Ceramide Metabolism Regulated by Sphingomyelin Synthase 2 Is Associated with Acquisition of Chemoresistance via Exosomes in Human Leukemia Cells
Abstract
:1. Introduction
2. Results
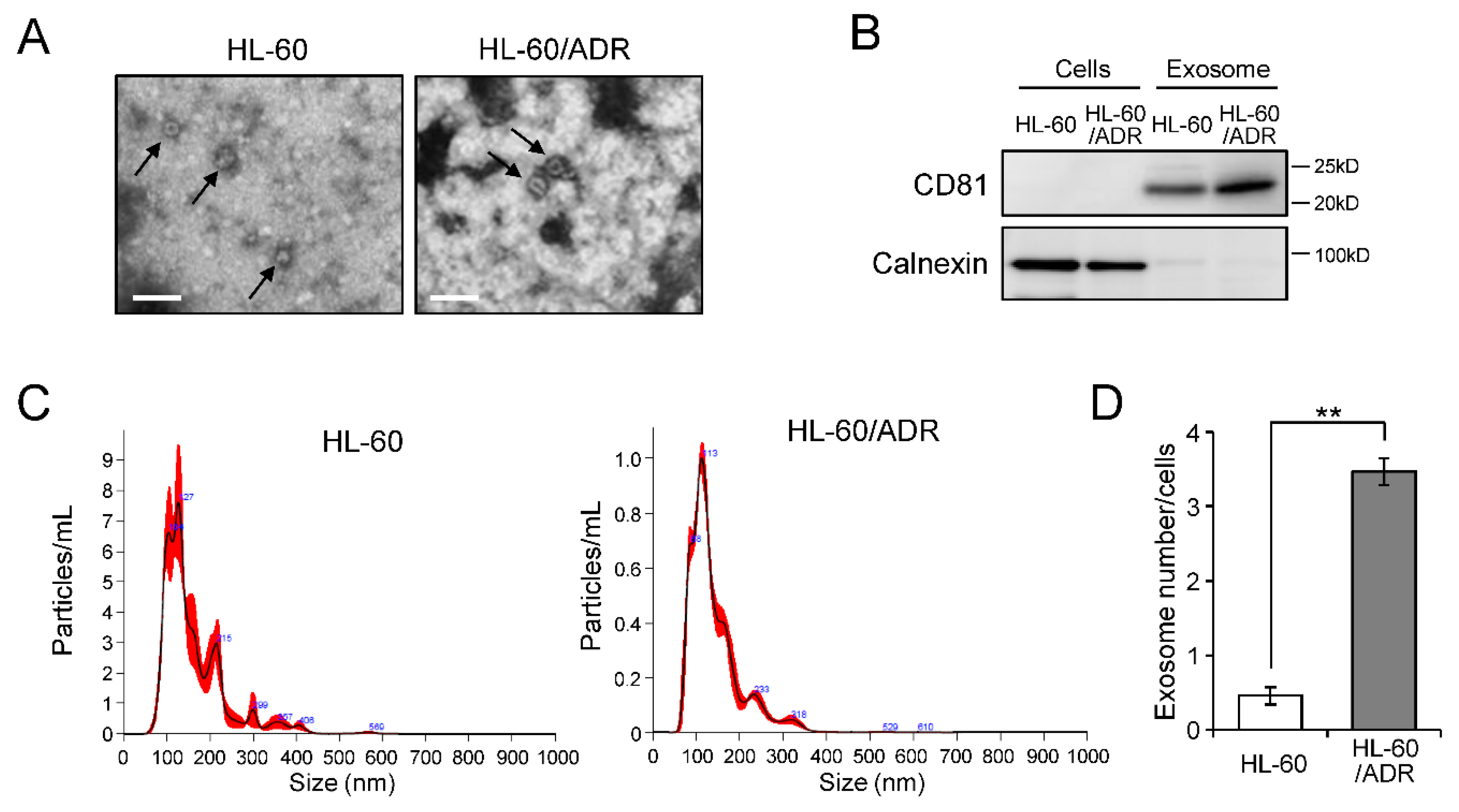
2.1. Exosome Release Is Promoted in Drug-Resistant Leukemia Cells
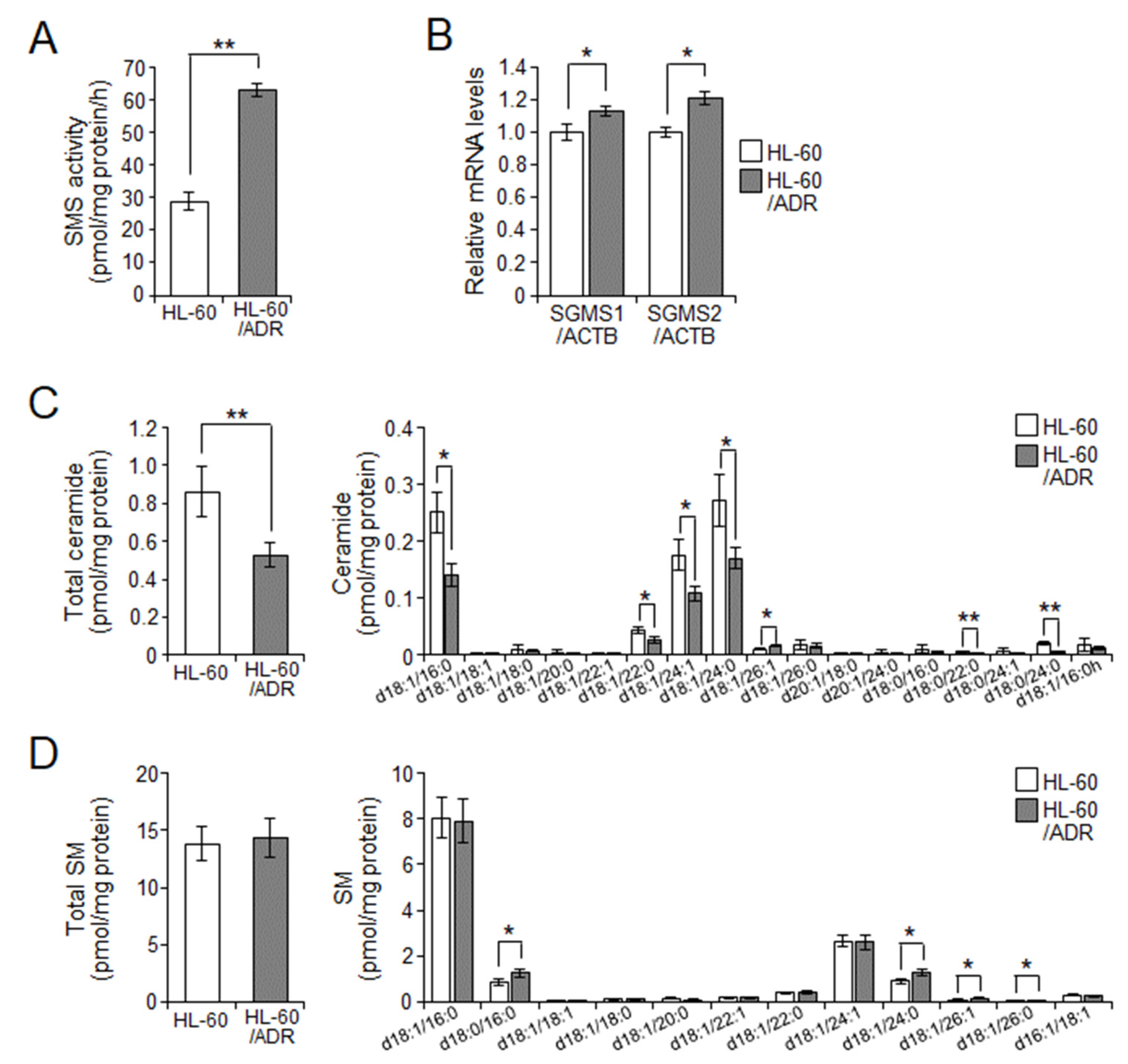
2.2. Cellular Ceramide Levels Are Related to Exosome Release in HL-60/ADR Cells
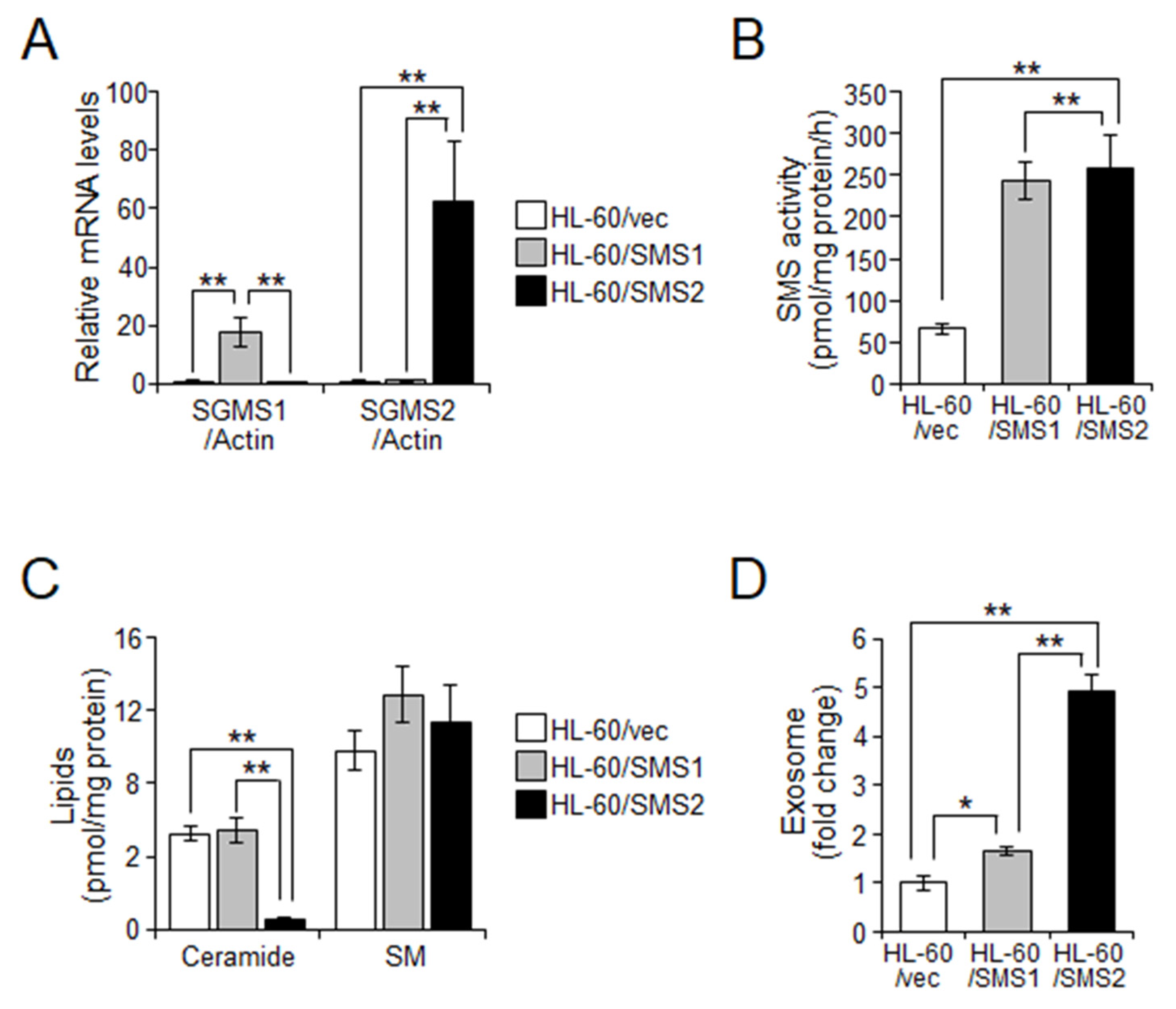
2.3. SMS2 Contributes to Exosome Release by Suppression of Ceramide Levels
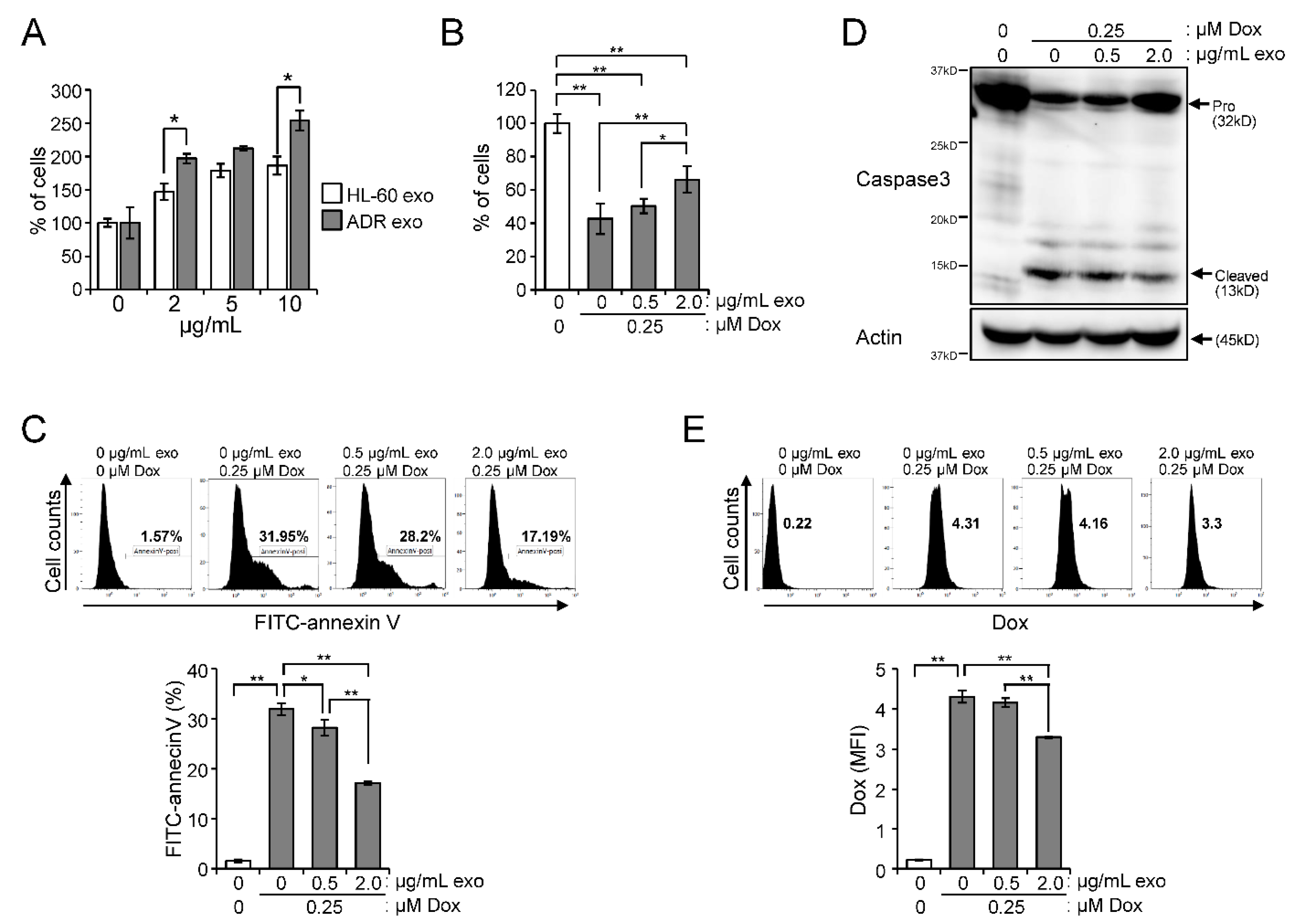
2.4. Effect of Exosomes Derived from HL-60/ADR Cells on Cell Proliferation and Drug Resistance
2.5. miR-484 Is Associated with HL-60 Cell Proliferation through Exosomes
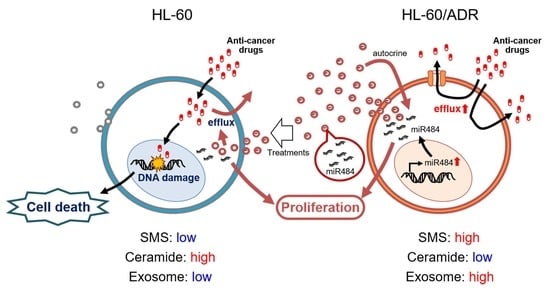
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Cell Culture
4.3. Exosome Preparation
4.4. Transmission Electron Microscopy
4.5. Analysis of Particle Number and Size
4.6. Western Blot Analysis
4.7. LC-MS/MS
4.8. Measurement of SMS and GCS Activity
4.9. Quantitative Real-Time PCR (qPCR)
4.10. Flow Cytometry
4.11. Microarray for miRNA
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ogretmen, B. Sphingolipid metabolism in cancer signalling and therapy. Nat. Rev. Cancer 2018, 18, 33–50. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, M.; Okazaki, T. Role of ceramide/sphingomyelin (SM) balance regulated through "SM cycle" in cancer. Cell. Signal. 2021, 87, 110119. [Google Scholar] [CrossRef] [PubMed]
- Hannun, Y.A.; Obeid, L.M. Sphingolipids and their metabolism in physiology and disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 175–191. [Google Scholar] [CrossRef] [PubMed]
- Itoh, M.; Kitano, T.; Watanabe, M.; Kondo, T.; Yabu, T.; Taguchi, Y.; Iwai, K.; Tashima, M.; Uchiyama, T.; Okazaki, T. Possible role of ceramide as an indicator of chemoresistance: Decrease of the ceramide content via activation of glucosylceramide synthase and sphingomyelin synthase in chemoresistant leukemia. Clin. Cancer Res. 2003, 9, 415–423. [Google Scholar]
- Maas, S.L.N.; Breakefield, X.O.; Weaver, A.M. Extracellular Vesicles: Unique Intercellular Delivery Vehicles. Trends Cell Biol. 2017, 27, 172–188. [Google Scholar] [CrossRef]
