Dharmadhikari, A.V.; Cooper, M.L.; Wiszniewska, J.; Zhang, F.; Withers, M.A.; et al. Chromosome catastrophes involve replication mechanisms generating complex genomic rearrangements. Cell 2011, 146, 889–903. [Google Scholar] [CrossRef]
- Maclachlan, K.H.; Rustad, E.H.; Derkach, A.; Zheng-Lin, B.; Yellapantula, V.; Diamond, B.; Hultcrantz, M.; Ziccheddu, B.; Boyle, E.M.; Blaney, P.; et al. Copy number signatures predict chromothripsis and clinical outcomes in newly diagnosed multiple myeloma. Nat. Commun. 2021, 12, 5172. [Google Scholar] [CrossRef] [PubMed]
- Rustad, E.H.; Yellapantula, V.D.; Glodzik, D.; Maclachlan, K.H.; Diamond, B.; Boyle, E.M.; Ashby, C.; Blaney, P.; Gundem, G.; Hultcrantz, M.; et al. Revealing the Impact of Structural Variants in Multiple Myeloma. Blood Cancer Discov. 2020, 1, 258–273. [Google Scholar] [CrossRef] [PubMed]
- Maura, F.; Boyle, E.M.; Rustad, E.H.; Ashby, C.; Kaminetzky, D.; Bruno, B.; Braunstein, M.; Bauer, M.; Blaney, P.; Wang, Y.; et al. Chromothripsis as a pathogenic driver of multiple myeloma. Semin Cell Dev. Biol. 2022, 123, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Ashby, C.; Boyle, E.M.; Bauer, M.A.; Mikulasova, A.; Wardell, C.P.; Williams, L.; Siegel, A.; Blaney, P.; Braunstein, M.; Kaminetsky, D.; et al. Structural variants shape the genomic landscape and clinical outcome of multiple myeloma. Blood Cancer J. 2022, 12, 85. [Google Scholar] [CrossRef] [PubMed]
- Magrangeas, F.; Avet-Loiseau, H.; Munshi, N.C.; Minvielle, S. Chromothripsis identifies a rare and aggressive entity among newly diagnosed multiple myeloma patients. Blood 2011, 118, 675–678. [Google Scholar] [CrossRef]
- Notta, F.; Chan-Seng-Yue, M.; Lemire, M.; Li, Y.; Wilson, G.W.; Connor, A.A.; Denroche, R.E.; Liang, S.B.; Brown, A.M.; Kim, J.C.; et al. A renewed model of pancreatic cancer evolution based on genomic rearrangement patterns. Nature 2016, 538, 378–382. [Google Scholar] [CrossRef]
- Chan-Seng-Yue, M.; Kim, J.C.; Wilson, G.W.; Ng, K.; Figueroa, E.F.; O’Kane, G.M.; Connor, A.A.; Denroche, R.E.; Grant, R.C.; McLeod, J.; et al. Transcription phenotypes of pancreatic cancer are driven by genomic events during tumor evolution. Nat. Genet. 2020, 52, 231–240. [Google Scholar] [CrossRef]
- Zhao, X.K.; Xing, P.; Song, X.; Zhao, M.; Zhao, L.; Dang, Y.; Lei, L.L.; Xu, R.H.; Han, W.L.; Wang, P.P.; et al. Focal amplifications are associated with chromothripsis events and diverse prognoses in gastric cardia adenocarcinoma. Nat. Commun. 2021, 12, 6489. [Google Scholar] [CrossRef]
- Vasmatzis, G.; Wang, X.; Smadbeck, J.B.; Murphy, S.J.; Geiersbach, K.B.; Johnson, S.H.; Gaitatzes, A.G.; Asmann, Y.W.; Kosari, F.; Borad, M.J.; et al. Chromoanasynthesis is a common mechanism that leads to ERBB2 amplifications in a cohort of early stage HER2+ breast cancer samples. BMC Cancer 2018, 18, 738. [Google Scholar] [CrossRef]
- Biermann, J.; Langen, B.; Nemes, S.; Holmberg, E.; Parris, T.Z.; Werner Rönnerman, E.; Engqvist, H.; Kovács, A.; Helou, K.; Karlsson, P. Radiation-induced genomic instability in breast carcinomas of the Swedish hemangioma cohort. Genes Chromosom. Cancer 2019, 58, 627–635. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, X.; Hou, C.; Zhou, Y.; Chen, J.; Cai, H.; Ye, Y.; Liu, J.; Huang, N. Comprehensive identification and characterization of somatic copy number alterations in triple-negative breast cancer. Int. J. Oncol. 2019, 56, 522–530. [Google Scholar] [CrossRef] [PubMed]
