The Role of microRNAs in the Gonocyte Theory as Target of Malignancy: Looking for Potential Diagnostic Biomarkers
Abstract
1. Introduction
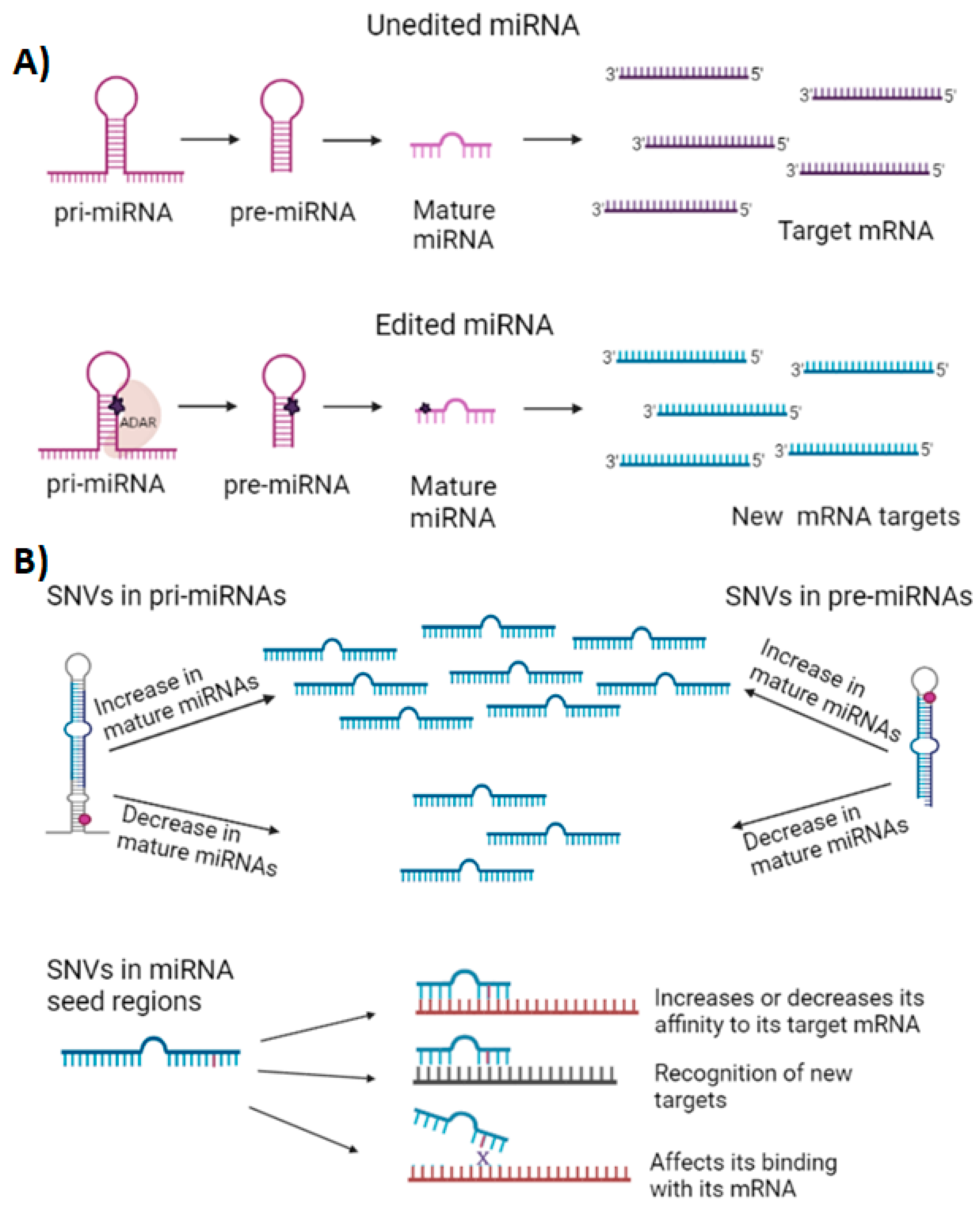
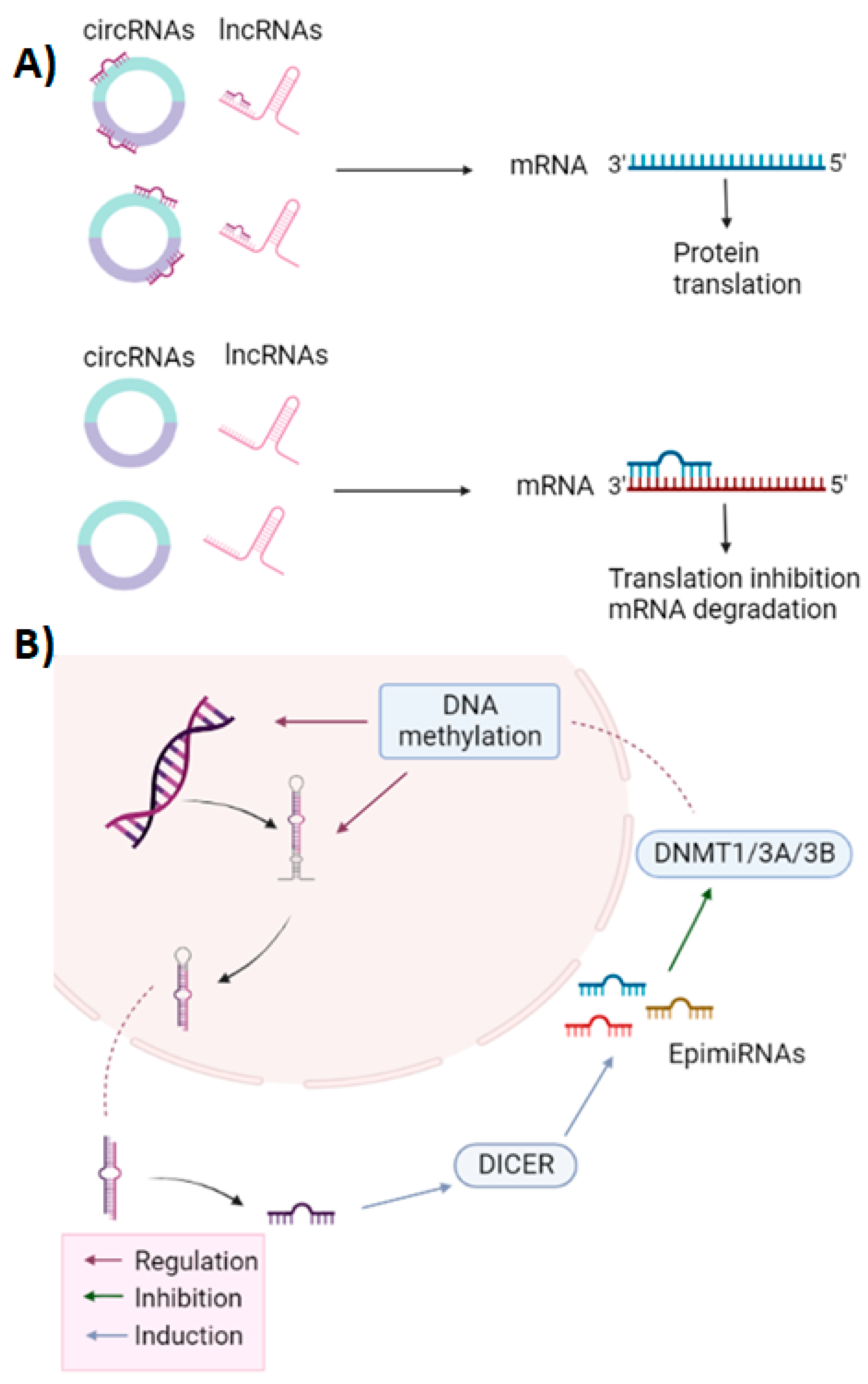
2. General Characteristics of miRNAs
3. From the Gonocyte to TGCT
3.1. miRNAs Expressed in Primordial Germinal Cells/Gonocytes
3.2. miRNAs Expressed in Cryptorchidism
3.3. miRNAs Expressed in Germ Cell Carcinoma In Situ
3.4. miRNAs Expressed in Testicular Germ Cell Tumors
4. miRNAs as Potential Serum Markers of TGCT
5. Application of Therapies Using miRNAs
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Palazzo, A.F.; Gregory, T.R. The Case for Junk DNA. PLoS Genet. 2014, 10, e1004351. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M. The Human Transcriptome: An Unfinished Story. Genes 2012, 3, 344–360. [Google Scholar] [CrossRef] [PubMed]
- Saliminejad, K.; Khorram Khorshid, H.R.; Soleymani Fard, S.; Ghaffari, S.H. An overview of microRNAs: Biology, functions, therapeutics, and analysis methods. J. Cell. Physiol. 2019, 234, 5451–5465. [Google Scholar] [CrossRef]
- Keller, A.; Gröger, L.; Tschernig, T.; Solomon, J.; Laham, O.; Schaum, N.; Wagner, V.; Kern, F.; Schmartz, G.P.; Li, Y.; et al. miRNATissueAtlas2: An update to the human miRNA tissue atlas. Nucleic Acids Res. 2021, 50, D211–D221. [Google Scholar] [CrossRef]
- Green, C.D.; Ma, Q.; Manske, G.L.; Shami, A.N.; Zheng, X.; Marini, S.; Moritz, L.; Sultan, C.; Gurczynski, S.J.; Moore, B.; et al. A Comprehensive Roadmap of Murine Spermatogenesis Defined by Single-Cell RNA-Seq. Dev. Cell 2018, 46, 651–667.e10. [Google Scholar] [CrossRef]
- Wang, Y.; Li, X.; Gong, X.; Zhao, Y.; Wu, J. MicroRNA-322 Regulates Self-renewal of Mouse Spermatogonial Stem Cells through Rassf8. Int. J. Biol. Sci. 2019, 15, 857–869. [Google Scholar] [CrossRef]
- Vigueras-Villaseñor, R.M.; Cortés-Trujillo, L.; Chávez-Saldaña, M.; Vázquez, F.G.; Carrasco-Daza, D.; Cuevas-Alpuche, O.; Rojas-Castañeda, J.C. Analysis of POU5F1, c-Kit, PLAP, AP2γ and SALL4 in gonocytes of patients with cryptorchidism. Acta Histochem. 2015, 117, 752–761. [Google Scholar] [CrossRef] [PubMed]
- Looijenga, L.H.J.; Gillis, A.J.M.; Stoop, H.; Biermann, K.; Oosterhuis, J.W. Dissecting the molecular pathways of (testicular) germ cell tumour pathogenesis; from initiation to treatment-resistance. Int. J. Androl. 2011, 34, e234–e251. [Google Scholar] [CrossRef]
- Meyts, E.R.-D. Developmental model for the pathogenesis of testicular carcinoma in situ: Genetic and environmental aspects. Hum. Reprod. Updat. 2006, 12, 303–323. [Google Scholar] [CrossRef]
- Rodríguez, P.Y.; Godoy, J.I. Tumor de células germinales. Revista Med. 2008, 16, 200–214. Available online: http://www.scielo.org.co/scielo.php?script=sci_arttext&pid=S0121-52562008000200008&lng=en&tlng=es (accessed on 12 April 2022).
- Almstrup, K.; Nielsen, J.E.; Mlynarska, O.; Jansen, M.T.; Jorgensen, A.; Skakkebæk, N.E.; Meyts, E.R.-D. Carcinoma in situ testis displays permissive chromatin modifications similar to immature foetal germ cells. Br. J. Cancer 2010, 103, 1269–1276. [Google Scholar] [CrossRef] [PubMed]
- Gegenschatz-Schmid, K.; Verkauskas, G.; Stadler, M.B.; Hadziselimovic, F. Genes located in Y-chromosomal regions important for male fertility show altered transcript levels in cryptorchidism and respond to curative hormone treatment. Basic Clin. Androl. 2019, 29, 8. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, D.G.; Skakkebk, N.E.; Meyts, E.R.-D.; Almstrup, K. Epigenetic features of testicular germ cell tumours in relation to epigenetic characteristics of foetal germ cells. Int. J. Dev. Biol. 2013, 57, 309–317. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.; Huang, Z.; He, X.; Wu, H.; Peng, D.; Zhang, L.; Zhang, X. Altered miRNA profile in testis of post-cryptorchidopexy patients with non-obstructive azoospermia. Reprod. Biol. Endocrinol. 2018, 16, 78. [Google Scholar] [CrossRef] [PubMed]
- Novotny, G.; Belling, K.; Bramsen, J.B.; Nielsen, J.E.; Bork-Jensen, J.; Almstrup, K.; Sonne, S.B.; Kjems, J.; Meyts, E.R.-D.; Leffers, H. MicroRNA expression profiling of carcinoma in situ cells of the testis. Endocr.-Relat. Cancer 2012, 19, 365–379. [Google Scholar] [CrossRef]
