Association of Polymorphic Variants in Argonaute Genes with Depression Risk in a Polish Population
Abstract
:1. Introduction
2. Results
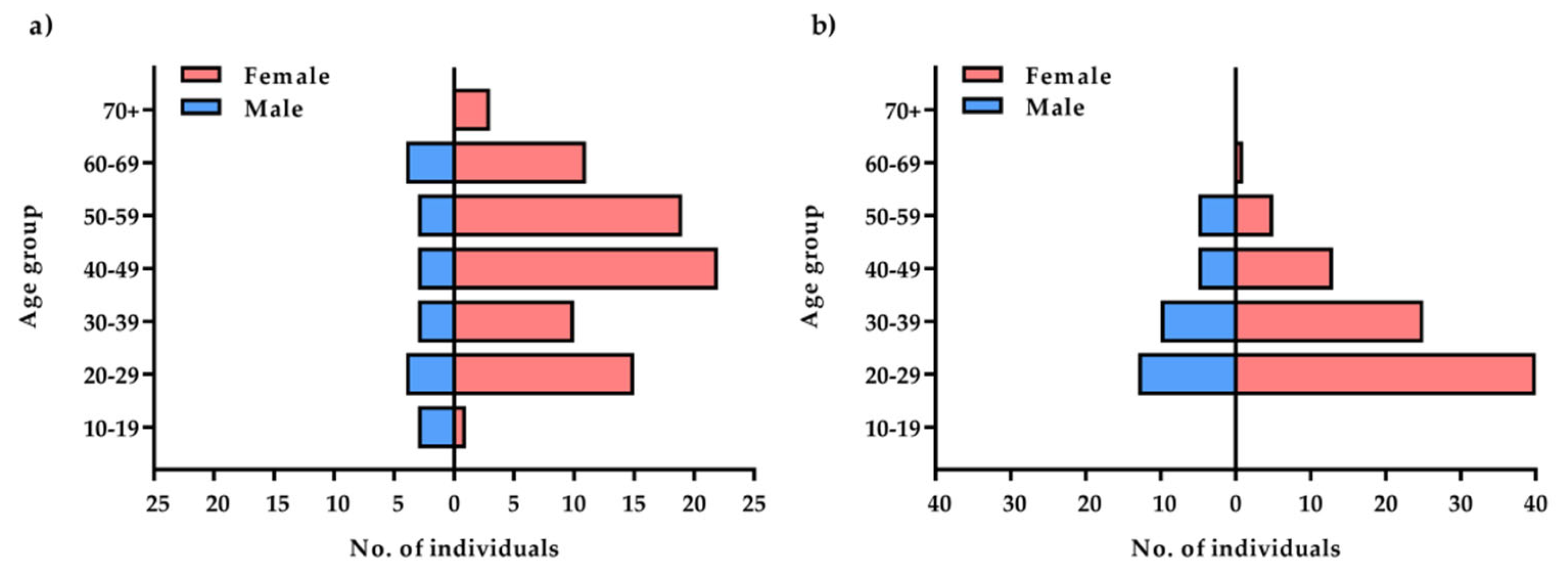
2.1. Characteristics of the Studied Groups
2.2. Analysis of a Relationship between the Occurrence of Depression and the Studied Polymorphic Variants of AGO Genes
3. Discussion
4. Materials and Methods
4.1. Study and Control Groups
4.2. Molecular Methods
4.2.1. DNA Isolation
4.2.2. Determination of Single-Nucleotide Polymorphisms (SNPs)
4.2.3. Statistical Analysis
5. Conclusions
Limitations and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Uher, R.; Payne, J.L.; Pavlova, B.; Perlis, R.H. Major depressive disorder in DSM-5: Implications for clinical practice and research of changes from DSM-IV. Depress. Anxiety 2014, 31, 459–471. [Google Scholar] [CrossRef] [PubMed]
- Friedrich, M.J. Depression is the leading cause of disability around the world. JAMA 2017, 317, 1517. [Google Scholar] [CrossRef] [PubMed]
- Malhi, G.S.; Mann, J.J. Depression. Lancet 2018, 392, 2299–2312. [Google Scholar] [CrossRef]
- Lin, C.-C.; Huang, T.-L. Brain-derived neurotrophic factor and mental disorders. Biomed. J. 2020, 43, 134–142. [Google Scholar] [CrossRef]
- Hsieh, M.-T.; Lin, C.-C.; Lee, C.-T.; Huang, T.-L. Abnormal brain-derived neurotrophic factor exon IX promoter methylation, protein, and mRNA levels in patients with major depressive disorder. J. Clin. Med. 2019, 8, 568. [Google Scholar] [CrossRef]
- Hammar, A.; Ardal, G. Cognitive functioning in major depression—A summary. Front. Hum. Neurosci. 2009, 3, 26. [Google Scholar] [CrossRef]
- Martos, D.; Tuka, B.; Tanaka, M.; Vécsei, L.; Telegdy, G. Memory enhancement with kynurenic acid and its mechanisms in neurotransmission. Biomedicines 2022, 10, 849. [Google Scholar] [CrossRef]
- Perini, G.; Cotta Ramusino, M.; Sinforiani, E.; Bernini, S.; Petrachi, R.; Costa, A. Cognitive impairment in depression: Recent advances and novel treatments. Neuropsychiatr. Dis. Treat. 2019, 15, 1249–1258. [Google Scholar] [CrossRef]
- Talarowska, M.; Galecki, P. Cognition and emotions in recurrent depressive disorders—The role of inflammation and the kynureninepathway. Curr. Pharm. Des. 2016, 22, 955–962. [Google Scholar] [CrossRef]
- Konarski, J.Z.; McIntyre, R.S.; Kennedy, S.H.; Rafi-Tari, S.; Soczynska, J.K.; Ketter, T.A. Volumetric neuroimaging investigations in mood disorders: Bipolar disorder versus major depressive disorder. Bipolar Disord. 2008, 10, 1–37. [Google Scholar] [CrossRef]
- Groenewold, N.A.; Opmeer, E.M.; de Jonge, P.; Aleman, A.; Costafreda, S.G. Emotional valence modulates brain functional abnormalities in depression: Evidence from a meta-analysis of fMRI studies. Neurosci. Biobehav. Rev. 2013, 37, 152–163. [Google Scholar] [CrossRef]