- Bolkestein, M.; Wong, J.K.L.; Thewes, V.; Körber, V.; Hlevnjak, M.; Elgaafary, S.; Schulze, M.; Kommoss, F.K.F.; Sinn, H.P.; Anzeneder, T.; et al. Chromothripsis in Human Breast Cancer. Cancer Res. 2020, 80, 4918–4931. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, Y.; Miyake, S.; Oka, M.; Kanai, A.; Kawai, Y.; Nagasawa, S.; Shiraishi, Y.; Tokunaga, K.; Kohno, T.; Seki, M.; et al. Phasing analysis of lung cancer genomes using a long read sequencer. Nat. Commun. 2022, 13, 3464. [Google Scholar] [CrossRef] [PubMed]
- Zavacka, K.; Plevova, K. Chromothripsis in Chronic Lymphocytic Leukemia: A Driving Force of Genome Instability. Front. Oncol. 2021, 11, 771664. [Google Scholar] [CrossRef] [PubMed]
- Baltus, C.; Toffoli, S.; London, F.; Delrée, P.; Gilliard, C.; Gustin, T. Chromothripsis in an Early Recurrent Chordoid Meningioma. World Neurosurg. 2019, 130, 380–385. [Google Scholar] [CrossRef] [PubMed]
- Cortés-Ciriano, I.; Lee, J.J.; Xi, R.; Jain, D.; Jung, Y.L.; Yang, L.; Gordenin, D.; Klimczak, L.J.; Zhang, C.Z.; Pellman, D.S.; et al. Comprehensive analysis of chromothripsis in 2,658 human cancers using whole-genome sequencing. Nat. Genet. 2020, 52, 331–341. [Google Scholar] [CrossRef]
- Carvalho CM, B.; Lupski, J.R. Mechanisms underlying structural variant formation in genomic disorders. Nat. Rev. Genet. 2016, 17, 224–238. [Google Scholar] [CrossRef]
- Tan, E.H.; Henry, I.M.; Ravi, M.; Bradnam, K.R.; Mandakova; Marimuthu, M.; Korf, I.; Lysak, M.; Comai, L.; Chan, S.W. Catastrophic chromosomal restructuring during genome elimination in plants. Elife 2015, 4, e06516. [Google Scholar] [CrossRef]
- Kloosterman, W.P.; Cuppen, E. Chromothripsis in congenital disorders and cancer: Similarities and differences. Curr. Opin. Cell Biol. 2013, 25, 341–348. [Google Scholar] [CrossRef]
- Henry, I.M.; Comai, L.; Tan, E.H. Detection of Chromothripsis in Plants. Methods Mol. Biol. 2018, 1769, 119–132. [Google Scholar] [CrossRef]
- Tan, E.; Comai, L.; Henry, I. Chromosome Dosage Analysis in Plants Using Whole Genome Sequencing. Bio. Protoc. 2016, 6, e1854. [Google Scholar] [CrossRef]
- Jones MJ, K.; Jallepalli, P.V. Chromothripsis: Chromosomes in Crisis. Dev. Cell 2012, 23, 908–917. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.Z.; Spektor, A.; Cornils, H.; Francis, J.M.; Jackson, E.K.; Liu, S.; Meyerson, M.; Pellman, D. Chromothripsis from DNA damage in micronuclei. Nature 2015, 522, 179–184. [Google Scholar] [CrossRef] [PubMed]
- Crasta, K.; Ganem, N.J.; Dagher, R.; Lantermann, A.B.; Ivanova, E.V.; Pan, Y.; Nezi, L.; Protopopov, A.; Chowdhury, D.; Pellman, D. DNA breaks and chromosome pulverization from errors in mitosis. Nature 2012, 482, 53–58. [Google Scholar] [CrossRef] [PubMed]
- Tubio, J.M.C.; Estivill, X. When catastrophe strikes a cell. Nature 2011, 470, 476–477. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.L.; Tang, H.M.; Mak, K.H.; Hu, S.; Wang, S.S.; Wong, K.M.; Wong, C.S.; Wu, H.Y.; Law, H.T.; Liu, K.; et al. Cell survival, DNA damage, and oncogenic transformation after a transient and reversible apoptotic response. Mol. Biol. Cell 2012, 23, 2240–2252. [Google Scholar] [CrossRef]