- van Balkom, B.W.; Eisele, A.S.; Pegtel, D.M.; Bervoets, S.; Verhaar, M.C. Quantitative and qualitative analysis of small RNAs in human endothelial cells and exosomes provides insights into localized RNA processing, degradation and sorting. J. Extracell. Vesicles 2015, 4, 26760. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Zomer, A.; van Rheenen, J. Implications of Extracellular Vesicle Transfer on Cellular Heterogeneity in Cancer: What Are the Potential Clinical Ramifications? Cancer Res. 2016, 76, 2071–2075. [Google Scholar] [CrossRef]
- Li, S.P.; Lin, Z.X.; Jiang, X.Y.; Yu, X.Y. Exosomal cargo-loading and synthetic exosome-mimics as potential therapeutic tools. Acta Pharmacol. Sin. 2018, 39, 542–551. [Google Scholar] [CrossRef]
- Skotland, T.; Sandvig, K.; Llorente, A. Lipids in exosomes: Current knowledge and the way forward. Prog. Lipid Res. 2017, 66, 30–41. [Google Scholar] [CrossRef]
- Skotland, T.; Hessvik, N.P.; Sandvig, K.; Llorente, A. Exosomal lipid composition and the role of ether lipids and phosphoinositides in exosome biology. J. Lipid Res. 2019, 60, 9–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trajkovic, K.; Hsu, C.; Chiantia, S.; Rajendran, L.; Wenzel, D.; Wieland, F.; Schwille, P.; Brugger, B.; Simons, M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 2008, 319, 1244–1247. [Google Scholar] [CrossRef] [PubMed]
- Kosaka, N.; Iguchi, H.; Yoshioka, Y.; Takeshita, F.; Matsuki, Y.; Ochiya, T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J. Biol. Chem. 2010, 285, 17442–17452. [Google Scholar] [CrossRef]
- Mittelbrunn, M.; Gutierrez-Vazquez, C.; Villarroya-Beltri, C.; Gonzalez, S.; Sanchez-Cabo, F.; Gonzalez, M.A.; Bernad, A.; Sanchez-Madrid, F. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat. Commun. 2011, 2, 282. [Google Scholar] [CrossRef]
- Yuyama, K.; Sun, H.; Mitsutake, S.; Igarashi, Y. Sphingolipid-modulated exosome secretion promotes clearance of amyloid-beta by microglia. J. Biol. Chem. 2012, 287, 10977–10989. [Google Scholar] [CrossRef] [PubMed]
- Kosaka, N.; Iguchi, H.; Hagiwara, K.; Yoshioka, Y.; Takeshita, F.; Ochiya, T. Neutral sphingomyelinase 2 (nSMase2)-dependent exosomal transfer of angiogenic microRNAs regulate cancer cell metastasis. J. Biol. Chem. 2013, 288, 10849–10859. [Google Scholar] [CrossRef]