- Gałecki, P.; Talarowska, M. The evolutionary theory of depression. Med. Sci. Monit. 2017, 23, 2267–2274. [Google Scholar] [CrossRef]
- Levy, M.J.F.; Boulle, F.; Steinbusch, H.W.; van den Hove, D.L.A.; Kenis, G.; Lanfumey, L. Neurotrophic factors and neuroplasticity pathways in the pathophysiology and treatment of depression. Psychopharmacology 2018, 235, 2195–2220. [Google Scholar] [CrossRef]
- Battaglia, S. Neurobiological advances of learned fear in humans. Adv. Clin. Exp. Med. 2022, 31, 217–221. [Google Scholar] [CrossRef]
- Battaglia, S.; Thayer, J.F. Functional interplay between central and autonomic nervous systems in human fear conditioning. Trends Neurosci. 2022, 45, 504–506. [Google Scholar] [CrossRef]
- Talarowska, M.; Wysiadecki, G.; Chodkiewicz, J. Affective neuroscience personality scales and early maladaptive schemas in depressive disorders. Int. J. Environ. Res. Public Health 2022, 19, 8062. [Google Scholar] [CrossRef]
- Liu, N.; Wang, Z.-Z.; Zhao, M.; Zhang, Y.; Chen, N.-H. Role of non-coding RNA in the pathogenesis of depression. Gene 2020, 735, 144276. [Google Scholar] [CrossRef]
- Serafini, G.; Pompili, M.; Hansen, K.F.; Obrietan, K.; Dwivedi, Y.; Shomron, N.; Girardi, P. The involvement of microRNAs in major depression, suicidal behavior, and related disorders: A focus on miR-185 and miR-491-3p. Cell. Mol. Neurobiol. 2014, 34, 17–30. [Google Scholar] [CrossRef]
- Serafini, G.; Pompili, M.; Innamorati, M.; Giordano, G.; Montebovi, F.; Sher, L.; Dwivedi, Y.; Girardi, P. The role of microRNAs in synaptic plasticity, major affective disorders and suicidal behavior. Neurosci. Res. 2012, 73, 179–190. [Google Scholar] [CrossRef]
- Dwivedi, Y. MicroRNAs in depression and suicide: Recent insights and future perspectives. J. Affect. Disord. 2018, 240, 146–154. [Google Scholar] [CrossRef]
- Mattick, J.S.; Makunin, I. V Non-coding RNA. Hum. Mol. Genet. 2006, 15, R17–R29. [Google Scholar] [CrossRef]
- Shi, Y.; Wang, Q.; Song, R.; Kong, Y.; Zhang, Z. Non-coding RNAs in depression: Promising diagnostic and therapeutic biomarkers. EBioMedicine 2021, 71, 103569. [Google Scholar] [CrossRef]
- Bayne, E.H.; Allshire, R.C. RNA-directed transcriptional gene silencing in mammals. Trends Genet. 2005, 21, 370–373. [Google Scholar] [CrossRef]
- Filipowicz, W. RNAi: The nuts and bolts of the RISC machine. Cell 2005, 122, 17–20. [Google Scholar] [CrossRef]
- Pratt, A.J.; MacRae, I.J. The RNA-induced silencing complex: A versatile gene-silencing machine. J. Biol. Chem. 2009, 284, 17897–17901. [Google Scholar] [CrossRef]
