Evaluation of Disability Progression in Multiple Sclerosis via Magnetic-Resonance-Based Deep Learning Techniques
Abstract
:1. Introduction
2. Results
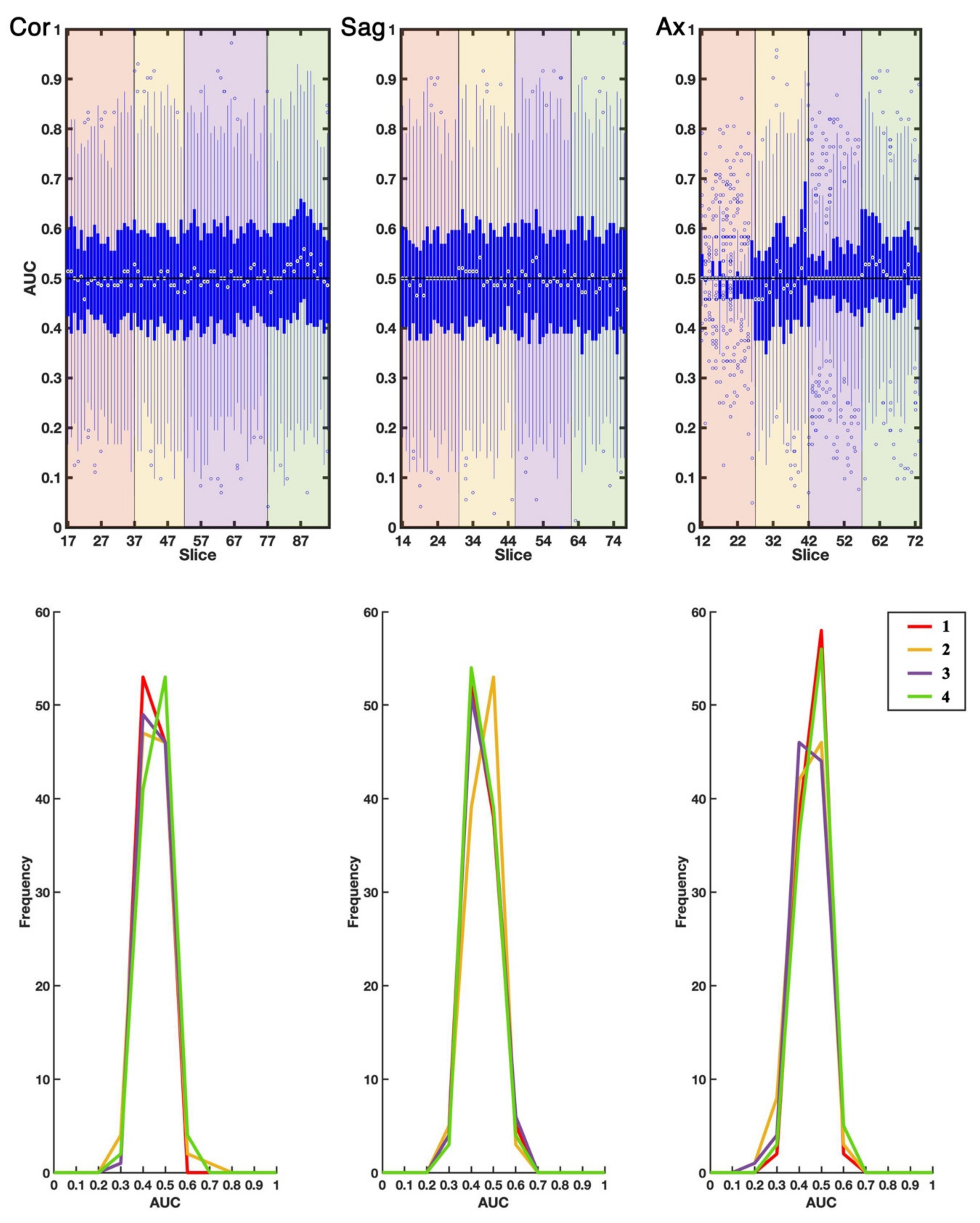
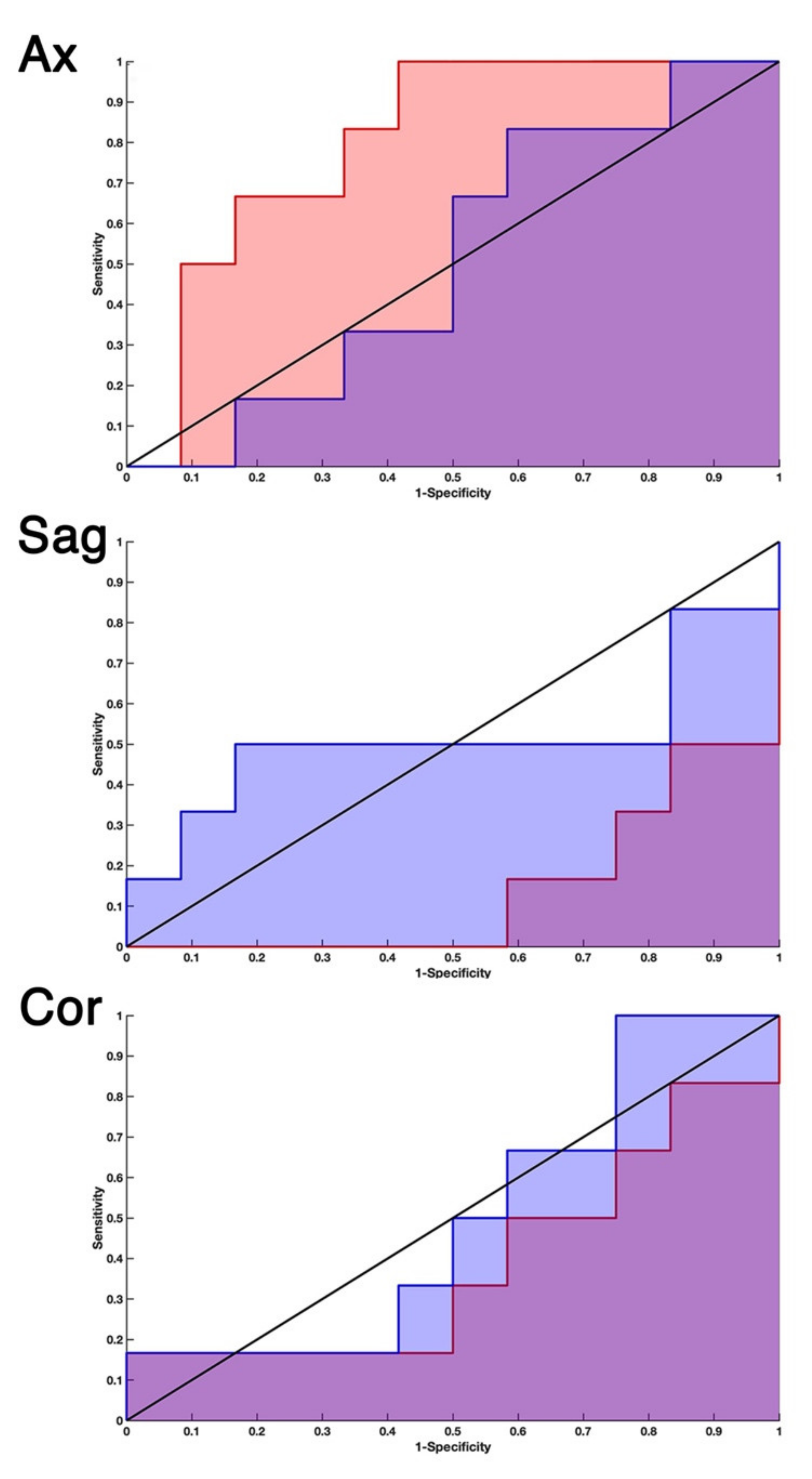
2.1. EDSS-Discriminators
2.2. Comparison with Null EDSS-Ds
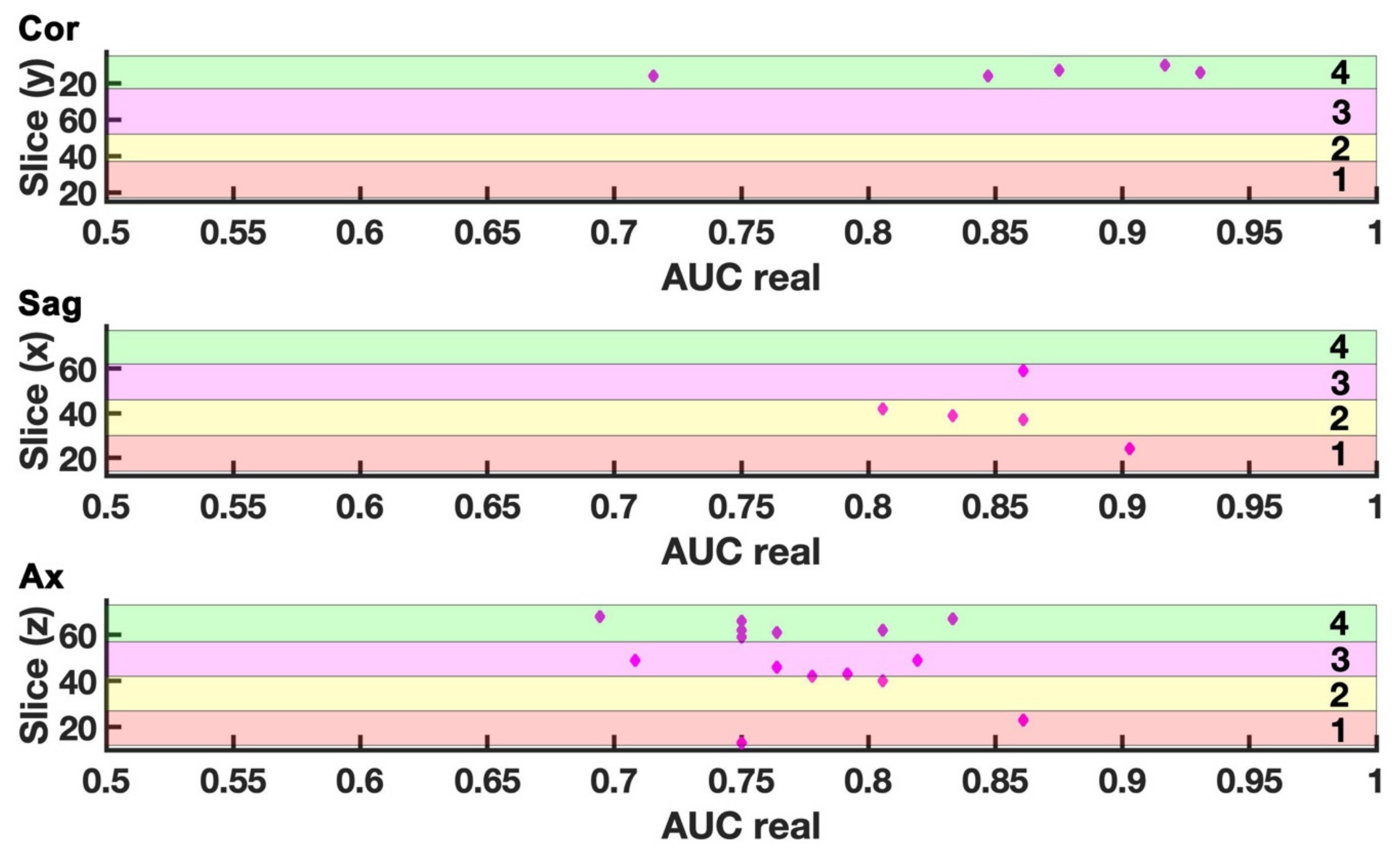
2.3. Three-Dimensional Representation
3. Discussion
3.1. Two-Step Training of Pre-Trained Deep Neural Network
3.2. Models Built on Either Single Slice or Slice Combination
3.3. 3-DT1 Images to Predict Disability Progression in MS
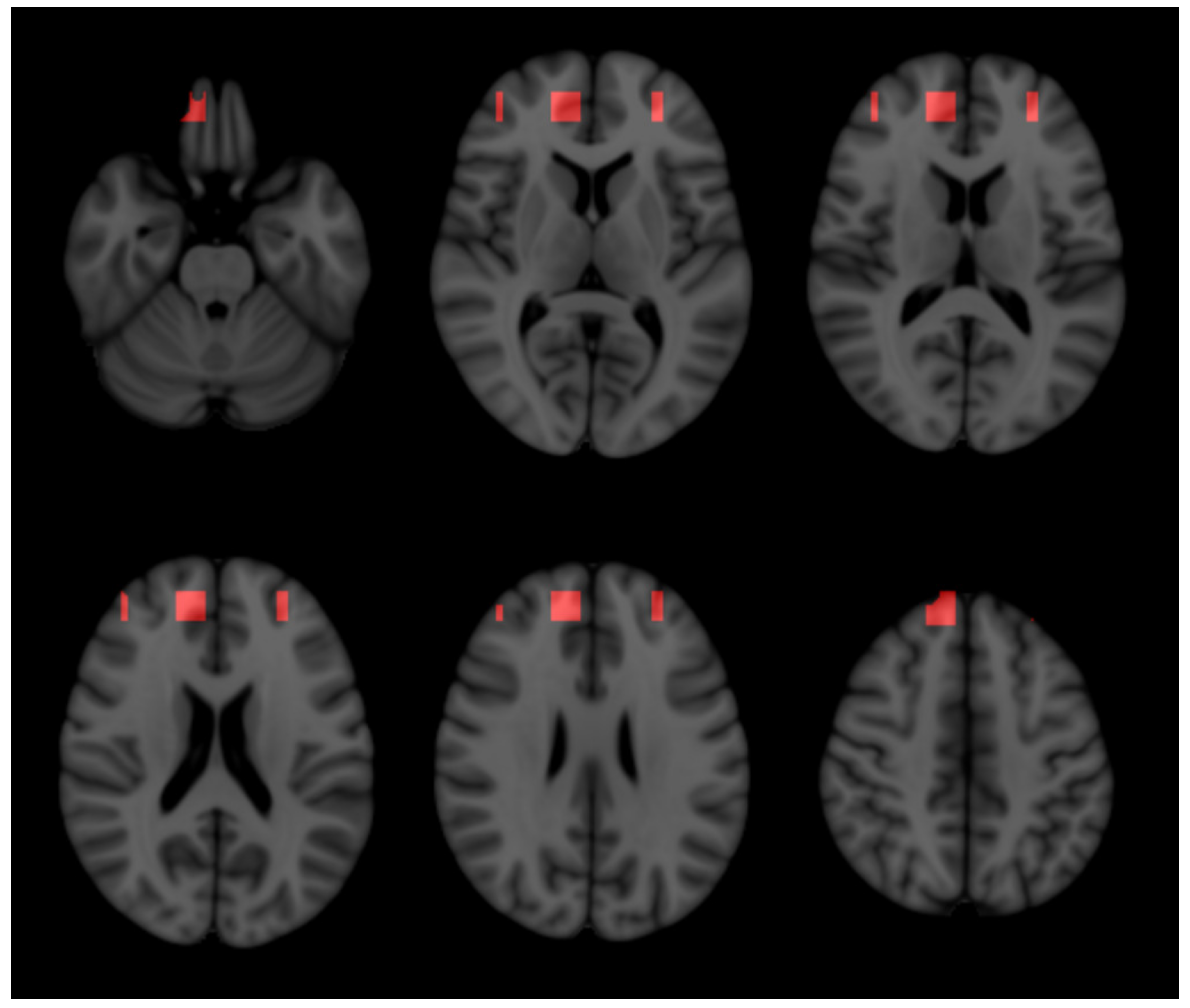
3.4. Anatomical Structures Relevant in Predicting Disability Progression
3.5. Model Generalization
3.6. Limits
4. Methods and Materials
4.1. Patients Selection and Disability Assessment
4.2. Population
4.3. Magnetic Resonance Imaging
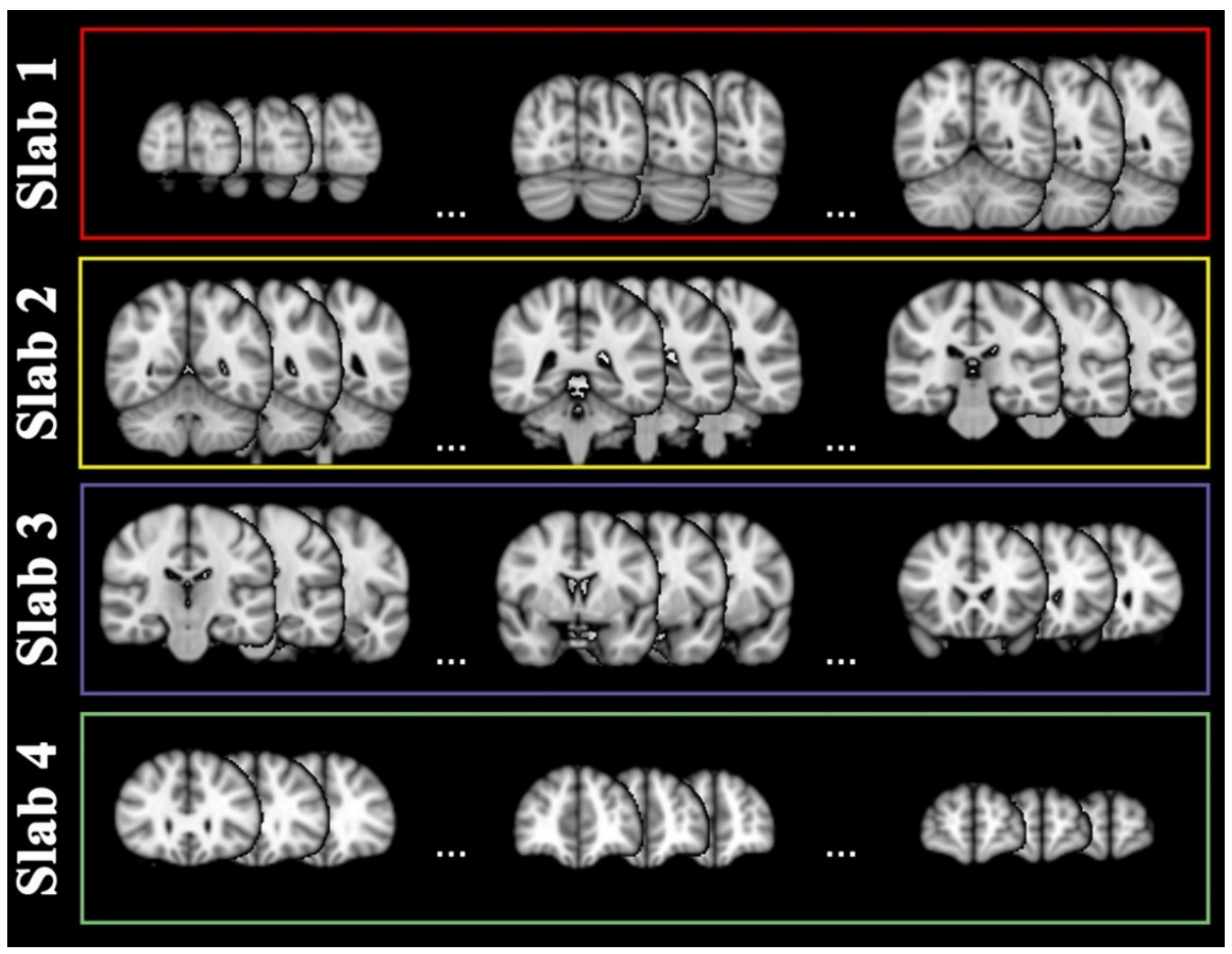
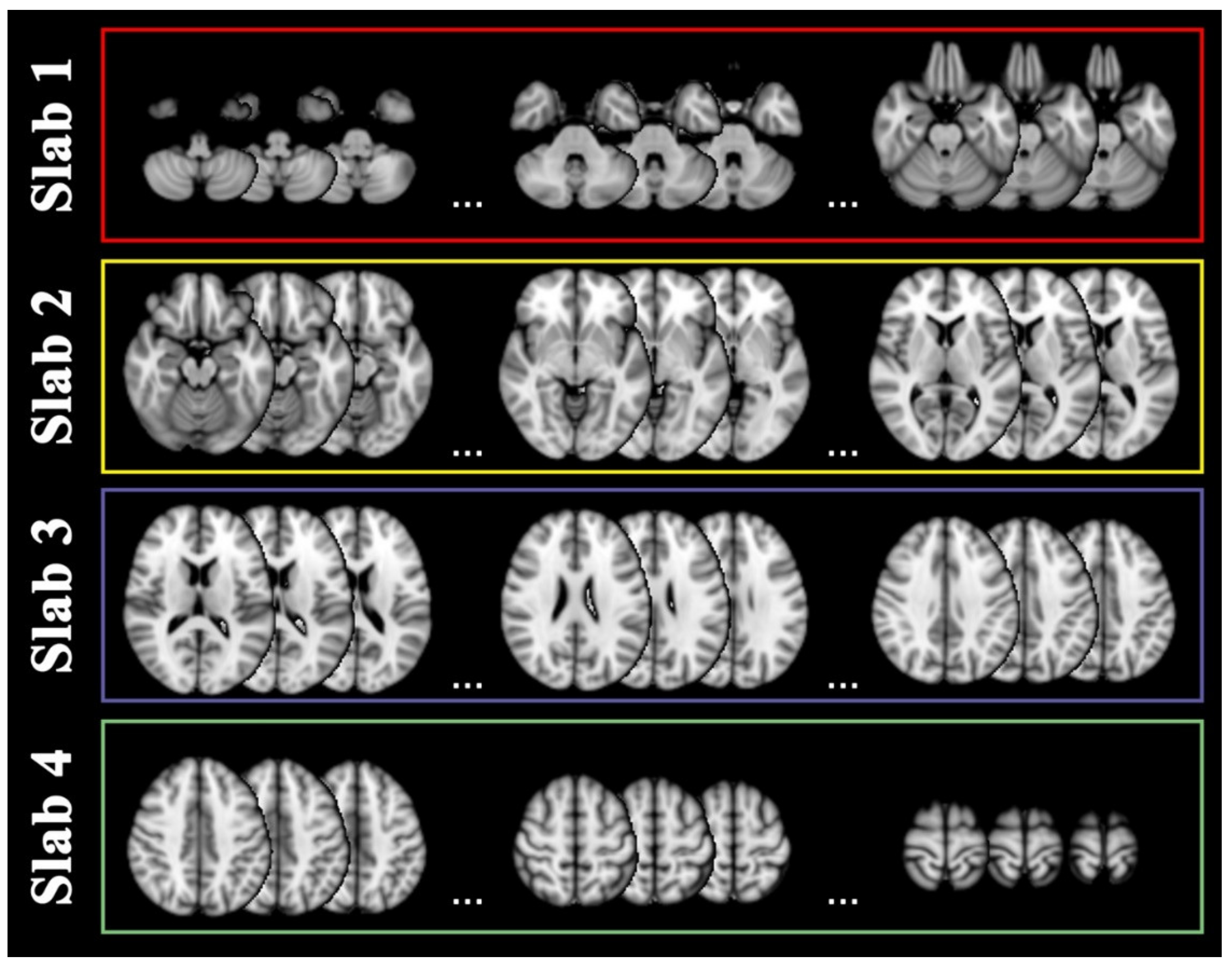
4.4. Slab Selection
4.5. Modelling Strategy
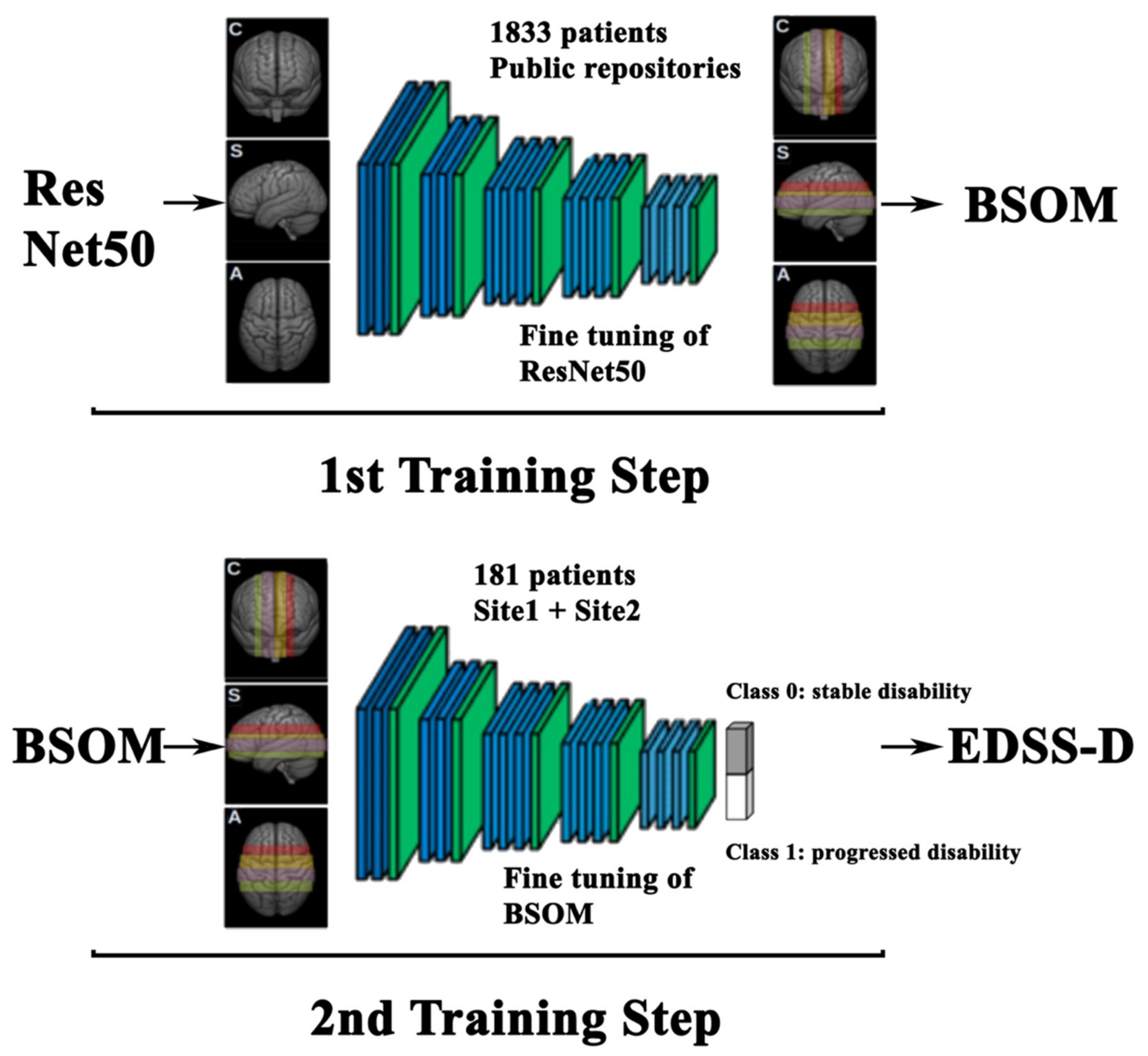
4.5.1. 1st Training Step: BSOM
4.5.2. 2nd Training Step: EDSS-Discriminators
4.5.3. AUC Statistical Analysis
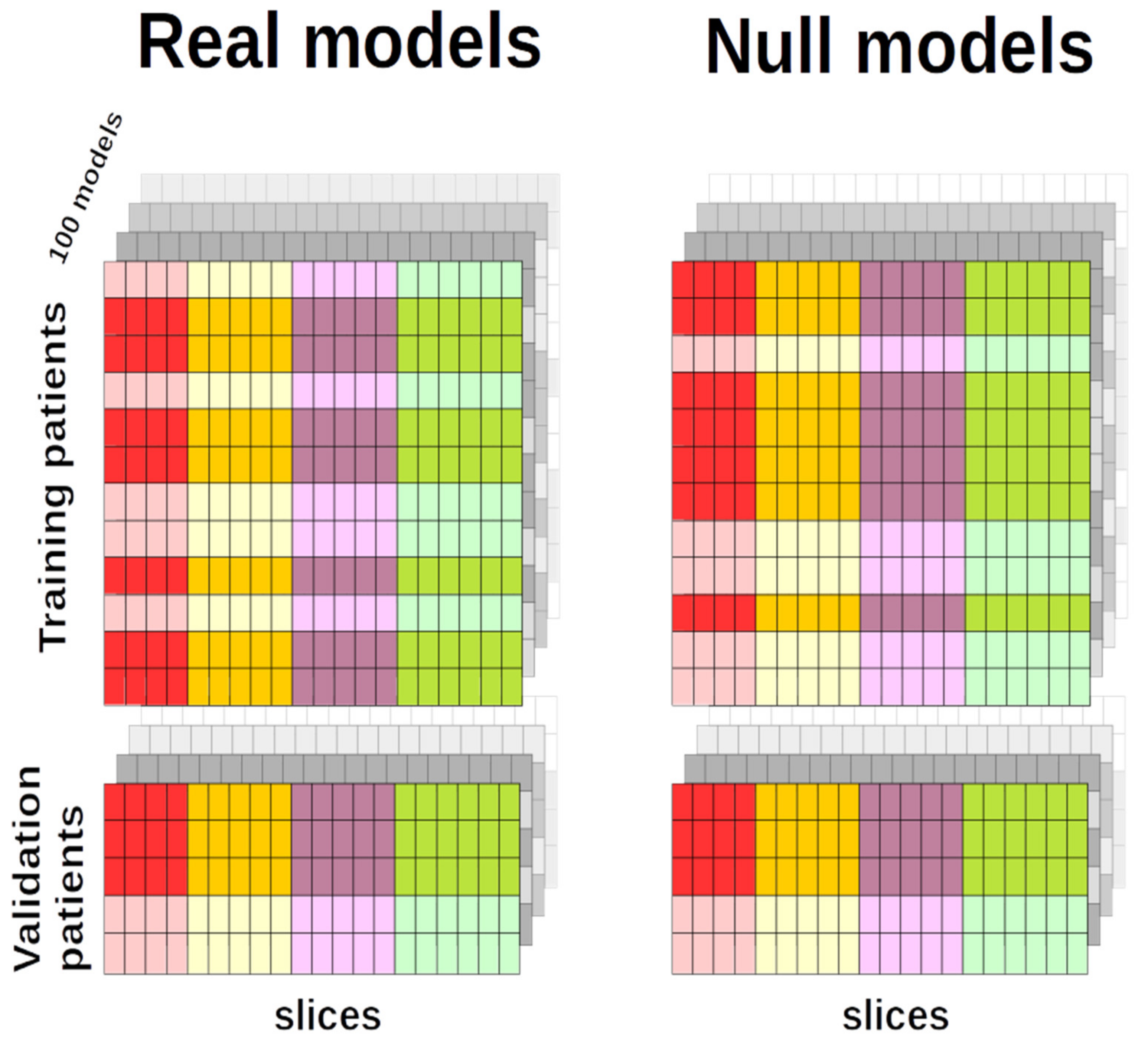
4.5.4. Null EDSS-Discriminators
4.5.5. Three-Dimensional Representation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Confavreux, C.; Vukusic, S. The Clinical Course of Multiple Sclerosis. Handb. Clin. Neurol. 2014, 122, 343–369. [Google Scholar] [CrossRef] [PubMed]
- Ciccarelli, O.; Barkhof, F.; Bodini, B.; Stefano, N.D.; Golay, X.; Nicolay, K.; Pelletier, D.; Pouwels, P.J.W.; Smith, S.A.; Wheeler-Kingshott, C.A.M.; et al. Pathogenesis of Multiple Sclerosis: Insights from Molecular and Metabolic Imaging. Lancet Neurol. 2014, 13, 807–822. [Google Scholar] [CrossRef]
- Rotstein, D.; Montalban, X. Reaching an Evidence-Based Prognosis for Personalized Treatment of Multiple Sclerosis. Nat. Rev. Neurol. 2019, 15, 287–300. [Google Scholar] [CrossRef] [PubMed]
- Gasperini, C.; Prosperini, L.; Tintoré, M.; Sormani, M.P.; Filippi, M.; Rio, J.; Palace, J.; Rocca, M.A.; Ciccarelli, O.; Barkhof, F.; et al. Unraveling Treatment Response in Multiple Sclerosis: A Clinical and MRI Challenge. Neurology 2019, 92, 180–192. [Google Scholar] [CrossRef]