- Cheng, Q.; Li, X.; Wang, Y.; Dong, M.; Zhan, F.H.; Liu, J. The ceramide pathway is involved in the survival, apoptosis and exosome functions of human multiple myeloma cells in vitro. Acta Pharmacol. Sin. 2018, 39, 561–568. [Google Scholar] [CrossRef]
- Liu, L.; Ye, Q.; Bihl, J.C.; Chen, Y.; Liu, J.; Cheng, Q. C6-ceramide treatment inhibits the proangiogenic activity of multiple myeloma exosomes via the miR-29b/Akt pathway. J. Transl. Med. 2020, 18, 298. [Google Scholar] [CrossRef]
- Elmallah, M.I.Y.; Ortega-Deballon, P.; Hermite, L.; Pais-De-Barros, J.P.; Gobbo, J.; Garrido, C. Lipidomic profiling of exosomes from colorectal cancer cells and patients reveals potential biomarkers. Mol. Oncol. 2022, 16, 2710–2718. [Google Scholar] [CrossRef]
- Taniguchi, M.; Okazaki, T. Ceramide/Sphingomyelin Rheostat Regulated by Sphingomyelin Synthases and Chronic Diseases in Murine Models. J. Lipid Atheroscler. 2020, 9, 380–405. [Google Scholar] [CrossRef]
- Kosaka, N.; Ochiya, T. Unraveling the Mystery of Cancer by Secretory microRNA: Horizontal microRNA Transfer between Living Cells. Front. Genet. 2011, 2, 97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melero-Fernandez de Mera, R.M.; Villasenor, A.; Rojo, D.; Carrion-Navarro, J.; Gradillas, A.; Ayuso-Sacido, A.; Barbas, C. Ceramide Composition in Exosomes for Characterization of Glioblastoma Stem-Like Cell Phenotypes. Front. Oncol. 2022, 11, 788100. [Google Scholar] [CrossRef] [PubMed]
- Senchenkov, A.; Litvak, D.A.; Cabot, M.C. Targeting ceramide metabolism--a strategy for overcoming drug resistance. J. Natl. Cancer Inst. 2001, 93, 347–357. [Google Scholar] [CrossRef] [PubMed]
- Shammout, O.D.A.; Ashmawy, N.S.; Shakartalla, S.B.; Altaie, A.M.; Semreen, M.H.; Omar, H.A.; Soliman, S.S.M. Comparative sphingolipidomic analysis reveals significant differences between doxorubicin-sensitive and -resistance MCF-7 cells. PLoS ONE 2021, 16, e0258363. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Tong, T.T.; Yau, L.F.; Chen, C.Y.; Mi, J.N.; Wang, J.R.; Jiang, Z.H. LC-MS Based Sphingolipidomic Study on A2780 Human Ovarian Cancer Cell Line and its Taxol-resistant Strain. Sci. Rep. 2016, 6, 34684. [Google Scholar] [CrossRef]
- Bataller, M.; Sanchez-Garcia, A.; Garcia-Mayea, Y.; Mir, C.; Rodriguez, I.; ME, L.L. The Role of Sphingolipids Metabolism in Cancer Drug Resistance. Front. Oncol. 2021, 11, 807636. [Google Scholar] [CrossRef]
