Development of Soft Rice Lines by Regulating Amylose Content via Editing the 5′UTR of the Wx Gene
Abstract
:1. Introduction
2. Results
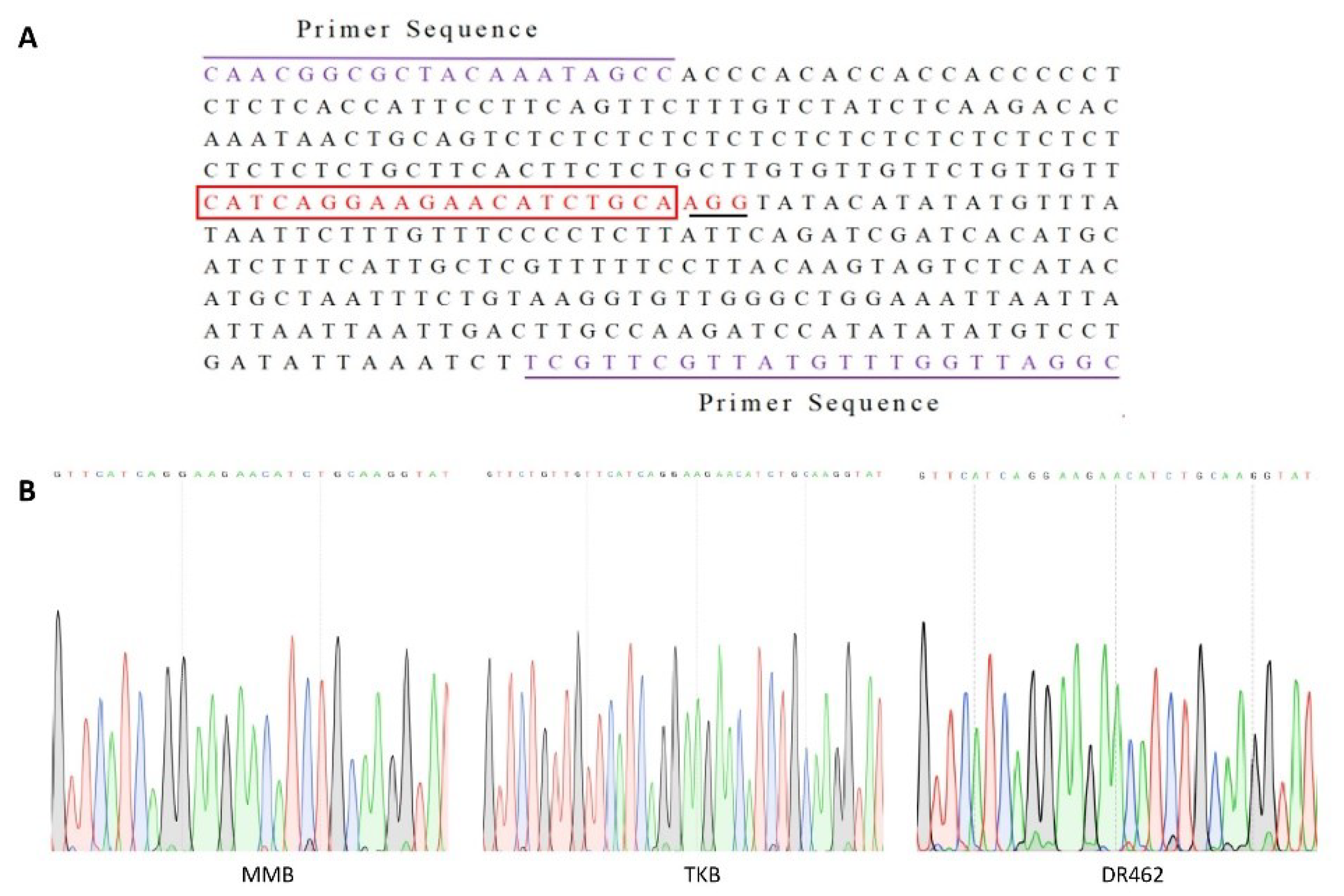