- McIver, S.C.; Stanger, S.J.; Santarelli, D.M.; Roman, S.D.; Nixon, B.; McLaughlin, E.A. A Unique Combination of Male Germ Cell miRNAs Coordinates Gonocyte Differentiation. PLoS ONE 2012, 7, e35553. [Google Scholar] [CrossRef]
- Gillis, A.J.M.; Stoop, H.J.; Hersmus, R.; Oosterhuis, J.W.; Sun, Y.; Chen, C.; Guenther, S.; Sherlock, J.; Veltman, I.; Baeten, J.; et al. High-throughput microRNAome analysis in human germ cell tumours. J. Pathol. 2007, 213, 319–328. [Google Scholar] [CrossRef]
- Murray, M.J.; Bell, E.; Raby, K.L.; Rijlaarsdam, M.; Gillis, A.J.M.; Looijenga, L.; Brown, H.; Destenaves, B.; Nicholson, J.C.; Coleman, N.S. A pipeline to quantify serum and cerebrospinal fluid microRNAs for diagnosis and detection of relapse in paediatric malignant germ-cell tumours. Br. J. Cancer 2015, 114, 151–162. [Google Scholar] [CrossRef]
- van Agthoven, T.; Looijenga, L.H. Accurate primary germ cell cancer diagnosis using serum based microRNA detection (ampTSmiR test). Oncotarget 2016, 8, 58037–58049. [Google Scholar] [CrossRef]
- Yao, S. MicroRNA biogenesis and their functions in regulating stem cell potency and differentiation. Biol. Proced. Online 2016, 18, 8. [Google Scholar] [CrossRef]
- Bartel, D.P. Metazoan MicroRNAs. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef] [PubMed]
- Harchegani, A.B.; Shafaghatian, H.; Tahmasbpour, E.; Shahriary, A. Regulatory Functions of MicroRNAs in Male Reproductive Health: A New Approach to Understanding Male Infertility; Reproductive Sciences: Thousand Oaks, CA, USA, 2018; pp. 1–11. [Google Scholar] [CrossRef]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef] [PubMed]
- Donati, S.; Ciuffi, S.; Brandi, M.L. Human Circulating miRNAs Real-time qRT-PCR-based Analysis: An Overview of Endogenous Reference Genes Used for Data Normalization. Int. J. Mol. Sci. 2019, 20, 4353. [Google Scholar] [CrossRef] [PubMed]
- Steiman-Shimony, A.; Shtrikman, O.; Margalit, H. Assessing the functional association of intronic miRNAs with their host genes. RNA 2018, 24, 991–1004. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Luo, J.; Zhang, H.; Lu, J. microRNAs in the Same Clusters Evolve to Coordinately Regulate Functionally Related Genes. Mol. Biol. Evol. 2016, 33, 2232–2247. [Google Scholar] [CrossRef] [PubMed]
- DeVeale, B.; Swindlehurst-Chan, J.; Blelloch, R. The roles of microRNAs in mouse development. Nat. Rev. Genet. 2021, 22, 307–323. [Google Scholar] [CrossRef]
- Catalanotto, C.; Cogoni, C.; Zardo, G. MicroRNA in Control of Gene Expression: An Overview of Nuclear Functions. Int. J. Mol. Sci. 2016, 17, 1712. [Google Scholar] [CrossRef]
- de Sousa, M.C.; Gjorgjieva, M.; Dolicka, D.; Sobolewski, C.; Foti, M. Deciphering miRNAs’ Action through miRNA Editing. Int. J. Mol. Sci. 2019, 20, 6249. [Google Scholar] [CrossRef]
- Wang, Y.; Ru, J.; Meng, X.; Song, J.; Jiang, Q.; Li, S.; Jiang, J.; Li, Y. Role of SNPs in the Biogenesis of Mature miRNAs. BioMed Res. Int. 2021, 2021, 2403418. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.; Li, J.-L.; Marchica, J.; Mercola, M.; Patel, V.; Perera, R.J. Mapping genetic variability in mature miRNAs and miRNA binding sites in prostate cancer. J. Hum. Genet. 2021, 66, 1127–1137. [Google Scholar] [CrossRef]
- Sun, X.; Liu, J.; Xu, C.; Tang, S.; Ren, H. The insights of Let-7 miRNAs in oncogenesis and stem cell potency. J. Cell. Mol. Med. 2016, 20, 1779–1788. [Google Scholar] [CrossRef] [PubMed]
- Pajares, M.; Alemany-Cosme, E.; Goñi, S.; Bandres, E.; Palanca-Ballester, C.; Sandoval, J. Epigenetic Regulation of microRNAs in Cancer: Shortening the Distance from Bench to Bedside. Int. J. Mol. Sci. 2021, 22, 7350. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.J.; Tay, Y. Noncoding RNA:RNA Regulatory Networks in Cancer. Int. J. Mol. Sci. 2018, 19, 1310. [Google Scholar] [CrossRef] [PubMed]
- Ravnik-Glavač, M.; Glavač, D. Circulating RNAs as Potential Biomarkers in Amyotrophic Lateral Sclerosis. Int. J. Mol. Sci. 2020, 21, 1714. [Google Scholar] [CrossRef]
- Jin, W.; Wang, J.; Liu, C.-P.; Wang, H.-W.; Xu, R.-M. Structural Basis for pri-miRNA Recognition by Drosha. Mol. Cell 2020, 78, 423–433.e5. [Google Scholar] [CrossRef]