- Jia, Y.Z.; Liu, J.; Wang, G.Q.; Song, Z.F. miR-484: A Potential Biomarker in Health and Disease. Front. Oncol. 2022, 12, 830420. [Google Scholar] [CrossRef]
- Li, Y.; Liu, Y.; Yao, J.; Li, R.; Fan, X. Downregulation of miR-484 is associated with poor prognosis and tumor progression of gastric cancer. Diagn. Pathol. 2020, 15, 25. [Google Scholar] [CrossRef]
- Liu, J.; Li, S.M. MiR-484 suppressed proliferation, migration, invasion and induced apoptosis of gastric cancer via targeting CCL-18. Int. J. Exp. Pathol. 2020, 101, 203–214. [Google Scholar] [CrossRef]
- Shen, Y.; Qi, L.; Li, Y.; Zhang, Y.; Gao, X.; Zhu, Y.; Wang, K. The Downregulation of lncRNA pgm5-as1 Inhibits the Proliferation and Metastasis Via Increasing miR-484 Expression in Colorectal Cancer. Cancer Biother. Radiopharm. 2021, 36, 220–229. [Google Scholar] [CrossRef]
- Xie, S.; Ge, Q.; Wang, X.; Sun, X.; Kang, Y. Long non-coding RNA ZFAS1 sponges miR-484 to promote cell proliferation and invasion in colorectal cancer. Cell Cycle 2018, 17, 154–161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, X.; Yue, M.; Li, C.; Sun, D.; Wang, L. Long Non-Coding RNA LINC00239 Functions as a Competitive Endogenous RNA by Sponging microRNA-484 and Enhancing KLF12 Expression to Promote the Oncogenicity of Colorectal Cancer. OncoTargets Ther. 2020, 13, 12067–12081. [Google Scholar] [CrossRef]
- Li, N.; Yang, G.; Luo, L.; Ling, L.; Wang, X.; Shi, L.; Lan, J.; Jia, X.; Zhang, Q.; Long, Z.; et al. lncRNA THAP9-AS1 Promotes Pancreatic Ductal Adenocarcinoma Growth and Leads to a Poor Clinical Outcome via Sponging miR-484 and Interacting with YAP. Clin. Cancer Res. 2020, 26, 1736–1748. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Han, T.; Hu, P.; Zhu, C.; Wang, Y.; Chang, S. Five microRNAs in serum as potential biomarkers for prostate cancer risk assessment and therapeutic intervention. Int. Urol. Nephrol. 2018, 50, 2193–2200. [Google Scholar] [CrossRef] [PubMed]
- Qiu, L.; Huang, Y.; Li, Z.; Dong, X.; Chen, G.; Xu, H.; Zeng, Y.; Cai, Z.; Liu, X.; Liu, J. Circular RNA profiling identifies circADAMTS13 as a miR-484 sponge which suppresses cell proliferation in hepatocellular carcinoma. Mol. Oncol. 2019, 13, 441–455. [Google Scholar] [CrossRef]
- Wang, S.; Wang, W.; Han, X.; Wang, Y.; Ge, Y.; Tan, Z. Dysregulation of miR484-TUSC5 axis takes part in the progression of hepatocellular carcinoma. J. Biochem. 2019, 166, 271–279. [Google Scholar] [CrossRef]