- Russakovsky, O.; Deng, J.; Su, H.; Krause, J.; Satheesh, S.; Ma, S.; Huang, Z.; Karpathy, A.; Khosla, A.; Bernstein, M.; et al. ImageNet Large Scale Visual Recognition Challenge. Int. J. Comput. Vis. 2015, 115, 211–252. [Google Scholar] [CrossRef]
- Sander, L.; Pezold, S.; Andermatt, S.; Amann, M.; Meier, D.; Wendebourg, M.J.; Sinnecker, T.; Radue, E.-W.; Naegelin, Y.; Granziera, C.; et al. Accurate, Rapid and Reliable, Fully Automated MRI Brainstem Segmentation for Application in Multiple Sclerosis and Neurodegenerative Diseases. Hum. Brain Mapp. 2019, 40, 4091–4104. [Google Scholar] [CrossRef]
- Narayana, P.A.; Coronado, I.; Sujit, S.J.; Wolinsky, J.S.; Lublin, F.D.; Gabr, R.E. Deep Learning for Predicting Enhancing Lesions in Multiple Sclerosis from Noncontrast MRI. Radiology 2019, 294, 398–404. [Google Scholar] [CrossRef]
- Gabr, R.E.; Coronado, I.; Robinson, M.; Sujit, S.J.; Datta, S.; Sun, X.; Allen, W.J.; Lublin, F.D.; Wolinsky, J.S.; Narayana, P.A. Brain and Lesion Segmentation in Multiple Sclerosis Using Fully Convolutional Neural Networks: A Large-Scale Study. Mult. Scler. 2020, 26, 1217–1226. [Google Scholar] [CrossRef]
- McKinley, R.; Wepfer, R.; Grunder, L.; Aschwanden, F.; Fischer, T.; Friedli, C.; Muri, R.; Rummel, C.; Verma, R.; Weisstanner, C.; et al. Automatic Detection of Lesion Load Change in Multiple Sclerosis Using Convolutional Neural Networks with Segmentation Confidence. Neuroimage Clin. 2020, 25, 102104. [Google Scholar] [CrossRef]
- Narayana, P.A.; Coronado, I.; Sujit, S.J.; Sun, X.; Wolinsky, J.S.; Gabr, R.E. Are Multi-Contrast Magnetic Resonance Images Necessary for Segmenting Multiple Sclerosis Brains? A Large Cohort Study Based on Deep Learning. Magn. Reson. Imaging 2020, 65, 8–14. [Google Scholar] [CrossRef]
- Salem, M.; Valverde, S.; Cabezas, M.; Pareto, D.; Oliver, A.; Salvi, J.; Rovira, À.; Lladó, X. A Fully Convolutional Neural Network for New T2-w Lesion Detection in Multiple Sclerosis. Neuroimage Clin. 2020, 25, 102149. [Google Scholar] [CrossRef]
- Coronado, I.; Gabr, R.E.; Narayana, P.A. Deep Learning Segmentation of Gadolinium-Enhancing Lesions in Multiple Sclerosis. Mult. Scler. 2020, 27, 519–527. [Google Scholar] [CrossRef]