- Ali Syeda, Z.; Langden, S.S.S.; Munkhzul, C.; Lee, M.; Song, S.J. Regulatory Mechanism of MicroRNA Expression in Cancer. Int. J. Mol. Sci. 2020, 21, 1723. [Google Scholar] [CrossRef]
- Esquela-Kerscher, A.; Slack, F. Oncomirs—MicroRNAs with a role in cancer. Nat. Cancer 2006, 6, 259–269. [Google Scholar] [CrossRef]
- Alečković, M.; Kang, Y. Regulation of cancer metastasis by cell-free miRNAs. Biochim. Biophys. Acta 2014, 1855, 24–42. [Google Scholar] [CrossRef]
- Chen, X.; Li, X.; Guo, J.; Zhang, P.; Zeng, W. The roles of microRNAs in regulation of mammalian spermatogenesis. J. Anim. Sci. Biotechnol. 2017, 8, 35. [Google Scholar] [CrossRef]
- Yin, Y.; Zhu, L.; Li, Q.; Zhou, P.; Ma, L. Cullin4 E3 Ubiquitin Ligases Regulate Male Gonocyte Migration, Proliferation and Blood-Testis Barrier Homeostasis. Cells 2021, 10, 2732. [Google Scholar] [CrossRef]
- Wang, R.; Liu, X.; Li, L.; Yang, M.; Yong, J.; Zhai, F.; Wen, L.; Yan, L.; Qiao, J.; Tang, F. Dissecting human gonadal cell lineage specification and sex determination using a single-cell RNA-seq approach. Genom. Proteom. Bioinform. 2022. [Google Scholar] [CrossRef]
- Hadziselimovic, F. Temperature is not a major factor in the differentiation of gonocytes into ad spermatogonia and fertility outcome in congenitally cryptorchid boys. Basic Clin. Androl. 2022, 32, 2. [Google Scholar] [CrossRef] [PubMed]
- Eini, R.; Dorssers, L.C.J.; Looijenga, L.H.J. Role of stem cell proteins and microRNAs in embryogenesis and germ cell cancer. Int. J. Dev. Biol. 2013, 57, 319–332. [Google Scholar] [CrossRef]
- Chen, L.; Heikkinen, L.; Knott, K.E.; Liang, Y.; Wong, G. Evolutionary conservation and function of the human embryonic stem cell specific miR-302/367 cluster. Comp. Biochem. Physiol. Part D Genom. Proteom. 2015, 16, 83–98. [Google Scholar] [CrossRef]
- Wu, S.; Aksoy, M.; Shi, J.; Houbaviy, H.B. Evolution of the miR-290–295/miR-371–373 Cluster Family Seed Repertoire. PLoS ONE 2014, 9, e108519. [Google Scholar] [CrossRef]
- Hamer, G.; De Rooij, D.G. Mutations causing specific arrests in the development of mouse primordial germ cells and gonocytes. Biol. Reprod. 2018, 99, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Lewis, B.P.; Shih, I.-H.; Jones-Rhoades, M.W.; Bartel, D.P.; Burge, C.B. Prediction of Mammalian MicroRNA Targets. Cell 2003, 115, 787–798. [Google Scholar] [CrossRef]
- Hayashi, K.; Lopes, S.M.C.D.S.; Kaneda, M.; Tang, F.; Hajkova, P.; Lao, K.; O’Carroll, D.; Das, P.P.; Tarakhovsky, A.; Miska, E.; et al. MicroRNA Biogenesis Is Required for Mouse Primordial Germ Cell Development and Spermatogenesis. PLoS ONE 2008, 3, e1738. [Google Scholar] [CrossRef]
- Rosa, A.; Brivanlou, A.H. A regulatory circuitry comprised of miR-302 and the transcription factors OCT4 and NR2F2 regulates human embryonic stem cell differentiation. EMBO J. 2010, 30, 237–248. [Google Scholar] [CrossRef]
- Tan, M.H.; Au, K.F.; Leong, D.E.; Foygel, K.; Wong, W.H.; Yao, M.W. An Oct4-Sall4-Nanog network controls developmental progression in the pre-implantation mouse embryo. Mol. Syst. Biol. 2013, 9, 632. [Google Scholar] [CrossRef]
- Chen, E.Y.Y.; Chen, J.S.; Ying, S.-Y. The microRNA and the perspectives of miR-302. Heliyon 2019, 5, e01167. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, N.; Ishii, H.; Nagano, H.; Haraguchi, N.; Dewi, D.L.; Kano, Y.; Nishikawa, S.; Tanemura, M.; Mimori, K.; Tanaka, F.; et al. Reprogramming of Mouse and Human Cells to Pluripotency Using Mature MicroRNAs. Cell Stem Cell 2011, 8, 633–638. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Ding, X.; Nie, S.; Li-Ling, J.; Zhang, Y.; Zhang, H.; Chen, L.; Li, L.; Ding, M. Association of hsa-miR-145 overexpression in human testicular cells with male infertility. Mol. Med. Rep. 2015, 11, 4365–4372. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tay, Y.; Zhang, J.; Thomson, A.M.; Lim, B.; Rigoutsos, I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature 2008, 455, 1124–1128, Erratum in Nature 2009, 458, 538. [Google Scholar] [CrossRef]
- Brieño-Enríquez, M.A.; García-López, J.; Cárdenas, D.B.; Guibert, S.; Cleroux, E.; Děd, L.; Hourcade, J.D.D.; Pěknicová, J.; Weber, M.; del Mazo, J. Exposure to Endocrine Disruptor Induces Transgenerational Epigenetic Deregulation of MicroRNAs in Primordial Germ Cells. PLoS ONE 2015, 10, e0124296. [Google Scholar] [CrossRef]