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef]
- Dueck, A.; Meister, G. Assembly and function of small RNA—Argonaute protein complexes. Biol. Chem. 2014, 395, 611–629. [Google Scholar] [CrossRef]
- Niaz, S. The AGO proteins: An overview. Biol. Chem. 2018, 399, 525–547. [Google Scholar] [CrossRef]
- Jonas, S.; Izaurralde, E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 2015, 16, 421–433. [Google Scholar] [CrossRef]
- Capitão, C.; Paiva, J.A.P.; Santos, D.M.; Fevereiro, P. In Medicago truncatula, water deficit modulates the transcript accumulation of components of small RNA pathways. BMC Plant Biol. 2011, 11, 79. [Google Scholar] [CrossRef] [Green Version]
- Xu, D.; Yang, H.; Zou, C.; Li, W.-X.; Xu, Y.; Xie, C. Identification and functional characterization of the AGO1 ortholog in maize. J. Integr. Plant Biol. 2016, 58, 749–758. [Google Scholar] [CrossRef] [PubMed]
- Rajman, M.; Schratt, G. MicroRNAs in neural development: From master regulators to fine-tuners. Development 2017, 144, 2310–2322. [Google Scholar] [CrossRef] [PubMed]
- Schratt, G. microRNAs at the synapse. Nat. Rev. Neurosci. 2009, 10, 842–849. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Carmell, M.A.; Rivas, F.V.; Marsden, C.G.; Thomson, J.M.; Song, J.-J.; Hammond, S.M.; Joshua-Tor, L.; Hannon, G.J. Argonaute2 is the catalytic engine of mammalian RNAi. Science 2004, 305, 1437–1441. [Google Scholar] [CrossRef]
- Lessel, D.; Zeitler, D.M.; Reijnders, M.R.F.; Kazantsev, A.; Hassani Nia, F.; Bartholomäus, A.; Martens, V.; Bruckmann, A.; Graus, V.; McConkie-Rosell, A.; et al. Germline AGO2 mutations impair RNA interference and human neurological development. Nat. Commun. 2020, 11, 5797. [Google Scholar] [CrossRef]
- Li, H.-F.; Yu, X.; Yang, K.; He, C.-Y.; Kou, S.-J.; Cao, S.-X.; Xie, G.-R. The relationship between single nucleotide polymorphisms in 5-HT2A signal transduction-related genes and the response efficacy to selective serotonin reuptake inhibitor treatments in Chinese patients with major depressive disorder. Genet. Test. Mol. Biomarkers 2012, 16, 667–671. [Google Scholar] [CrossRef]
- Katsuki, A.; Kakeda, S.; Watanabe, K.; Igata, R.; Otsuka, Y.; Kishi, T.; Nguyen, L.; Ueda, I.; Iwata, N.; Korogi, Y.; et al. A single-nucleotide polymorphism influences brain morphology in drug-naïve patients with major depressive disorder. Neuropsychiatr. Dis. Treat. 2019, 15, 2425–2432. [Google Scholar] [CrossRef]
- Wigner, P.; Czarny, P.; Synowiec, E.; Bijak, M.; Białek, K.; Talarowska, M.; Galecki, P.; Szemraj, J.; Sliwinski, T. Association between single nucleotide polymorphisms of TPH1 and TPH2 genes, and depressive disorders. J. Cell. Mol. Med. 2018, 22, 1778–1791. [Google Scholar] [CrossRef]