- Rocca, M.A.; Anzalone, N.; Storelli, L.; Del Poggio, A.; Cacciaguerra, L.; Manfredi, A.A.; Meani, A.; Filippi, M. Deep Learning on Conventional Magnetic Resonance Imaging Improves the Diagnosis of Multiple Sclerosis Mimics. Investig. Radiol. 2020, 56, 252–260. [Google Scholar] [CrossRef]
- Zurita, M.; Montalba, C.; Labbé, T.; Cruz, J.P.; Dalboni da Rocha, J.; Tejos, C.; Ciampi, E.; Cárcamo, C.; Sitaram, R.; Uribe, S. Characterization of Relapsing-Remitting Multiple Sclerosis Patients Using Support Vector Machine Classifications of Functional and Diffusion MRI Data. Neuroimage Clin. 2018, 20, 724–730. [Google Scholar] [CrossRef]
- Pontillo, G.; Tommasin, S.; Cuocolo, R.; Petracca, M.; Petsas, N.; Ugga, L.; Carotenuto, A.; Pozzilli, C.; Iodice, R.; Lanzillo, R.; et al. A Combined Radiomics and Machine Learning Approach to Overcome the Clinicoradiologic Paradox in Multiple Sclerosis. AJNR Am. J. Neuroradiol. 2021, 42, 1927–1933. [Google Scholar] [CrossRef]
- Tommasin, S.; Cocozza, S.; Taloni, A.; Giannì, C.; Petsas, N.; Pontillo, G.; Petracca, M.; Ruggieri, S.; De Giglio, L.; Pozzilli, C.; et al. Machine Learning Classifier to Identify Clinical and Radiological Features Relevant to Disability Progression in Multiple Sclerosis. J. Neurol. 2021, 268, 4834–4845. [Google Scholar] [CrossRef] [PubMed]
- Storelli, L.; Azzimonti, M.; Gueye, M.; Vizzino, C.; Preziosa, P.; Tedeschi, G.; De Stefano, N.; Pantano, P.; Filippi, M.; Rocca, M.A. A Deep Learning Approach to Predicting Disease Progression in Multiple Sclerosis Using Magnetic Resonance Imaging. Investig. Radiol. 2022, 57, 423–432. [Google Scholar] [CrossRef]
- Peng, Y.; Zheng, Y.; Tan, Z.; Liu, J.; Xiang, Y.; Liu, H.; Dai, L.; Xie, Y.; Wang, J.; Zeng, C.; et al. Prediction of Unenhanced Lesion Evolution in Multiple Sclerosis Using Radiomics-Based Models: A Machine Learning Approach. Mult. Scler. Relat. Disord. 2021, 53, 102989. [Google Scholar] [CrossRef]
- Law, M.T.; Traboulsee, A.L.; Li, D.K.; Carruthers, R.L.; Freedman, M.S.; Kolind, S.H.; Tam, R. Machine Learning in Secondary Progressive Multiple Sclerosis: An Improved Predictive Model for Short-Term Disability Progression. Mult. Scler. J. Exp. Transl. Clin. 2019, 5, 2055217319885983. [Google Scholar] [CrossRef]