- Verkauskas, G.; Malcius, D.; Dasevicius, D.; Hadziselimovic, F. Histopathology of Unilateral Cryptorchidism. Pediatr. Dev. Pathol. 2018, 22, 53–58. [Google Scholar] [CrossRef]
- Sarila, G.; Hutson, J.M.; Vikraman, J. Testicular descent: A review of a complex, multistaged process to identify potential hidden causes of UDT. J. Pediatr. Surg. 2021, 57, 479–487. [Google Scholar] [CrossRef]
- Boisen, K.; Kaleva, M.; Main, K.; Virtanen, H.; Haavisto, A.-M.; Schmidt, I.; Chellakooty, M.; Damgaard, I.; Mau, C.; Reunanen, M.; et al. Difference in prevalence of congenital cryptorchidism in infants between two Nordic countries. Lancet 2004, 363, 1264–1269. [Google Scholar] [CrossRef]
- Fénichel, P.; Chevalier, N.; Lahlou, N.; Coquillard, P.; Wagner-Mahler, K.; Pugeat, M.; Panaïa-Ferrari, P.; Brucker-Davis, F. Endocrine Disrupting Chemicals Interfere With Leydig Cell Hormone Pathways During Testicular Descent in Idiopathic Cryptorchidism. Front. Endocrinol. 2019, 9, 786. [Google Scholar] [CrossRef]
- Gurney, J.K.; McGlynn, K.A.; Stanley, J.; Merriman, T.; Signal, V.; Shaw, C.; Edwards, R.; Richiardi, L.; Hutson, J.; Sarfati, D. Risk factors for cryptorchidism. Nat. Rev. Urol. 2017, 14, 534–548. [Google Scholar] [CrossRef]
- Loebenstein, M.; Thorup, J.; Cortes, D.; Clasen-Linde, E.; Hutson, J.M.; Li, R. Cryptorchidism, gonocyte development, and the risks of germ cell malignancy and infertility: A systematic review. J. Pediatr. Surg. 2020, 55, 1201–1210. [Google Scholar] [CrossRef] [PubMed]
- Dieckmann, K.-P.; Pichlmeier, U. Clinical epidemiology of testicular germ cell tumors. World J. Urol. 2004, 22, 2–14. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, J.A.; Siegmund, H.I.; Roesch, W.; Hadziselimovic, F.; Hofstaedter, F. Male Infertility: Assessment of Juvenile Testicular Dysfunction and Risk for Malignancy in Cryptorchid Boys Based on Resin Section Evaluation. Ultrastruct. Pathol. 2013, 37, 373–377. [Google Scholar] [CrossRef] [PubMed]
- Hadziselimović, F. Cryptorchidism. Ultrastructure of normal and cryptorchid testis development. Adv. Anat. Embryol. Cell Biol. 1977, 53, 3–71. [Google Scholar] [CrossRef] [PubMed]
- Yue, S.S.; Hutson, J.M.; Li, R. Gene expression during gonocyte transformation into spermatogonial stem cells is not androgen dependent. J. Pediatr. Surg. 2015, 50, 2090–2093. [Google Scholar] [CrossRef]
- Su, S.; Szarek, M.; Vooght, A.; Hutson, J.; Li, R. Gonocyte transformation to spermatogonial stem cells occurs earlier in patients with undervirilisation syndromes. J. Pediatr. Surg. 2014, 49, 323–327. [Google Scholar] [CrossRef]
- Wikström, A.M.; Hoei-Hansen, C.E.; Dunkel, L.; Meyts, E.R.-D. Immunoexpression of Androgen Receptor and Nine Markers of Maturation in the Testes of Adolescent Boys with Klinefelter Syndrome: Evidence for Degeneration of Germ Cells at the Onset of Meiosis. J. Clin. Endocrinol. Metab. 2006, 92, 714–719. [Google Scholar] [CrossRef]
- Hadziselimovic, F.; Verkauskas, G.; Vincel, B.; Stadler, M.B. Testicular expression of long non-coding RNAs is affected by curative GnRHa treatment of cryptorchidism. Basic Clin. Androl. 2019, 29, 18. [Google Scholar] [CrossRef]
- Zheng, F.; Li, J.; Ma, C.; Tang, X.; Tang, Q.; Wu, J.; Chai, X.; Xie, J.; Yang, X.; Hann, S.S. Novel regulation of miR-34a-5p and HOTAIR by the combination of berberine and gefitinib leading to inhibition of EMT in human lung cancer. J. Cell. Mol. Med. 2020, 24, 5578–5592. [Google Scholar] [CrossRef]
- Zhao, W.; Geng, D.; Li, S.; Chen, Z.; Sun, M. LncRNA HOTAIR influences cell growth, migration, invasion, and apoptosis via the miR-20a-5p/HMGA2 axis in breast cancer. Cancer Med. 2018, 7, 842–855. [Google Scholar] [CrossRef]
- Yuan, S.; Luan, X.; Chen, H.; Shi, X.; Zhang, X. Long non-coding RNA EGFR-AS1 sponges micorRNA-381 to upregulate ROCK2 in bladder cancer. Oncol. Lett. 2020, 19, 1899–1905. [Google Scholar] [CrossRef] [PubMed]
- Hoei-Hansen, C.E.; Meyts, E.R.-D.; Daugaard, G.; Skakkebaek, N.E. Carcinoma in situ testis, the progenitor of testicular germ cell tumours: A clinical review. Ann. Oncol. 2005, 16, 863–868. [Google Scholar] [CrossRef] [PubMed]