- Morishita, M.; Muramatsu, T.; Suto, Y.; Hirai, M.; Konishi, T.; Hayashi, S.; Shigemizu, D.; Tsunoda, T.; Moriyama, K.; Inazawa, J. Chromothripsis-like chromosomal rearrangements induced by ionizing radiation using proton microbeam irradiation system. Oncotarget 2016, 7, 10182–10192. [Google Scholar] [CrossRef]
- Schütze, D.M.; Krijgsman, O.; Snijders, P.J.; Ylstra, B.; Weischenfeldt, J.; Mardin, B.R.; Stütz, A.M.; Korbel, J.O.; de Winter, J.P.; Meijer, C.J.; et al. Immortalization capacity of HPV types is inversely related to chromosomal instability. Oncotarget 2016, 7, 37608–37621. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, J.L.; Rotmensch, J.; Giovanazzi, S.; Cohen, M.B.; Weichselbaum, R.R. Faster repair of DNA double-strand breaks in radioresistant human tumor cells. Int. J. Radiat Oncol. Biol. Phys. 1988, 15, 907–912. [Google Scholar] [CrossRef]
- Wurm, R.; Burnet, N.G.; Duggal, N.; Yarnold, J.R.; Peacock, J.H. Cellular radiosensitivity and dna damage in primary human fibroblasts. Int. J. Radiat Oncol. Biol. Phys. 1994, 30, 625–633. [Google Scholar] [CrossRef]
- Bohgaki, T.; Bohgaki, M.; Hakem, R. DNA double-strand break signaling and human disorders. Genome Integr. 2010, 1, 15. [Google Scholar] [CrossRef] [PubMed]
- Jackson, S.P.; Bartek, J. The DNA-damage response in human biology and disease. Nature 2009, 461, 1071–1078. [Google Scholar] [CrossRef] [PubMed]
- Vilenchik, M.M.; Knudson, A.G. Endogenous DNA double-strand breaks: Production, fidelity of repair, and induction of cancer. Proc. Natl. Acad. Sci. USA 2003, 100, 12871–12876. [Google Scholar] [CrossRef] [PubMed]
- Hogenbirk, M.A.; Heideman, M.; de Rink, I.; Velds, A.; Kerkhoven, R.M.; Wessels, L.F.; Jacobs, H. Defining chromosomal translocation risks in cancer. Proc. Natl. Acad. Sci. USA 2016, 113, E3649–E3656. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.; Gordenin, D.A. Clusters of Multiple Mutations: Incidence and Molecular Mechanisms. Ann. Rev. Genet. 2015, 49, 243–267. [Google Scholar] [CrossRef]
- Cairns, J. Mutation selection and the natural history of cancer. Nature 1975, 255, 197–200. [Google Scholar] [CrossRef]
- Martincorena, I.; Campbell, P.J. Somatic mutation in cancer and normal cells. Science 2015, 349, 1483–1489. [Google Scholar] [CrossRef]
- Kinsella, M.; Patel, A.; Bafna, V. The elusive evidence for chromothripsis. Nucleic Acids Res. 2014, 42, 8231–8242. [Google Scholar] [CrossRef]
- Ly, P.; Teitz, L.S.; Kim, D.H.; Shoshani, O.; Skaletsky, H.; Fachinetti, D.; Page, D.C.; Cleveland, D.W. Selective y centromere inactivation triggers chromosome shattering in micronuclei and repair by non-homologous end joining. Nat. Cell Biol. 2017, 19, 68–75. [Google Scholar] [CrossRef]
- Willis, N.A.; Rass, E.; Scully, R. Deciphering the Code of the Cancer Genome: Mechanisms of Chromosome Rearrangement. Trends Cancer 2015, 1, 217–230. [Google Scholar] [CrossRef]
- So, A.; le Guen, T.; Lopez, B.S.; Guirouilh-Barbat, J. Genomic rearrangements induced by unscheduled DNA double strand breaks in somatic mammalian cells. FEBS J. 2017, 284, 2324–2344. [Google Scholar] [CrossRef] [PubMed]
- Chiang, C.; Jacobsen, J.C.; Ernst, C.; Hanscom, C.; Heilbut, A.; Blumenthal, I.; Mills, R.E.; Kirby, A.; Lindgren, A.M.; Rudiger, S.R.; et al. Complex reorganization and predominant non-homologous repair following chromosomal breakage in karyotypically balanced germline rearrangements and transgenic integration. Nat. Genet. 2012, 44, 390–397. [Google Scholar] [CrossRef] [PubMed]