- De Brouwer, E.; Becker, T.; Moreau, Y.; Havrdova, E.K.; Trojano, M.; Eichau, S.; Ozakbas, S.; Onofrj, M.; Grammond, P.; Kuhle, J.; et al. Longitudinal Machine Learning Modeling of MS Patient Trajectories Improves Predictions of Disability Progression. Comput. Methods Programs Biomed. 2021, 208, 106180. [Google Scholar] [CrossRef] [PubMed]
- Eshaghi, A.; Young, A.L.; Wijeratne, P.A.; Prados, F.; Arnold, D.L.; Narayanan, S.; Guttmann, C.R.G.; Barkhof, F.; Alexander, D.C.; Thompson, A.J.; et al. Identifying Multiple Sclerosis Subtypes Using Unsupervised Machine Learning and MRI Data. Nat. Commun. 2021, 12, 2078. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Healy, B.C.; Rotstein, D.; Guttmann, C.R.G.; Bakshi, R.; Weiner, H.L.; Brodley, C.E.; Chitnis, T. Exploration of Machine Learning Techniques in Predicting Multiple Sclerosis Disease Course. PLoS ONE 2017, 12, e0174866. [Google Scholar] [CrossRef]
- Roca, P.; Attye, A.; Colas, L.; Tucholka, A.; Rubini, P.; Cackowski, S.; Ding, J.; Budzik, J.-F.; Renard, F.; Doyle, S.; et al. Artificial Intelligence to Predict Clinical Disability in Patients with Multiple Sclerosis Using FLAIR MRI. Diagn. Interv. Imaging 2020, 101, 795–802. [Google Scholar] [CrossRef]
- Pinto, M.F.; Oliveira, H.; Batista, S.; Cruz, L.; Pinto, M.; Correia, I.; Martins, P.; Teixeira, C. Prediction of Disease Progression and Outcomes in Multiple Sclerosis with Machine Learning. Sci. Rep. 2020, 10, 21038. [Google Scholar] [CrossRef]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.W.M.; van Ginneken, B.; Sánchez, C.I. A Survey on Deep Learning in Medical Image Analysis. Medical. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef]
- Torrey, L.; Shavlik, J.; Walker, T.; Maclin, R. Transfer Learning via Advice Taking. In Advances in Machine Learning I: Dedicated to the Memory of Professor Ryszard S. Michalski; Koronacki, J., Raś, Z.W., Wierzchoń, S.T., Kacprzyk, J., Eds.; Studies in Computational Intelligence; Springer: Berlin/Heidelberg, Germany, 2010; pp. 147–170. ISBN 978-3-642-05177-7. [Google Scholar]