- Skakkebæk, N.E.; Berthelsen, J.G.; Giwercman, A.; Müller, J. Carcinoma-in-situ of the testis: Possible origin from gonocytes and precursor of all types of germ cell tumours except spermatocytoma. Int. J. Androl. 1987, 10, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Nowak, R.; Grzybowska, E.A.; Wilczynska, A.; Pykało, R.; Siedlecki, J. Low Expression of DNA Polymerase β in Human Testicular Germ Cell Tumours: Impact on Foetal Gonocytic Origin Theory. Acta Oncol. 2002, 41, 188–191. [Google Scholar] [CrossRef]
- Skakkebaek, N.E.; Rajpert-De Meyts, E.; Buck Louis, G.M.; Toppari, J.; Andersson, A.-M.; Eisenberg, M.L.; Jensen, T.K.; Jørgensen, N.; Swan, S.H.; Sapra, K.J.; et al. Male Reproductive Disorders and Fertility Trends: Influences of Environment and Genetic Susceptibility. Physiol. Rev. 2016, 96, 55–97. [Google Scholar] [CrossRef]
- Huang, Z.; Tang, D.; Gao, J.; Dou, X.; Cheng, P.; Peng, D.; Zhang, Y.; Mao, J.; Zhang, L.; Zhang, X. miR-34c disrupts spermatogonial stem cell homeostasis in cryptorchid testes by targeting Nanos2. Reprod. Biol. Endocrinol. RB&E 2018, 16, 97. [Google Scholar] [CrossRef]
- Duan, Z.; Huang, H.; Sun, F. The functional and predictive roles of miR-210 in cryptorchidism. Sci. Rep. 2016, 6, 32265. [Google Scholar] [CrossRef]
- Jia, H.; Ma, T.; Jia, S.; Ouyang, Y. AKT3 and related molecules as potential biomarkers responsible for cryptorchidism and cryptorchidism-induced azoospermia. Transl. Pediatr. 2021, 10, 1805–1817. [Google Scholar] [CrossRef]
- Lv, W.; Yu, M.; Su, Y. miR-22-5p regulates the self-renewal of spermatogonial stem cells by targeting EZH2. Open Med. 2022, 17, 556–565. [Google Scholar] [CrossRef]
- Iida, N.; Takemura, K.; Ito, M.; Funata, N.; Yonese, I.; Suzuki, H.; Tsuzuki, T.; Koga, F. Primary Testicular Neuroendocrine Tumor Coexisting With Seminoma Sharing Germ Cell Origin. Int. J. Surg. Pathol. 2021, 29, 887–891. [Google Scholar] [CrossRef]
- Fink, C.; Baal, N.; Wilhelm, J.; Sarode, P.; Weigel, R.; Schumacher, V.; Nettersheim, D.; Schorle, H.; Schröck, C.; Bergmann, M.; et al. On the origin of germ cell neoplasia in situ: Dedifferentiation of human adult Sertoli cells in cross talk with seminoma cells in vitro. Neoplasia 2021, 23, 731–742. [Google Scholar] [CrossRef] [PubMed]
- Lobo, J.; van Zogchel, L.M.J.; Nuru, M.G.; Gillis, A.J.M.; van der Schoot, C.E.; Tytgat, G.A.M.; Looijenga, L.H.J. Combining Hypermethylated RASSF1A Detection Using ddPCR with miR-371a-3p Testing: An Improved Panel of Liquid Biopsy Biomarkers for Testicular Germ Cell Tumor Patients. Cancers 2021, 13, 5228. [Google Scholar] [CrossRef] [PubMed]
- Novotny, G.; Sonne, S.B.; Nielsen, J.E.; Jonstrup, S.P.; Hansen, M.A.; Skakkebaek, N.E.; Meyts, E.R.-D.; Kjems, J.; Leffers, H. Translational repression of E2F1 mRNA in carcinoma in situ and normal testis correlates with expression of the miR-17-92 cluster. Cell Death Differ. 2007, 14, 879–882. [Google Scholar] [CrossRef] [PubMed]
- Batool, A.; Karimi, N.; Wu, X.-N.; Chen, S.-R.; Liu, Y.-X. Testicular germ cell tumor: A comprehensive review. Cell. Mol. Life Sci. 2019, 76, 1713–1727. [Google Scholar] [CrossRef] [PubMed]
- Huerta, D.A.L.; Vigueras-Villaseñor, R.M.; Yokoyama-Rebollar, E.; García-Andrade, F.; Rojas-Castañeda, J.C.; Herrera-Montalvo, L.A.; Díaz-Chávez, J.; Pérez-Añorve, I.X.; Aréchaga-Ocampo, E.; Chávez-Saldaña, M.D. Cryptorchidism and Testicular Tumor: Comprehensive Analysis of Common Clinical Features and Search of SNVs in the KIT and AR Genes. Front. Cell Dev. Biol. 2020, 8, 762. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Devesa, S.S.; McGlynn, K.A.; Moller, H. Interpreting the international trends in testicular seminoma and nonseminoma incidence. Nat. Clin. Pract. Urol. 2006, 3, 532–543. [Google Scholar] [CrossRef]
- Bumbasirevic, U.; Bojanic, N.; Pljesa-Ercegovac, M.; Zivkovic, M.; Djukic, T.; Zekovic, M.; Milojevic, B.; Kajmakovic, B.; Janicic, A.; Simic, T.; et al. The Polymorphisms of Genes Encoding Catalytic Antioxidant Proteins Modulate the Susceptibility and Progression of Testicular Germ Cell Tumor. Cancers 2022, 14, 1068. [Google Scholar] [CrossRef]
- Global Cancer Observatory. Available online: https://gco.iarc.fr/ (accessed on 16 May 2021).