- Fokkema, R.W.; Korsten, P.; Schmoll, T.; Wilson, A.J. Social competition as a driver of phenotype-environment correlations: Implications for ecology and evolution. Biol. Rev. Camb. Philos. Soc. 2021, 96, 2561–2572. [Google Scholar] [CrossRef]
- He, Y.; Zhou, Y.; Xi, Q.; Cui, H.; Luo, T.; Song, H.; Nie, X.; Wang, L.; Ying, B. Genetic variations in microRNA processing genes are associated with susceptibility in depression. DNA Cell Biol. 2012, 31, 1499–1506. [Google Scholar] [CrossRef]
- Bodak, M.; Cirera-Salinas, D.; Luitz, J.; Ciaudo, C. The Role of RNA interference in stem cell biology: Beyond the mutant phenotypes. J. Mol. Biol. 2017, 429, 1532–1543. [Google Scholar] [CrossRef]
- Bartel, D.P. Metazoan microRNAs. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef]
- Djuranovic, S.; Nahvi, A.; Green, R. A parsimonious model for gene regulation by miRNAs. Science 2011, 331, 550–553. [Google Scholar] [CrossRef]
- Nawalpuri, B.; Ravindran, S.; Muddashetty, R.S. The role of dynamic miRISC during neuronal development. Front. Mol. Biosci. 2020, 7, 8. [Google Scholar] [CrossRef]
- Ferreira, R.; Santos, T.; Amar, A.; Gong, A.; Chen, T.C.; Tahara, S.M.; Giannotta, S.L.; Hofman, F.M. Argonaute-2 promotes miR-18a entry in human brain endothelial cells. J. Am. Heart Assoc. 2014, 3, e000968. [Google Scholar] [CrossRef]
- Lopez, J.P.; Fiori, L.M.; Gross, J.A.; Labonte, B.; Yerko, V.; Mechawar, N.; Turecki, G. Regulatory role of miRNAs in polyamine gene expression in the prefrontal cortex of depressed suicide completers. Int. J. Neuropsychopharmacol. 2014, 17, 23–32. [Google Scholar] [CrossRef]
- Li, J.; Meng, H.; Cao, W.; Qiu, T. MiR-335 is involved in major depression disorder and antidepressant treatment through targeting GRM4. Neurosci. Lett. 2015, 606, 167–172. [Google Scholar] [CrossRef]
- Li, Y.-J.; Xu, M.; Gao, Z.-H.; Wang, Y.-Q.; Yue, Z.; Zhang, Y.-X.; Li, X.-X.; Zhang, C.; Xie, S.-Y.; Wang, P.-Y. Alterations of serum levels of BDNF-related miRNAs in patients with depression. PLoS ONE 2013, 8, e63648. [Google Scholar]
- Lian, N.; Niu, Q.; Lei, Y.; Li, X.; Li, Y.; Song, X. MiR-221 is involved in depression by regulating Wnt2/CREB/BDNF axis in hippocampal neurons. Cell Cycle 2018, 17, 2745–2755. [Google Scholar] [CrossRef]
- Bai, M.; Zhu, X.; Zhang, Y.; Zhang, S.; Zhang, L.; Xue, L.; Yi, J.; Yao, S.; Zhang, X. Abnormal hippocampal BDNF and miR-16 expression is associated with depression-like behaviors induced by stress during early life. PLoS ONE 2012, 7, e46921. [Google Scholar] [CrossRef]