- Tajbakhsh, N.; Shin, J.Y.; Gurudu, S.R.; Hurst, R.T.; Kendall, C.B.; Gotway, M.B.; Liang, J. Convolutional Neural Networks for Medical Image Analysis: Full Training or Fine Tuning? IEEE Trans. Med. Imaging 2016, 35, 1299–1312. [Google Scholar] [CrossRef]
- Chartrand, G.; Cheng, P.M.; Vorontsov, E.; Drozdzal, M.; Turcotte, S.; Pal, C.J.; Kadoury, S.; Tang, A. Deep Learning: A Primer for Radiologists. RadioGraphics 2017, 37, 2113–2131. [Google Scholar] [CrossRef]
- Wong, K.C.L.; Syeda-Mahmood, T.; Moradi, M. Building Medical Image Classifiers with Very Limited Data Using Segmentation Networks. Med. Image Anal. 2018, 49, 105–116. [Google Scholar] [CrossRef]
- Heaton, J. Ian Goodfellow, Yoshua Bengio, and Aaron Courville: Deep Learning. Genet. Program Evolvable Mach. 2018, 19, 305–307. [Google Scholar] [CrossRef]
- Eitel, F.; Soehler, E.; Bellmann-Strobl, J.; Brandt, A.U.; Ruprecht, K.; Giess, R.M.; Kuchling, J.; Asseyer, S.; Weygandt, M.; Haynes, J.-D.; et al. Uncovering Convolutional Neural Network Decisions for Diagnosing Multiple Sclerosis on Conventional MRI Using Layer-Wise Relevance Propagation. Neuroimage Clin. 2019, 24, 102003. [Google Scholar] [CrossRef]
- Tommasin, S.; Giannì, C.; De Giglio, L.; Pantano, P. Neuroimaging Techniques to Assess Inflammation in Multiple Sclerosis. Neuroscience 2019, 403, 4–16. [Google Scholar] [CrossRef]
- Maggi, P.; Fartaria, M.J.; Jorge, J.; La Rosa, F.; Absinta, M.; Sati, P.; Meuli, R.; Du Pasquier, R.; Reich, D.S.; Cuadra, M.B.; et al. CVSnet: A Machine Learning Approach for Automated Central Vein Sign Assessment in Multiple Sclerosis. NMR Biomed. 2020, 33, e4283. [Google Scholar] [CrossRef]
- Eshaghi, A.; Marinescu, R.V.; Young, A.L.; Firth, N.C.; Prados, F.; Jorge Cardoso, M.; Tur, C.; De Angelis, F.; Cawley, N.; Brownlee, W.J.; et al. Progression of Regional Grey Matter Atrophy in Multiple Sclerosis. Brain 2018, 141, 1665–1677. [Google Scholar] [CrossRef] [PubMed]
- Haider, L.; Zrzavy, T.; Hametner, S.; Höftberger, R.; Bagnato, F.; Grabner, G.; Trattnig, S.; Pfeifenbring, S.; Brück, W.; Lassmann, H. The Topograpy of Demyelination and Neurodegeneration in the Multiple Sclerosis Brain. Brain 2016, 139, 807–815. [Google Scholar] [CrossRef]