- Pinto, M.; Cárcano, F.; Vieira, A.; Cabral, E.; Lopes, L. Molecular Biology of Pediatric and Adult Male Germ Cell Tumors. Cancers 2021, 13, 2349. [Google Scholar] [CrossRef]
- Reilley, M.J.; Pagliaro, L.C. Testicular Choriocarcinoma: A Rare Variant that Requires a Unique Treatment Approach. Curr. Oncol. Rep. 2015, 17, 2. [Google Scholar] [CrossRef]
- Baroni, T.; Arato, I.; Mancuso, F.; Calafiore, R.; Luca, G. On the Origin of Testicular Germ Cell Tumors: From Gonocytes to Testicular Cancer. Front. Endocrinol. 2019, 10, 343. [Google Scholar] [CrossRef]
- Murray, M.J.; Huddart, R.A.; Coleman, N. The present and future of serum diagnostic tests for testicular germ cell tumours. Nature reviews. Urology 2016, 13, 715–725. [Google Scholar] [CrossRef] [PubMed]
- Regouc, M.; Belge, G.; Lorch, A.; Dieckmann, K.-P.; Pichler, M. Non-Coding microRNAs as Novel Potential Tumor Markers in Testicular Cancer. Cancers 2020, 12, 749. [Google Scholar] [CrossRef] [PubMed]
- Bing, Z.; Master, S.R.; Tobias, J.W.; Baldwin, D.A.; Xu, X.W.; Tomaszewski, J.E. MicroRNA expression profiles of seminoma from paraffin-embedded formalin-fixed tissue. Virchows Arch. Int. J. Pathol. 2012, 461, 663–668. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.; Li, N.; Liang, S.; Huang, Q.; Coukos, G.; Zhang, L. Identification of MicroRNAs Regulating Reprogramming Factor LIN28 in Embryonic Stem Cells and Cancer Cells. J. Biol. Chem. 2010, 285, 41961–41971. [Google Scholar] [CrossRef] [PubMed]
- Dieckmann, K.-P.; Radtke, A.; Geczi, L.; Matthies, C.; Anheuser, P.; Eckardt, U.; Sommer, J.; Zengerling, F.; Trenti, E.; Pichler, R.; et al. Serum Levels of MicroRNA-371a-3p (M371 Test) as a New Biomarker of Testicular Germ Cell Tumors: Results of a Prospective Multicentric Study. J. Clin. Oncol. 2019, 37, 1412–1423. [Google Scholar] [CrossRef]
- Voorhoeve, P.M.; le Sage, C.; Schrier, M.; Gillis, A.J.; Stoop, H.; Nagel, R.; Liu, Y.-P.; van Duijse, J.; Drost, J.; Griekspoor, A.; et al. A Genetic Screen Implicates miRNA-372 and miRNA-373 As Oncogenes in Testicular Germ Cell Tumors. Cell 2006, 124, 1169–1181. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K.; Kondo, K.; Kitajima, K.; Muraoka, M.; Nozawa, A.; Hara, T. Tumor-suppressive Function of Protein-tyrosine Phosphatase Non-receptor Type 23 in Testicular Germ Cell Tumors Is Lost upon Overexpression of miR142-3p microRNA. J. Biol. Chem. 2013, 288, 23990–23999. [Google Scholar] [CrossRef]
- Batool, A.; Wang, Y.-Q.; Hao, X.-X.; Chen, S.-R.; Liu, Y.-X. A miR-125b/CSF1-CX3CL1/tumor-associated macrophage recruitment axis controls testicular germ cell tumor growth. Cell Death Dis. 2018, 9, 962. [Google Scholar] [CrossRef]
- Liu, J.; Shi, H.; Li, X.; Chen, G.; Larsson, C.; Lui, W.-O. miR-223-3p regulates cell growth and apoptosis via FBXW7 suggesting an oncogenic role in human testicular germ cell tumors. Int. J. Oncol. 2016, 50, 356–364. [Google Scholar] [CrossRef]
- Cheung, H.-H.; Davis, A.J.; Lee, T.-L.; Pang, A.L.; Nagrani, S.; Rennert, O.M.; Chan, W.-Y. Methylation of an intronic region regulates miR-199a in testicular tumor malignancy. Oncogene 2011, 30, 3404–3415. [Google Scholar] [CrossRef]
- Lian, J.; Tian, H.; Liu, L.; Zhang, X.-S.; Li, W.-Q.; Deng, Y.-M.; Yao, G.-D.; Yin, M.-M.; Sun, F. Downregulation of microRNA-383 is associated with male infertility and promotes testicular embryonal carcinoma cell proliferation by targeting IRF1. Cell Death Dis. 2010, 1, e94. [Google Scholar] [CrossRef] [PubMed]
- De Martino, M.; Esposito, F.; Pellecchia, S.; Penha, R.C.C.; Botti, G.; Fusco, A.; Chieffi, P. HMGA1-Regulating microRNAs Let-7a and miR-26a are Downregulated in Human Seminomas. Int. J. Mol. Sci. 2020, 21, 3014. [Google Scholar] [CrossRef] [PubMed]
- Lizé, M.; Pilarski, S.; Dobbelstein, M.; Liz, M. E2F1-inducible microRNA 449a/b suppresses cell proliferation and promotes apoptosis. Cell Death Differ. 2009, 17, 452–458. [Google Scholar] [CrossRef]
- Özata, D.M.; Li, X.; Lee, L.; Liu, J.; Warsito, D.; Hajeri, P.; Hultman, I.; Fotouhi, O.; Marklund, S.; Ährlund-Richter, L.; et al. Loss of miR-514a-3p regulation of PEG3 activates the NF-kappa B pathway in human testicular germ cell tumors. Cell Death Dis. 2017, 8, e2759. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Duan, H.; Zhou, S.; Liu, Z.; Wu, D.; Zhao, T.; Xu, S.; Yang, L.; Li, D. microRNA-199a-3p functions as tumor suppressor by regulating glucose metabolism in testicular germ cell tumors. Mol. Med. Rep. 2016, 14, 2311–2320. [Google Scholar] [CrossRef]
- Plaza, X.R.; van Agthoven, T.; Meijer, C.; van Vugt, M.A.T.M.; de Jong, S.; Gietema, J.A.; Looijenga, L.H.J. miR-371a-3p, miR-373-3p and miR-367-3p as Serum Biomarkers in Metastatic Testicular Germ Cell Cancers Before, During and After Chemotherapy. Cells 2019, 8, 1221. [Google Scholar] [CrossRef]
- Port, M.; Glaesener, S.; Ruf, C.; Riecke, A.; Bokemeyer, C.; Meineke, V.; Honecker, F.; Abend, M. Micro-RNA expression in cisplatin resistant germ cell tumor cell lines. Mol. Cancer 2011, 10, 52. [Google Scholar] [CrossRef]