- Yang, Y.; Lin, X.; Lu, X.; Luo, G.; Zeng, T.; Tang, J.; Jiang, F.; Li, L.; Cui, X.; Huang, W.; et al. Interferon-microRNA signalling drives liver precancerous lesion formation and hepatocarcinogenesis. Gut 2016, 65, 1186–1201. [Google Scholar] [CrossRef]
- Zhuang, Z.; Sun, C.; Gong, H. High serum miR-484 expression is associated with the diagnosis and prognosis of patients with non-small cell lung cancer. Exp. Ther. Med. 2019, 18, 4095–4102. [Google Scholar] [CrossRef]
- Li, T.; Ding, Z.L.; Zheng, Y.L.; Wang, W. MiR-484 promotes non-small-cell lung cancer (NSCLC) progression through inhibiting Apaf-1 associated with the suppression of apoptosis. Biomed. Pharmacother. 2017, 96, 153–164. [Google Scholar] [CrossRef]
- Xue, X.; Wang, C.; Xue, Z.; Wen, J.; Han, J.; Ma, X.; Zang, X.; Deng, H.; Guo, R.; Asuquo, I.P.; et al. Exosomal miRNA profiling before and after surgery revealed potential diagnostic and prognostic markers for lung adenocarcinoma. Acta Biochim. Biophys. Sin. 2020, 52, 281–293. [Google Scholar] [CrossRef]
- Yi, R.; Feng, J.; Yang, S.; Huang, X.; Liao, Y.; Hu, Z.; Luo, M. miR-484/MAP2/c-Myc-positive regulatory loop in glioma promotes tumor-initiating properties through ERK1/2 signaling. J. Mol. Histol. 2018, 49, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Wu, A.; Liu, J.; Zhang, X.; Niu, C.; Shu, G.; Yin, G. Comprehensive network analysis of dysregulated genes revealed MNX1-AS1/hsa-miR-4697-3p/HOXB13 axis in ovarian cancer chemotherapy response. Cancer Sci. 2022, 113, 2627. [Google Scholar] [CrossRef] [PubMed]
- Li, D.F.; Yang, M.F.; Shi, S.L.; Du, Y.L.; Wang, H.; Zhou, Y.L.; Luo, Y.L.; Ren, L.L.; Nie, Y.Q. TM4SF5-CTD-2354A18.1-miR-4697-3P may play a key role in the pathogenesis of gastric cancer. Bratisl. Lek. Listy 2015, 116, 608–615. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Miao, Y.; Xiao, B.; Huan, R.; Jiang, Z.; Meng, D.; Wang, Y. Differential expression of microRNA species in human gastric cancer versus non-tumorous tissues. J. Gastroenterol. Hepatol. 2009, 24, 652–657. [Google Scholar] [CrossRef]
- Wu, Y.; Wan, X.; Ji, F.; Song, Z.; Fang, X. Serum miR-658 induces metastasis of gastric cancer by activating PAX3-MET pathway: A population-based study. Cancer Biomark. 2018, 22, 111–118. [Google Scholar] [CrossRef]