- Steenwijk, M.D.; Geurts, J.J.G.; Daams, M.; Tijms, B.M.; Wink, A.M.; Balk, L.J.; Tewarie, P.K.; Uitdehaag, B.M.J.; Barkhof, F.; Vrenken, H.; et al. Cortical Atrophy Patterns in Multiple Sclerosis Are Non-Random and Clinically Relevant. Brain 2016, 139, 115–126. [Google Scholar] [CrossRef]
- Favaretto, A.; Lazzarotto, A.; Poggiali, D.; Rolma, G.; Causin, F.; Rinaldi, F.; Perini, P.; Gallo, P. MRI-Detectable Cortical Lesions in the Cerebellum and Their Clinical Relevance in Multiple Sclerosis. Mult. Scler. 2016, 22, 494–501. [Google Scholar] [CrossRef]
- Weier, K.; Banwell, B.; Cerasa, A.; Collins, D.L.; Dogonowski, A.-M.; Lassmann, H.; Quattrone, A.; Sahraian, M.A.; Siebner, H.R.; Sprenger, T. The Role of the Cerebellum in Multiple Sclerosis. Cerebellum 2015, 14, 364–374. [Google Scholar] [CrossRef]
- Ruggieri, S.; Bharti, K.; Prosperini, L.; Giannì, C.; Petsas, N.; Tommasin, S.; Giglio, L.D.; Pozzilli, C.; Pantano, P. A Comprehensive Approach to Disentangle the Effect of Cerebellar Damage on Physical Disability in Multiple Sclerosis. Front. Neurol. 2020, 11, 529. [Google Scholar] [CrossRef]
- Leray, E.; Yaouanq, J.; Le Page, E.; Coustans, M.; Laplaud, D.; Oger, J.; Edan, G. Evidence for a Two-Stage Disability Progression in Multiple Sclerosis. Brain 2010, 133, 1900–1913. [Google Scholar] [CrossRef]
- Bluemke, D.A.; Moy, L.; Bredella, M.A.; Ertl-Wagner, B.B.; Fowler, K.J.; Goh, V.J.; Halpern, E.F.; Hess, C.P.; Schiebler, M.L.; Weiss, C.R. Assessing Radiology Research on Artificial Intelligence: A Brief Guide for Authors, Reviewers, and Readers-From the Radiology Editorial Board. Radiology 2020, 294, 487–489. [Google Scholar] [CrossRef] [Green Version]
- Cohen, J.A.; Reingold, S.C.; Polman, C.H.; Wolinsky, J.S. International Advisory Committee on Clinical Trials in Multiple Sclerosis Disability Outcome Measures in Multiple Sclerosis Clinical Trials: Current Status and Future Prospects. Lancet Neurol. 2012, 11, 467–476. [Google Scholar] [CrossRef]
- Polman, C.H.; Reingold, S.C.; Banwell, B.; Clanet, M.; Cohen, J.A.; Filippi, M.; Fujihara, K.; Havrdova, E.; Hutchinson, M.; Kappos, L.; et al. Diagnostic Criteria for Multiple Sclerosis: 2010 Revisions to the McDonald Criteria. Ann. Neurol. 2011, 69, 292–302. [Google Scholar] [CrossRef]