- Fang, Y.; Qiu, Q.; Zhang, S.; Sun, L.; Li, G.; Xiao, S.; Li, X. Changes in miRNA-132 and miR-124 levels in non-treated and citalopram-treated patients with depression. J. Affect. Disord. 2018, 227, 745–751. [Google Scholar] [CrossRef] [PubMed]
- Lopez, J.P.; Fiori, L.M.; Cruceanu, C.; Lin, R.; Labonte, B.; Cates, H.M.; Heller, E.A.; Vialou, V.; Ku, S.M.; Gerald, C.; et al. MicroRNAs 146a/b-5 and 425-3p and 24-3p are markers of antidepressant response and regulate MAPK/Wnt-system genes. Nat. Commun. 2017, 8, 15497. [Google Scholar] [CrossRef]
- Liu, J.; Valencia-Sanchez, M.A.; Hannon, G.J.; Parker, R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 2005, 7, 719–723. [Google Scholar] [CrossRef]
- Sen, G.L.; Blau, H.M. Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat. Cell Biol. 2005, 7, 633–636. [Google Scholar] [CrossRef]
- Oh, J.-Y.; Kwon, A.; Jo, A.; Kim, H.; Goo, Y.-S.; Lee, J.-A.; Kim, H.K. Activity-dependent synaptic localization of processing bodies and their role in dendritic structural plasticity. J. Cell Sci. 2013, 126, 2114–2123. [Google Scholar] [CrossRef]
- Cougot, N.; Bhattacharyya, S.N.; Tapia-Arancibia, L.; Bordonné, R.; Filipowicz, W.; Bertrand, E.; Rage, F. Dendrites of mammalian neurons contain specialized P-body-like structures that respond to neuronal activation. J. Neurosci. 2008, 28, 13793–13804. [Google Scholar] [CrossRef]
- Zeitelhofer, M.; Karra, D.; Macchi, P.; Tolino, M.; Thomas, S.; Schwarz, M.; Kiebler, M.; Dahm, R. Dynamic interaction between P-bodies and transport ribonucleoprotein particles in dendrites of mature hippocampal neurons. J. Neurosci. 2008, 28, 7555–7562. [Google Scholar] [CrossRef]
- Patranabis, S.; Bhattacharyya, S.N. Phosphorylation of Ago2 and subsequent inactivation of let-7a RNP-specific microRNAs control differentiation of mammalian sympathetic neurons. Mol. Cell. Biol. 2016, 36, 1260–1271. [Google Scholar] [CrossRef]
- Da Silva, S.K.; Wiener, C.; Ghisleni, G.; Oses, J.P.; Jansen, K.; Molina, M.L.; Silva, R.; Souza, L.D. Effects of cognitive-behavioral therapy on neurotrophic factors in patients with major depressive disorder. Braz. J. Psychiatry 2018, 40, 361–366. [Google Scholar] [CrossRef]
- Gershoni-Emek, N.; Altman, T.; Ionescu, A.; Costa, C.J.; Gradus-Pery, T.; Willis, D.E.; Perlson, E. Localization of RNAi machinery to axonal branch points and growth cones is facilitated by mitochondria and is disrupted in ALS. Front. Mol. Neurosci. 2018, 11, 311. [Google Scholar] [CrossRef]
- Faul, F.; Erdfelder, E.; Lang, A.-G.; Buchner, A. G*Power 3: A flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav. Res. Methods 2007, 39, 175–191. [Google Scholar] [CrossRef] [PubMed]
- WHO. ICD-10 Classification of Mental and Behavioural Disorders; WHO: Geneva, Switzerland, 1992.