- Azuma, Y.; Yokobori, T.; Mogi, A.; Yajima, T.; Kosaka, T.; Iijima, M.; Shimizu, K.; Shirabe, K.; Kuwano, H. Cancer exosomal microRNAs from gefitinib-resistant lung cancer cells cause therapeutic resistance in gefitinib-sensitive cells. Surg. Today 2020, 50, 1099–1106. [Google Scholar] [CrossRef]
- Yang, G.; He, F.; Duan, H.; Shen, J.; Dong, Q. lncRNA FLVCR-AS1 promotes osteosarcoma growth by targeting miR381-3p/CCND1. OncoTargets Ther. 2020, 13, 163–172. [Google Scholar] [CrossRef]
- Kalhori, M.R.; Arefian, E.; Fallah Atanaki, F.; Kavousi, K.; Soleimani, M. miR-548x and miR-4698 controlled cell proliferation by affecting the PI3K/AKT signaling pathway in Glioblastoma cell lines. Sci. Rep. 2020, 10, 1558. [Google Scholar] [CrossRef]
- Yu, D.; Zhu, L.; Tu, H.; Wu, L.; Yang, H.; Xu, C. miR-4698-Trim59 axis plays a suppressive role in hepatocellular carcinoma. Front. Biosci. 2020, 25, 1120–1131. [Google Scholar]
- Zhao, J.; Li, X.; Fu, L.; Zhang, N.; Yang, J.; Cai, J. lncRNA LIFRAS1 inhibits gastric carcinoma cell proliferation, migration and invasion by sponging miR4698. Mol. Med. Rep. 2020, 23, 153. [Google Scholar] [CrossRef]
- Zhang, G.; Zheng, H.; Wang, L. miR4913p functions as a tumor suppressor in nonsmall cell lung cancer by targeting fibroblast growth factor 5. Oncol. Rep. 2022, 48, 164. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Luo, S. Low Expression of miR-491-3p Is Correlated with Lymph Node Metastasis in Gastric Cancer. Evid.-Based Complement. Altern. Med. 2022, 2022, 7807956. [Google Scholar] [CrossRef] [PubMed]
- Sueta, A.; Fujiki, Y.; Goto-Yamaguchi, L.; Tomiguchi, M.; Yamamoto-Ibusuki, M.; Iwase, H.; Yamamoto, Y. Exosomal miRNA profiles of triple-negative breast cancer in neoadjuvant treatment. Oncol. Lett. 2021, 22, 819. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Li, C.; Li, H.; Changyong, E. LncRNA CACNA1G-AS1 facilitates hepatocellular carcinoma progression through the miR-2392/C1orf61 pathway. J. Cell. Physiol. 2019, 234, 18415–18422. [Google Scholar] [CrossRef]
- Sun, B.; Ji, W.; Liu, C.; Lin, X.; Chen, L.; Qian, H.; Su, C. miR-2392 functions as tumour suppressor and inhibits malignant progression of hepatocellular carcinoma via directly targeting JAG2. Liver Int. 2022, 42, 1658–1673. [Google Scholar] [CrossRef]