- Thompson, A.J.; Banwell, B.L.; Barkhof, F.; Carroll, W.M.; Coetzee, T.; Comi, G.; Correale, J.; Fazekas, F.; Filippi, M.; Freedman, M.S.; et al. Diagnosis of Multiple Sclerosis: 2017 Revisions of the McDonald Criteria. Lancet Neurol. 2018, 17, 162–173. [Google Scholar] [CrossRef]
- Kurtzke, J.F. Rating Neurologic Impairment in Multiple Sclerosis: An Expanded Disability Status Scale (EDSS). Neurology 1983, 33, 1444–1452. [Google Scholar] [CrossRef]
- Río, J.; Rovira, À.; Tintoré, M.; Otero-Romero, S.; Comabella, M.; Vidal-Jordana, Á.; Galán, I.; Castilló, J.; Arrambide, G.; Nos, C.; et al. Disability Progression Markers over 6-12 Years in Interferon-β-Treated Multiple Sclerosis Patients. Mult. Scler. 2018, 24, 322–330. [Google Scholar] [CrossRef]
- Collins, D.L.; Zijdenbos, A.P.; Kollokian, V.; Sled, J.G.; Kabani, N.J.; Holmes, C.J.; Evans, A.C. Design and Construction of a Realistic Digital Brain Phantom. IEEE Trans. Med. Imaging 1998, 17, 463–468. [Google Scholar] [CrossRef]
- DeLong, E.R.; DeLong, D.M.; Clarke-Pearson, D.L. Comparing the Areas under Two or More Correlated Receiver Operating Characteristic Curves: A Nonparametric Approach. Biometrics 1988, 44, 837–845. [Google Scholar] [CrossRef]
- Sun, X.; Xu, W. Fast Implementation of DeLong’s Algorithm for Comparing the Areas Under Correlated Receiver Operating Characteristic Curves. IEEE Signal Processing Lett. 2014, 21, 1389–1393. [Google Scholar] [CrossRef]
- Robin, X.; Turck, N.; Hainard, A.; Tiberti, N.; Lisacek, F.; Sanchez, J.-C.; Müller, M. PROC: An Open-Source Package for R and S+ to Analyze and Compare ROC Curves. BMC Bioinform. 2011, 12, 77. [Google Scholar] [CrossRef]

| All Subjects (Site 1 + Site 2) | Subjects at Site 1 | Subjects at Site 2 | Between Sites Comparison | |
|---|---|---|---|---|
| Average (std) | Average (std) | Average (std) | z-(p-Value) | |
| Number | 181 | 105 | 76 | - |
| Age [years] | 39.57 (10.46) | 38.29 (9.75) | 41.33 (11.20) | −1.90(0.06) |
| Sex (F/M) | 112/69 | 80/25 | 32/44 | 21.71 (0.001) * |
| Phenotype (RR/P) | 136/45 | 85/20 | 51/25 | 4.53 (0.04) * |
| Disease duration [years] | 9.90 (8.06) | 8.27 (7.97) | 12.06 (7.36) | −3.31 (0.001) |
| EDSS at baseline | 3.0 [0.0–7.5] ** | 2.0 [0.0–7.5] ** | 3.5 [2.0–7.5] ** | −5.06 (0.001) |