- Martin, C.; Hunter, L.; Vinood Patel, V.; Preedy, V.; Rajendram, R. The Neuroscience of Depression. Features, Diagnosis, and Treatment; Elsevier: Amsterdam, The Netherlands, 2021. [Google Scholar]
| Polymorphism/Gene | p-Value * | ||
|---|---|---|---|
| Totality | Control Group | Study Group | |
| rs636882/AGO1 | 0.15 | 0.45 | 0.11 |
| rs2292779/AGO2 | 0.00071 | 0. 0097 | 0.03 |
| rs2977490/AGO2 | 0.87 | 0.66 | 0.36 |
| rs4961280/AGO2 | 0.87 | 0.84 | 0.59 |
| Polymorphism/Gene | Model | Genotype | Control Group | Study Group | OR (95% CI) | p-Value * |
|---|---|---|---|---|---|---|
| rs636882/AGO1 | Codominant | G/G | 87 (75%) | 77 (75.5%) | 1.00 | 0.82 |
| G/C | 26 (22.4%) | 21 (20.6%) | 0.91 (0.48–1.75) | |||
| C/C | 3 (2.6%) | 4 (3.9%) | 1.51 (0.33–6.94) | |||
| Dominant | G/G | 87 (75%) | 77 (75.5%) | 1.00 | 0.93 | |
| G/C-C/C | 29 (25%) | 25 (24.5%) | 0.97 (0.53–1.80) | |||
| Recessive | G/G-G/C | 113 (97.4%) | 98 (96.1%) | 1.00 | 0.58 | |
| C/C | 3 (2.6%) | 4 (3.9%) | 1.54 (0.34–7.04) | |||
| Overdominant | G/G-C/C | 90 (77.6%) | 81 (79.4%) | 1.00 | 0.74 | |
| G/C | 26 (22.4%) | 21 (20.6%) | 0.90 (0.47–1.72) | |||
| rs2292779/AGO2 | Codominant | G/G | 39 (33.6%) | 30 (29.4%) | 1.00 | 0.79 |
| C/G | 44 (37.9%) | 40 (39.2%) | 1.18 (0.62–2.24) | |||
| C/C | 33 (28.4%) | 32 (31.4%) | 1.26 (0.64–2.49) | |||
| Dominant | C/G | 39 (33.6%) | 30 (29.4%) | 1.00 | 0.5 | |
| C/G-C/C | 77 (66.4%) | 72 (70.6%) | 1.22 (0.68–2.16) | |||
| Recessive | G/G-C/G | 83 (71.5%) | 70 (68.6%) | 1.00 | 0.64 | |
| C/C | 33 (28.4%) | 32 (31.4%) | 1.15 (0.64–2.06) | |||
| Overdominant | G/G-C/C | 72 (62.1%) | 62 (60.8%) | 1.00 | 0.85 | |
| C/G | 44 (37.9%) | 40 (39.2%) | 1.06 (0.61–1.82) | |||
| rs2977490/AGO2 | Codominant | G/G | 55 (47.4%) | 49 (48%) | 1.00 | 0.42 |
| G/A | 52 (44.8%) | 40 (39.2%) | 0.68 (0.49–1.52) | |||
| A/A | 9 (7.8%) | 13 (12.8%) | 1.62 (0.64–4.12) | |||
| Dominant | G/G | 55 (47.4%) | 49 (48%) | 1.00 | 0.93 | |
| G/A-A/A | 61 (52.6%) | 53 (52%) | 0.98 (0.57–1.66) | |||
| Recessive | G/G-G/A | 107 (92.2%) | 89 (87.2%) | 1.00 | 0.22 | |
| A/A | 9 (7.8%) | 13 (12.8%) | 1.74 (0.71–4.25) | |||
| Overdominant | G/G-A/A | 64 (55.2%) | 62 (60.8%) | 1.00 | 0.4 | |
| G/A | 52 (44.8%) | 40 (39.2%) | 0.79 (0.46–1.36) | |||