- Li, J.; Li, T.; Lu, Y.; Shen, G.; Guo, H.; Wu, J.; Lei, C.; Du, F.; Zhou, F.; Zhao, X.; et al. MiR-2392 suppresses metastasis and epithelial-mesenchymal transition by targeting MAML3 and WHSC1 in gastric cancer. FASEB J. 2017, 31, 3774–3786. [Google Scholar] [CrossRef]
- Moffat, J.; Grueneberg, D.A.; Yang, X.; Kim, S.Y.; Kloepfer, A.M.; Hinkle, G.; Piqani, B.; Eisenhaure, T.M.; Luo, B.; Grenier, J.K.; et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell 2006, 124, 1283–1298. [Google Scholar] [CrossRef]
- Uchida, Y.; Itoh, M.; Taguchi, Y.; Yamaoka, S.; Umehara, H.; Ichikawa, S.; Hirabayashi, Y.; Holleran, W.M.; Okazaki, T. Ceramide reduction and transcriptional up-regulation of glucosylceramide synthase through doxorubicin-activated Sp1 in drug-resistant HL-60/ADR cells. Cancer Res. 2004, 64, 6271–6279. [Google Scholar] [CrossRef]
- Shakor, A.B.A.; Taniguchi, M.; Kitatani, K.; Hashimoto, M.; Asano, S.; Hayashi, A.; Nomura, K.; Bielawski, J.; Bielawska, A.; Watanabe, K.; et al. Sphingomyelin synthase 1-generated sphingomyelin plays an important role in transferrin trafficking and cell proliferation. J. Biol. Chem. 2011, 286, 36053–36062. [Google Scholar] [CrossRef]
- Yamanaka, T.; Furukawa, T.; Matsumoto-Mashimo, C.; Yamane, K.; Sugimori, C.; Nambu, T.; Mori, N.; Nishikawa, H.; Walker, C.B.; Leung, K.P.; et al. Gene expression profile and pathogenicity of biofilm-forming Prevotella intermedia strain 17. BMC Microbiol. 2009, 9, 11. [Google Scholar] [CrossRef]
- Ogiso, H.; Taniguchi, M.; Araya, S.; Aoki, S.; Wardhani, L.O.; Yamashita, Y.; Ueda, Y.; Okazaki, T. Comparative Analysis of Biological Sphingolipids with Glycerophospholipids and Diacylglycerol by LC-MS/MS. Metabolites 2014, 4, 98–114. [Google Scholar] [CrossRef] [PubMed]
- Ogiso, H.; Taniguchi, M.; Okazaki, T. Analysis of lipid-composition changes in plasma membrane microdomains. J. Lipid Res. 2015, 56, 1594–1605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taniguchi, M.; Ogiso, H.; Takeuchi, T.; Kitatani, K.; Umehara, H.; Okazaki, T. Lysosomal ceramide generated by acid sphingomyelinase triggers cytosolic cathepsin B-mediated degradation of X-linked inhibitor of apoptosis protein in natural killer/T lymphoma cell apoptosis. Cell Death Dis. 2015, 6, e1717. [Google