| Time to follow up [years] | 3.94 (0.91) | 4.2 (0.93) | 3.53 (0.69) | 5.87 (0.001) |
| Therapy at baseline (1st line, 2nd line, none) | 58, 75, 48 | 32, 31, 42 | 26, 44, 6 | - |
| Therapy switch (no switch, switch to same line treatment, none to 1st line, none to 2nd line, 1st to 2nd line, 2nd to 1st line, 1st line to none, 2nd line to none) | 80, 32, 15, 14, 18, 13, 8, 1 | 46, 10, 13, 13, 9, 6, 7, 1 | 34, 22, 2, 1, 9, 7, 1, - | - |
| Relapse | 0 [0–5] ** | 0 [0–5] ** | 0 [0–4] ** | 0.82 (0.41) |
| Disability progression (Yes/No) | 62/119 | 36/69 | 26/50 | 0.0001 (0.99) * |
| Progressed patients (%) | 34 | 34 | 34 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Taloni, A.; Farrelly, F.A.; Pontillo, G.; Petsas, N.; Giannì, C.; Ruggieri, S.; Petracca, M.; Brunetti, A.; Pozzilli, C.; Pantano, P.; et al. Evaluation of Disability Progression in Multiple Sclerosis via Magnetic-Resonance-Based Deep Learning Techniques. Int. J. Mol. Sci. 2022, 23, 10651. https://doi.org/10.3390/ijms231810651
Taloni A, Farrelly FA, Pontillo G, Petsas N, Giannì C, Ruggieri S, Petracca M, Brunetti A, Pozzilli C, Pantano P, et al. Evaluation of Disability Progression in Multiple Sclerosis via Magnetic-Resonance-Based Deep Learning Techniques. International Journal of Molecular Sciences. 2022; 23(18):10651. https://doi.org/10.3390/ijms231810651
Chicago/Turabian StyleTaloni, Alessandro, Francis Allen Farrelly, Giuseppe Pontillo, Nikolaos Petsas, Costanza Giannì, Serena Ruggieri, Maria Petracca, Arturo Brunetti, Carlo Pozzilli, Patrizia Pantano, and et al. 2022. "Evaluation of Disability Progression in Multiple Sclerosis via Magnetic-Resonance-Based Deep Learning Techniques" International Journal of Molecular Sciences 23, no. 18: 10651. https://doi.org/10.3390/ijms231810651
APA StyleTaloni, A., Farrelly, F. A., Pontillo, G., Petsas, N., Giannì, C., Ruggieri, S., Petracca, M., Brunetti, A., Pozzilli, C., Pantano, P., & Tommasin, S. (2022). Evaluation of Disability Progression in Multiple Sclerosis via Magnetic-Resonance-Based Deep Learning Techniques. International Journal of Molecular Sciences, 23(18), 10651. https://doi.org/10.3390/ijms231810651