- Ratnaparkhe, M.; Wong, J.K.L.; Wei, P.C.; Hlevnjak, M.; Kolb, T.; Simovic, M.; Haag, D.; Paul, Y.; Devens, F.; Northcott, P.; et al. Defective DNA damage repair leads to frequent catastrophic genomic events in murine and human tumors. Nat. Commun. 2018, 9, 4760. [Google Scholar] [CrossRef]
- Kloosterman, W.P.; Tavakoli-Yaraki, M.; van Roosmalen, M.J.; van Binsbergen, E.; Renkens, I.; Duran, K.; Ballarati, L.; Vergult, S.; Giardino, D.; Hansson, K.; et al. Constitutional Chromothripsis Rearrangements Involve Clustered Double-Stranded DNA Breaks and Nonhomologous Repair Mechanisms. Cell Rep. 2012, 1, 648–655. [Google Scholar] [CrossRef] [PubMed]
- Kloosterman, W.P.; Koster, J.; Molenaar, J.J. Prevalence and clinical implications of chromothripsis in cancer genomes. Curr. Opin. Oncol. 2014, 26, 64–72. [Google Scholar] [CrossRef]
- Righolt, C.; Mai, S. Shattered and stitched chromosomes-chromothripsis and chromoanasynthesis-manifestations of a new chromosome crisis? Genes Chromosom. Cancer 2012, 51, 975–981. [Google Scholar] [CrossRef] [PubMed]
- Forment, J.V.; Kaidi, A.; Jackson, S.P. Chromothripsis and cancer: Causes and consequences of chromosome shattering. Nat. Rev. Cancer 2012, 12, 663–670. [Google Scholar] [CrossRef]
- Meyerson, M.; Pellman, D. Cancer genomes evolve by pulverizing single chromosomes. Cell 2011, 144, 9–10. [Google Scholar] [CrossRef] [PubMed]
- Maciejowski, J.; Li, Y.; Bosco, N.; Campbell, P.J.; de Lange, T. Chromothripsis and Kataegis Induced by Telomere Crisis. Cell 2015, 163, 1641–1654. [Google Scholar] [CrossRef]
- Umbreit, N.T.; Zhang, C.Z.; Lynch, L.D.; Blaine, L.J.; Cheng, A.M.; Tourdot, R.; Sun, L.; Almubarak, H.F.; Judge, K.; Mitchell, T.J.; et al. Mechanisms generating cancer genome complexity from a single cell division error. Science 2020, 368, eaba0712. [Google Scholar] [CrossRef]
- McClintock, B. The Stability of Broken Ends of Chromosomes in Zea Mays. Genetics 1941, 26, 234–282. [Google Scholar] [CrossRef] [PubMed]
- Kamranvar, S.A.; Gruhne, B.; Szeles, A.; Masucci, M.G. Epstein-Barr virus promotes genomic instability in Burkitt’s lymphoma. Oncogene 2007, 26, 5115–5123. [Google Scholar] [CrossRef] [PubMed]
- Mardin, B.R.; Drainas, A.P.; Waszak, S.M.; Weischenfeldt, J.; Isokane, M.; Stütz, A.M.; Raeder, B.; Efthymiopoulos, T.; Buccitelli, C.; Segura-Wang, M.; et al. A cell-based model system links chromothripsis with hyperploidy. Mol. Syst. Biol. 2015, 11, 828. [Google Scholar] [CrossRef] [PubMed]
- Maher, C.A.; Wilson, R.K. Chromothripsis and Human Disease: Piecing Together the Shattering Process. Cell 2012, 148, 29–32. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.Z.; Leibowitz, M.L.; Pellman, D. Chromothripsis and beyond: Rapid genome evolution from complex chromosomal rearrangements. Genes Dev. 2013, 27, 2513–2530. [Google Scholar] [CrossRef]
- Cimini, D. Merotelic kinetochore orientation, aneuploidy, and cancer. Biochim Biophys. Acta 2008, 1786, 32–40. [Google Scholar] [CrossRef]
- Terradas, M.; Martín, M.; Tusell, L.; Genescà, A. Genetic activities in micronuclei: Is the DNA entrapped in micronuclei lost for the cell? Mutat Res. 2010, 705, 60–67. [Google Scholar] [CrossRef]
- Thompson, S.L.; Compton, D.A. Chromosome missegregation in human cells arises through specific types of kinetochore-microtubule attachment errors. Proc. Natl. Acad. Sci. USA 2011, 108, 17974–17978. [Google Scholar] [CrossRef]