Scholar] [CrossRef] [PubMed]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, M.; Ueda, Y.; Matsushita, M.; Nagaya, S.; Hashizume, C.; Arai, K.; Kabayama, K.; Fukase, K.; Watanabe, K.; Wardhani, L.O.; et al. Deficiency of sphingomyelin synthase 2 prolongs survival by the inhibition of lymphoma infiltration through ICAM-1 reduction. FASEB J. 2020, 34, 3838–3854. [Google Scholar] [CrossRef]
- Fujita, J.; Taniguchi, M.; Hashizume, C.; Ueda, Y.; Sakai, S.; Kondo, T.; Hashimoto-Nishimura, M.; Hanada, K.; Kosaka, T.; Okazaki, T. Nuclear Ceramide Is Associated with Ataxia Telangiectasia Mutated Activation in the Neocarzinostatin-Induced Apoptosis of Lymphoblastoid Cells. Mol. Pharmacol. 2022, 101, 322–333. [Google Scholar] [CrossRef]
- Sampson, A.; Peterson, B.G.; Tan, K.W.; Iram, S.H. Doxorubicin as a fluorescent reporter identifies novel MRP1 (ABCC1) inhibitors missed by calcein-based high content screening of anticancer agents. Biomed. Pharmacother. 2019, 118, 109289. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Taniguchi, M.; Nagaya, S.; Yuyama, K.; Kotani, A.; Igarashi, Y.; Okazaki, T. Ceramide Metabolism Regulated by Sphingomyelin Synthase 2 Is Associated with Acquisition of Chemoresistance via Exosomes in Human Leukemia Cells. Int. J. Mol. Sci. 2022, 23, 10648. https://doi.org/10.3390/ijms231810648
Taniguchi M, Nagaya S, Yuyama K, Kotani A, Igarashi Y, Okazaki T. Ceramide Metabolism Regulated by Sphingomyelin Synthase 2 Is Associated with Acquisition of Chemoresistance via Exosomes in Human Leukemia Cells. International Journal of Molecular Sciences. 2022; 23(18):10648. https://doi.org/10.3390/ijms231810648
Chicago/Turabian StyleTaniguchi, Makoto, Shingo Nagaya, Kohei Yuyama, Ai Kotani, Yasuyuki Igarashi, and Toshiro Okazaki. 2022. "Ceramide Metabolism Regulated by Sphingomyelin Synthase 2 Is Associated with Acquisition of Chemoresistance via Exosomes in Human Leukemia Cells" International Journal of Molecular Sciences 23, no. 18: 10648. https://doi.org/10.3390/ijms231810648
APA StyleTaniguchi, M., Nagaya, S., Yuyama, K., Kotani, A., Igarashi, Y., & Okazaki, T. (2022). Ceramide Metabolism Regulated by Sphingomyelin Synthase 2 Is Associated with Acquisition of Chemoresistance via Exosomes in Human Leukemia Cells. International Journal of Molecular Sciences, 23(18), 10648. https://doi.org/10.3390/ijms231810648