- Kremer, L.; von Brandenstein, M.; Wittersheim, M.; Koeditz, B.; Paffenholz, P.; Hellmich, M.; Pfister, D.; Heidenreich, A.; Nestler, T. The combination of microRNA-371a-3p and 375-5p can distinguish viable germ cell tumor and teratoma from necrosis in postchemotherapy retroperitoneal lymph node dissection specimens. Transl. Androl. Urol. 2021, 10, 1647–1655. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Moreno, A.; López-Domínguez, R.; Villatoro-García, J.A.; Ramirez-Mena, A.; Aparicio-Puerta, E.; Hackenberg, M.; Pascual-Montano, A.; Carmona-Saez, P. Functional Enrichment Analysis of Regulatory Elements. Biomedicines 2022, 10, 590. [Google Scholar] [CrossRef]
- Larrea, E.; Sole, C.; Manterola, L.; Goicoechea, I.; Armesto, M.; Arestin, M.; Caffarel, M.M.; Araujo, A.M.; Araiz, M.; Fernandez-Mercado, M.; et al. New Concepts in Cancer Biomarkers: Circulating miRNAs in Liquid Biopsies. Int. J. Mol. Sci. 2016, 17, 627. [Google Scholar] [CrossRef]
- Gilad, S.; Meiri, E.; Yogev, Y.; Benjamin, S.; Lebanony, D.; Yerushalmi, N.; Benjamin, H.; Kushnir, M.; Cholakh, H.; Melamed, N.; et al. Serum MicroRNAs Are Promising Novel Biomarkers. PLoS ONE 2008, 3, e3148. [Google Scholar] [CrossRef] [PubMed]
- Condrat, C.E.; Thompson, D.C.; Barbu, M.G.; Bugnar, O.L.; Boboc, A.; Cretoiu, D.; Suciu, N.; Cretoiu, S.M.; Voinea, S.C. miRNAs as Biomarkers in Disease: Latest Findings Regarding Their Role in Diagnosis and Prognosis. Cells 2020, 9, 276. [Google Scholar] [CrossRef] [PubMed]
- Dieckmann, K.-P.; Spiekermann, M.; Balks, T.; Ikogho, R.; Anheuser, P.; Wosniok, W.; Loening, T.; Bullerdiek, J.; Belge, G. MicroRNA miR-371a-3p - A Novel Serum Biomarker of Testicular Germ Cell Tumors: Evidence for Specificity from Measurements in Testicular Vein Blood and in Neoplastic Hydrocele Fluid. Urol. Int. 2016, 97, 76–83. [Google Scholar] [CrossRef]
- Gillis, A.J.; Rijlaarsdam, M.; Eini, R.; Dorssers, L.C.; Biermann, K.; Murray, M.J.; Nicholson, J.C.; Coleman, N.; Dieckmann, K.-P.; Belge, G.; et al. Targeted serum miRNA (TSmiR) test for diagnosis and follow-up of (testicular) germ cell cancer patients: A proof of principle. Mol. Oncol. 2013, 7, 1083–1092. [Google Scholar] [CrossRef] [PubMed]
- Wang, V.; Wu, W. MicroRNA-based therapeutics for cancer. BioDrugs 2009, 23, 15–23. [Google Scholar] [CrossRef]
- Eichelmann, A.; Matuszcak, C.; Hummel, R.; Haier, J. Role of miRNAs in cell signaling of cancer associated fibroblasts. Int. J. Biochem. Cell Biol. 2018, 101, 94–102. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Andresen, J.L.; Manan, R.S.; Langer, R. Nucleic acid delivery for therapeutic applications. Adv. Drug Deliv. Rev. 2021, 178, 113834. [Google Scholar] [CrossRef]
- Dasgupta, I.; Chatterjee, A. Recent Advances in miRNA Delivery Systems. Methods Protoc. 2021, 4, 10. [Google Scholar] [CrossRef]
- Yoshioka, K.; Kunieda, T.; Asami, Y.; Guo, H.; Miyata, H.; Yoshida-Tanaka, K.; Sujino, Y.; Piao, W.; Kuwahara, H.; Nishina, K.; et al. Highly efficient silencing of microRNA by heteroduplex oligonucleotides. Nucleic Acids Res. 2019, 47, 7321–7332. [Google Scholar] [CrossRef]
- Bernardo, B.C.; Ooi, J.Y.Y.; Lin, R.C.Y.; McMullen, J.R. miRNA therapeutics: A new class of drugs with potential therapeutic applications in the heart. Future Med. Chem. 2015, 7, 1771–1792. [Google Scholar] [CrossRef]
- Smolle, M.A.; Calin, H.N.; Pichler, M.; Calin, G.A. Noncoding RNAs and immune checkpoints-clinical implications as cancer therapeutics. FEBS J. 2017, 284, 1952–1966. [Google Scholar] [CrossRef] [PubMed]
- To, K.K.; Fong, W.; Tong, C.W.; Wu, M.; Yan, W.; Cho, W.C. Advances in the discovery of microRNA-based anticancer therapeutics: Latest tools and developments. Expert Opin. Drug Discov. 2019, 15, 63–83. [Google Scholar] [CrossRef] [PubMed]

| Downregulated | ||
| miRNA | log2FoldChange | p adj |
| hsa-miR-3663-5p | −4.426 | 2.39 × 10−10 |
| hsa-miR-1233-3p | −4.228 | 1.84 × 10−8 |
| hsa-miR-552-5p | −4.056 | 1.21 × 10−10 |
| hsa-miR-449b-5p | −3.973 | 5.26 × 10−13 |
| hsa-miR-7153-5p | −3.813 | 5.18 × 10−8 |
| hsa-miR-122-5p | −3.791 | 1.60 × 10−9 |
| hsa-miR-552-3p | −3.761 | 2.31 × 10−9 |
| hsa-miR-449a | −3.741 | 5.97 × 10−11 |
| hsa-miR-122-3p | −3.722 | 0.0017 |
| hsa-miR-34b-5p | −3.688 | 3.56 × 10−9 |
| hsa-miR-449c-5p | −3.638 | 1.93 × 10−12 |
| hsa-miR-34c-5p | −3.554 | 5.26 × 10−13 |
| hsa-miR-449c-3p | −3.441 | 0.0011 |
| hsa-miR-375 | −3.409 | 9.99 × 10−18 |
| hsa-miR-3663-3p | −3.385 | 9.63 × 10−6 |
| hsa-miR-7159-5p | −3.259 | 5.29 × 10−5 |
| hsa-miR-449b-3p | −3.212 | 2.75 × 10−6 |
| hsa-miR-4700-5p | −3.209 | 0.0043 |
| hsa-miR-522-3p | −3.153 | 1.46 × 10−9 |
| hsa-miR-1273a | −3.118 | 2.44 × 10−8 |
| hsa-miR-1295a | −3.076 | 0.0005 |
| hsa-miR-34b-3p | −2.971 | 2.16 × 10−7 |
| hsa-miR-1283 | −2.798 | 2.41 × 10−7 |
| hsa-miR-3150b-3p | −2.768 | 0.0203 |
| hsa-miR-4423-3p | −2.703 | 0.0023 |
| hsa-miR-6507-5p | −2.699 | 0.0049 |
| hsa-miR-7154-5p | −2.647 | 0.0253 |
| hsa-miR-517c-3p | −2.639 | 9.92 × 10−10 |
| hsa-miR-3925-3p | −2.613 | 0.0025 |
| hsa-miR-515-5p | −2.600 | 8.84 × 10−10 |
| Upregulated | ||
| hsa-miR-7151-3p | 2.634 | 0.0137 |
| hsa-miR-376a-2-5p | 2.202 | 0.0109 |
| hsa-miR-1224-5p | 2.193 | 0.0024 |