- Koltsova, A.S.; Pendina, A.A.; Efimova, O.A.; Chiryaeva, O.G.; Kuznetzova, T.V.; Baranov, V.S. On the complexity of mechanisms and consequences of chromothripsis: An update. Front. Genet. 2019, 10, 393. [Google Scholar] [CrossRef]
- Tang, S.; Stokasimov, E.; Cui, Y.; Pellman, D. Breakage of cytoplasmic chromosomes by pathological DNA base excision repair. Nature 2022, 606, 930–936. [Google Scholar] [CrossRef]
- Strumberg, D.; Pilon, A.A.; Smith, M.; Hickey, R.; Malkas, L.; Pommier, Y. Conversion of Topoisomerase I Cleavage Complexes on the Leading Strand of Ribosomal DNA into 5′-Phosphorylated DNA Double-Strand Breaks by Replication Runoff. Mol. Cell Biol. 2000, 20, 3977–3987. [Google Scholar] [CrossRef] [PubMed]
- Yu, K.; Lieber, M.R. Current insights into the mechanism of mammalian immunoglobulin class switch recombination. Crit Rev. Biochem. Mol. Biol. 2019, 54, 333–351. [Google Scholar] [CrossRef] [PubMed]
- Kneissig, M.; Keuper, K.; de Pagter, M.S.; van Roosmalen, M.J.; Martin, J.; Otto, H.; Passerini, V.; Campos Sparr, A.; Renkens, I.; Kropveld, F.; et al. Micronuclei-based model system reveals functional consequences of chromothripsis in human cells. Elife 2019, 8, e50292. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; McCord, R.P.; Ho, Y.J.; Lajoie, B.R.; Hildebrand, D.G.; Simon, A.C.; Becker, M.S.; Alt, F.W.; Dekker, J. Spatial organization of the mouse genome and its role in recurrent chromosomal translocations. Cell 2012, 148, 908–921. [Google Scholar] [CrossRef] [PubMed]
- Chiarle, R.; Zhang, Y.; Frock, R.L.; Lewis, S.M.; Molinie, B.; Ho, Y.J.; Myers, D.R.; Choi, V.W.; Compagno, M.; Malkin, D.J.; et al. Genome-wide translocation sequencing reveals mechanisms of chromosome breaks and rearrangements in B cells. Cell 2011, 147, 107–119. [Google Scholar] [CrossRef]
- Jankovic, M.; Feldhahn, N.; Oliveira, T.Y.; Silva, I.T.; Kieffer-Kwon, K.R.; Yamane, A.; Resch, W.; Klein, I.; Robbiani, D.F.; Casellas, R.; et al. 53BP1 Alters the Landscape of DNA Rearrangements and Suppresses AID-Induced B Cell Lymphoma. Mol. Cell 2013, 49, 623–631. [Google Scholar] [CrossRef]
- Oliveira, T.Y.; Resch, W.; Jankovic, M.; Casellas, R.; Nussenzweig, M.C.; Klein, I.A. Translocation capture sequencing: A method for high throughput mapping of chromosomal rearrangements. J. Immunol. Methods 2012, 375, 176–181. [Google Scholar] [CrossRef]
- Klein, I.A.; Resch, W.; Jankovic, M.; Oliveira, T.; Yamane, A.; Nakahashi, H.; Di Virgilio, M.; Bothmer, A.; Nussenzweig, A.; Robbiani, D.F.; et al. Translocation-capture sequencing reveals the extent and nature of chromosomal rearrangements in B lymphocytes. Cell 2011, 147, 95–106. [Google Scholar] [CrossRef]
- Richardson, C.; Jasin, M. Frequent chromosomal translocations induced by DNA double-strand breaks. Nature 2000, 405, 697–700. [Google Scholar] [CrossRef]
- Godthelp, B.C.; van Buul, P.P.; Jaspers, N.G.; Elghalbzouri-Maghrani, E.; van Duijn-Goedhart, A.; Arwert, F.; Joenje, H.; Zdzienicka, M.Z. Cellular characterization of cells from the Fanconi anemia complementation group, FA-D1/BRCA2. Mutat Res. 2006, 601, 191–201. [Google Scholar] [CrossRef]
- Loucas, B.D.; Eberle, R.; Bailey, S.M.; Cornforth, M.N. Influence of dose rate on the induction of simple and complex chromosome exchanges by gamma rays. Radiat Res. 2004, 162, 339–349. [Google Scholar] [CrossRef] [PubMed]