| rs4961280/AGO2 | Codominant | C/C | 48 (41.4%) | 60 (58.4%) | 1.00 | 0.034 |
| C/A | 55 (47.4%) | 35 (34.3%) | 0.51 (0.29–0.90) | |||
| A/A | 13 (11.2%) | 7 (6.9%) | 0.43 (0.16–1.16) | |||
| Dominant | C/C | 48 (41.4%) | 60 (58.8%) | 1.00 | 0.01 | |
| C/A-A/A | 68 (58.6%) | 42 (41.2%) | 0.49 (0.25–0.85) | |||
| Recessive | C/C-C/A | 103 (88.8%) | 95 (93.1%) | 1.00 | 0.26 | |
| A/A | 13 (11.2%) | 7 (6.9%) | 0.58 (0.22–1.53) | |||
| Overdominant | C/C-A/A | 61 (52.6%) | 67 (65.7%) | 1.00 | 0.049 | |
| C/A | 55 (47.4%) | 35 (34.3%) | 0.58 (0.34–1.00) |
| Groups | Variables | ||
|---|---|---|---|
| Age (Years) Mean ± SD | Sex n (%) | ||
| Female | Male | ||
| Study group (n = 101) | 44.3 ± 19.1 | 81 (80.2) | 20 (19.8) |
|
Control group (n = 117) | 33.2 ± 9.1 | 83 (70.9) | 34 (29.1) |
|
All (n = 218) | 39.8 ± 14.0 | 164 (75.2) | 54 (24.8) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kowalczyk, M.; Kowalczyk, E.; Galita, G.; Majsterek, I.; Talarowska, M.; Popławski, T.; Kwiatkowski, P.; Lichota, A.; Sienkiewicz, M. Association of Polymorphic Variants in Argonaute Genes with Depression Risk in a Polish Population. Int. J. Mol. Sci. 2022, 23, 10586. https://doi.org/10.3390/ijms231810586
Kowalczyk M, Kowalczyk E, Galita G, Majsterek I, Talarowska M, Popławski T, Kwiatkowski P, Lichota A, Sienkiewicz M. Association of Polymorphic Variants in Argonaute Genes with Depression Risk in a Polish Population. International Journal of Molecular Sciences. 2022; 23(18):10586. https://doi.org/10.3390/ijms231810586
Chicago/Turabian StyleKowalczyk, Mateusz, Edward Kowalczyk, Grzegorz Galita, Ireneusz Majsterek, Monika Talarowska, Tomasz Popławski, Paweł Kwiatkowski, Anna Lichota, and Monika Sienkiewicz. 2022. "Association of Polymorphic Variants in Argonaute Genes with Depression Risk in a Polish Population" International Journal of Molecular Sciences 23, no. 18: 10586. https://doi.org/10.3390/ijms231810586
APA StyleKowalczyk, M., Kowalczyk, E., Galita, G., Majsterek, I., Talarowska, M., Popławski, T., Kwiatkowski, P., Lichota, A., & Sienkiewicz, M. (2022). Association of Polymorphic Variants in Argonaute Genes with Depression Risk in a Polish Population. International Journal of Molecular Sciences, 23(18), 10586. https://doi.org/10.3390/ijms231810586