| hsa-miR-1299 | 1.958 | 5.73 × 10−5 |
| hsa-miR-142-5p | 1.898 | 0.0060 |
| hsa-miR-543 | 1.869 | 0.0004 |
| hsa-miR-487a-3p | 1.865 | 0.0079 |
| hsa-miR-584-3p | 1.830 | 0.0060 |
| hsa-miR-665 | 1.799 | 0.0362 |
| hsa-miR-134-3p | 1.778 | 0.0130 |
| hsa-miR-369-3p | 1.692 | 0.0008 |
| hsa-miR-377-3p | 1.665 | 0.0113 |
| hsa-miR-33a-5p | 1.665 | 0.0113 |
| hsa-miR-376a-3p | 1.602 | 0.0016 |
| hsa-miR-758-3p | 1.589 | 0.0020 |
| hsa-miR-654-3p | 1.588 | 0.0004 |
| hsa-miR-134-5p | 1.558 | 0.0017 |
| hsa-miR-889-3p | 1.552 | 0.0052 |
| hsa-miR-127-3p | 1.549 | 0.0007 |
| hsa-miR-1185-1-3p | 1.539 | 0.0110 |
| hsa-miR-1185-2-3p | 1.534 | 0.0305 |
| hsa-miR-154-5p | 1.516 | 0.0001 |
| hsa-miR-381-3p | 1.511 | 0.0007 |
| hsa-miR-127-5p | 1.511 | 0.0013 |
| hsa-miR-337-5p | 1.510 | 0.0036 |
| hsa-miR-379-3p | 1.508 | 0.0013 |
| hsa-miR-136-3p | 1.506 | 0.0010 |
| hsa-miR-376c-3p | 1.492 | 0.0015 |
| hsa-miR-495-3p | 1.443 | 0.0016 |
| hsa-miR-376b-5p | 1.442 | 0.0449 |
| miRNA | Target Genes | Function | References |
|---|---|---|---|
| cluster-miR-302 | Akt SPRY4 NR2F2 CDK2/4 CyclinD1 | Participates in the maintenance of embryonic stem cell pluripotency and is highly expressed in seminoma. Regulates Akt, which inhibits the expression of other cell cycle inhibitors such as CDK2 and CDK4, and thus accelerates the transition from G1 to S phase. Inhibition of Spry4 in TGCT decreases cell growth and invasion. | [50,94] |
| hsa-miR-21 | They act as oncomiRs and are found at high levels in seminoma and spermatocytic seminoma. | [95] | |
| hsa-miR-221 hsa-miR-222 | Elevated in seminoma. | [95] | |
| hsa-miR-146 | Downregulated in seminoma and embryonal carcinoma. | [17] | |
| hsa-Let-7 | Lin28 | Downregulated in TGCT. | [96] |
| hsa-miR-371a-3p | Upregulated in TGCT. | [97] | |
| hsa-miR-372-3p hsa-miR-373-3p | LATS2 | They act as oncomiRs, inhibiting p53-mediated cyclin-dependent kinase (CDK) by regulating LATS2, a tumor suppressor, allowing tumor growth in the presence of WT p53. They participate in tumorigenesis. | [98] |
| hsa-miR-142-3p | PTPN23 | Participates in the pathogenesis of TGCT. | [99] |
| hsa-miR-125b | CSF1 CX3CL1 | Participates as a tumor suppressor in various types of tumors and has functions in proliferation and apoptosis. Found at low levels in TGCT. | [100] |
| hsa-miR-223-3p | FBXW7 | It acts as oncomiR, promoting cell growth and inhibiting apoptosis. | [101] |
| hsa-miR-199a-5p | PODXL | Participates in the development of TGCT. | [102] |
| hsa-miR-383 | IRF1 | High expression in embryonal carcinoma. Regulates cell proliferation by reducing the levels of cyclin D1, CDK2, and p21. | [103] |
| hsa-miR-26a hsa-Let-7a | HMGA1 | They inhibit proliferation and motility in seminoma. | [104] |
| hsa-miR-449a | Found at low levels in TGCT. | [105] | |
| hsa-miR-514a-3p | PEG3 | They have low expression in seminoma and embryonal carcinoma. They slow down the apoptotic mechanisms of TGCT. | [106] |
| hsa-miR-199a-3p | SP1 DNMT3A | Participates in aerobic glycolysis, DNA methylation, and low expression in TGCT. | [107] |
| has-miR-367-3p, | Highly expressed in TGCT. | [108] | |
| hsa-miR-512-3p, hsa-miR-515-518 hsa-miR-525 | Highly expressed in embryonal carcinoma | [109] | |
| hsa-miR-301 | Expressed in spermatocytic seminoma, yolk sac tumors, and teratoma. | [17] | |
| hsa-miR-17-5p | Expressed in embryonal carcinoma. | [17] | |
| hsa-miR-375-5p | Expressed in teratoma. | [110] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
García-Andrade, F.; Vigueras-Villaseñor, R.M.; Chávez-Saldaña, M.D.; Rojas-Castañeda, J.C.; Bahena-Ocampo, I.U.; Aréchaga-Ocampo, E.; Díaz-Chávez, J.; Landero-Huerta, D.A. The Role of microRNAs in the Gonocyte Theory as Target of Malignancy: Looking for Potential Diagnostic Biomarkers. Int. J. Mol. Sci. 2022, 23, 10526. https://doi.org/10.3390/ijms231810526
García-Andrade F, Vigueras-Villaseñor RM, Chávez-Saldaña MD, Rojas-Castañeda JC, Bahena-Ocampo IU, Aréchaga-Ocampo E, Díaz-Chávez J, Landero-Huerta DA. The Role of microRNAs in the Gonocyte Theory as Target of Malignancy: Looking for Potential Diagnostic Biomarkers. International Journal of Molecular Sciences. 2022; 23(18):10526. https://doi.org/10.3390/ijms231810526
Chicago/Turabian StyleGarcía-Andrade, Fabiola, Rosa María Vigueras-Villaseñor, Margarita Dolores Chávez-Saldaña, Julio César Rojas-Castañeda, Iván Uriel Bahena-Ocampo, Elena Aréchaga-Ocampo, José Díaz-Chávez, and Daniel Adrian Landero-Huerta. 2022. "The Role of microRNAs in the Gonocyte Theory as Target of Malignancy: Looking for Potential Diagnostic Biomarkers" International Journal of Molecular Sciences 23, no. 18: 10526. https://doi.org/10.3390/ijms231810526
APA StyleGarcía-Andrade, F., Vigueras-Villaseñor, R. M., Chávez-Saldaña, M. D., Rojas-Castañeda, J. C., Bahena-Ocampo, I. U., Aréchaga-Ocampo, E., Díaz-Chávez, J., & Landero-Huerta, D. A. (2022). The Role of microRNAs in the Gonocyte Theory as Target of Malignancy: Looking for Potential Diagnostic Biomarkers. International Journal of Molecular Sciences, 23(18), 10526. https://doi.org/10.3390/ijms231810526