- Bryant, P.E. Origin of Chromosome Aberrations: Mechanisms. In Chromosomal Alterations; Springer: Berlin, Heidelberg, 2007; pp. 177–199. [Google Scholar] [CrossRef]
- Hogenbirk, M.A.; Velds, A.; Kerkhoven, R.M.; Jacobs, H. Reassessing genomic targeting of AID. Nat. Immunol. 2012, 13, 797–798. [Google Scholar] [CrossRef]
- Rocha, P.P.; Micsinai, M.; Kim, J.R.; Hewitt, S.L.; Souza, P.P.; Trimarchi, T.; Strino, F.; Parisi, F.; Kluger, Y.; Skok, J.A. Close Proximity to Igh Is a Contributing Factor to AID-Mediated Translocations. Mol. Cell 2012, 47, 873–885. [Google Scholar] [CrossRef] [PubMed]
- Sidiropoulos, N.; Mardin, B.R.; Rodríguez-González, F.G.; Bochkov, I.D.; Garg, S.; Stütz, A.M.; Korbel, J.O.; Aiden, E.L.; Weischenfeldt, J. Somatic structural variant formation is guided by and influences genome architecture. Genome Res. 2022, 32, 643–655. [Google Scholar] [CrossRef] [PubMed]
- Scully, R.; Panday, A.; Elango, R.; Willis, N.A. DNA double-strand break repair-pathway choice in somatic mammalian cells. Nat. Rev. Mol. Cell Biol. 2019, 11, 698–714. [Google Scholar] [CrossRef] [PubMed]
- Aparicio, T.; Baer, R.; Gottesman, M.; Gautier, J. MRN, CtIP, and BRCA1 mediate repair of topoisomerase II-DNA adducts. J. Cell Biol. 2016, 212, 399–408. [Google Scholar] [CrossRef] [PubMed]
- Schellenberg, M.J.; Appel, C.D.; Adhikari, S.; Robertson, P.D.; Ramsden, D.A.; Williams, R.S. Mechanism of repair of 5’-topoisomerase II-DNA adducts by mammalian tyrosyl-DNA phosphodiesterase 2. Nat. Struct. Mol. Biol. 2012, 19, 1363–1371. [Google Scholar] [CrossRef] [PubMed]
- Collins, R.L.; Brand, H.; Redin, C.E.; Hanscom, C.; Antolik, C.; Stone, M.R.; Glessner, J.T.; Mason, T.; Pregno, G.; Dorrani, N.; et al. Defining the diverse spectrum of inversions, complex structural variation, and chromothripsis in the morbid human genome. Genome Biol. 2017, 36, 6. [Google Scholar] [CrossRef]
- Escaramís, G.; Docampo, E.; Rabionet, R. A decade of structural variants: Description, history and methods to detect structural variation. Brief. Funct. Genom. 2015, 14, 305–314. [Google Scholar] [CrossRef]
- Simovic, M.; Ernst, A. Chromothripsis, DNA repair and checkpoints defects. Semin Cell Dev. Biol. 2022, 123, 110–114. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Groot, D.; Spanjaard, A.; Hogenbirk, M.A.; Jacobs, H. Chromosomal Rearrangements and Chromothripsis: The Alternative End Generation Model. Int. J. Mol. Sci. 2023, 24, 794. https://doi.org/10.3390/ijms24010794
de Groot D, Spanjaard A, Hogenbirk MA, Jacobs H. Chromosomal Rearrangements and Chromothripsis: The Alternative End Generation Model. International Journal of Molecular Sciences. 2023; 24(1):794. https://doi.org/10.3390/ijms24010794
Chicago/Turabian Stylede Groot, Daniel, Aldo Spanjaard, Marc A. Hogenbirk, and Heinz Jacobs. 2023. "Chromosomal Rearrangements and Chromothripsis: The Alternative End Generation Model" International Journal of Molecular Sciences 24, no. 1: 794. https://doi.org/10.3390/ijms24010794
APA Stylede Groot, D., Spanjaard, A., Hogenbirk, M. A., & Jacobs, H. (2023). Chromosomal Rearrangements and Chromothripsis: The Alternative End Generation Model. International Journal of Molecular Sciences, 24(1), 794. https://doi.org/10.3390/ijms24010794