2.1. Construction of CRISPR/Cas9-Wx Vector
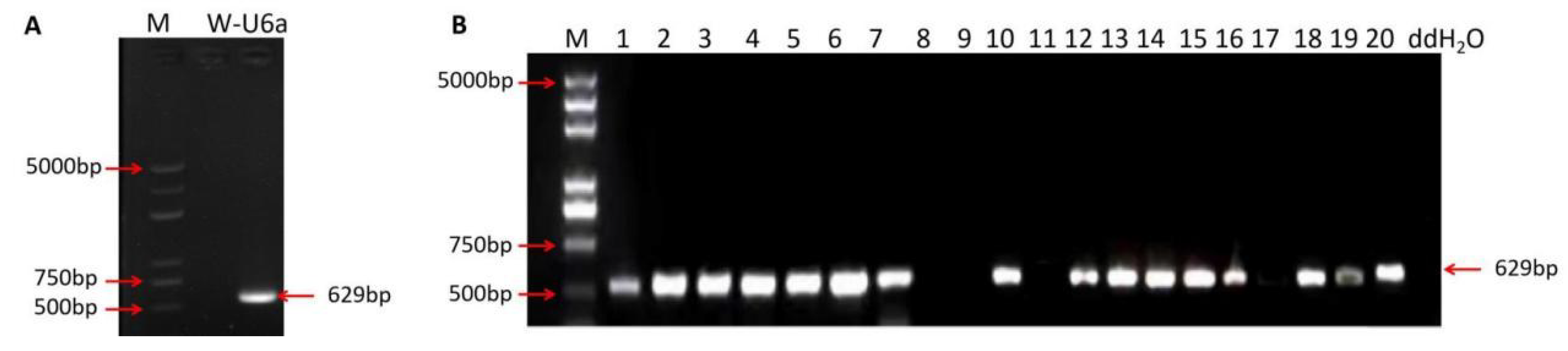
2.2. Genetic Transformation of MMB, TKB, and DR462
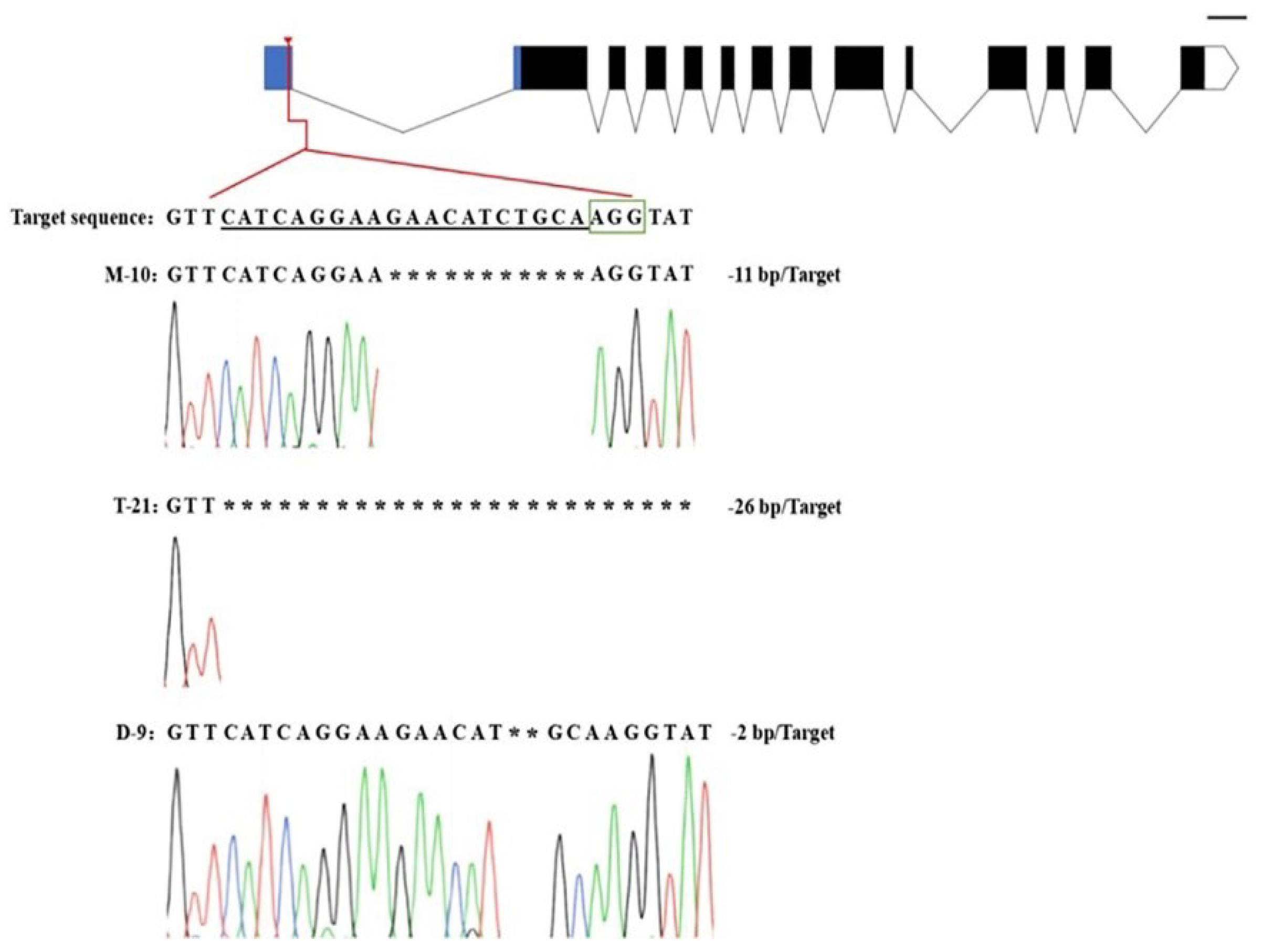
2.3. Mutation Analysis of T0 Transformants
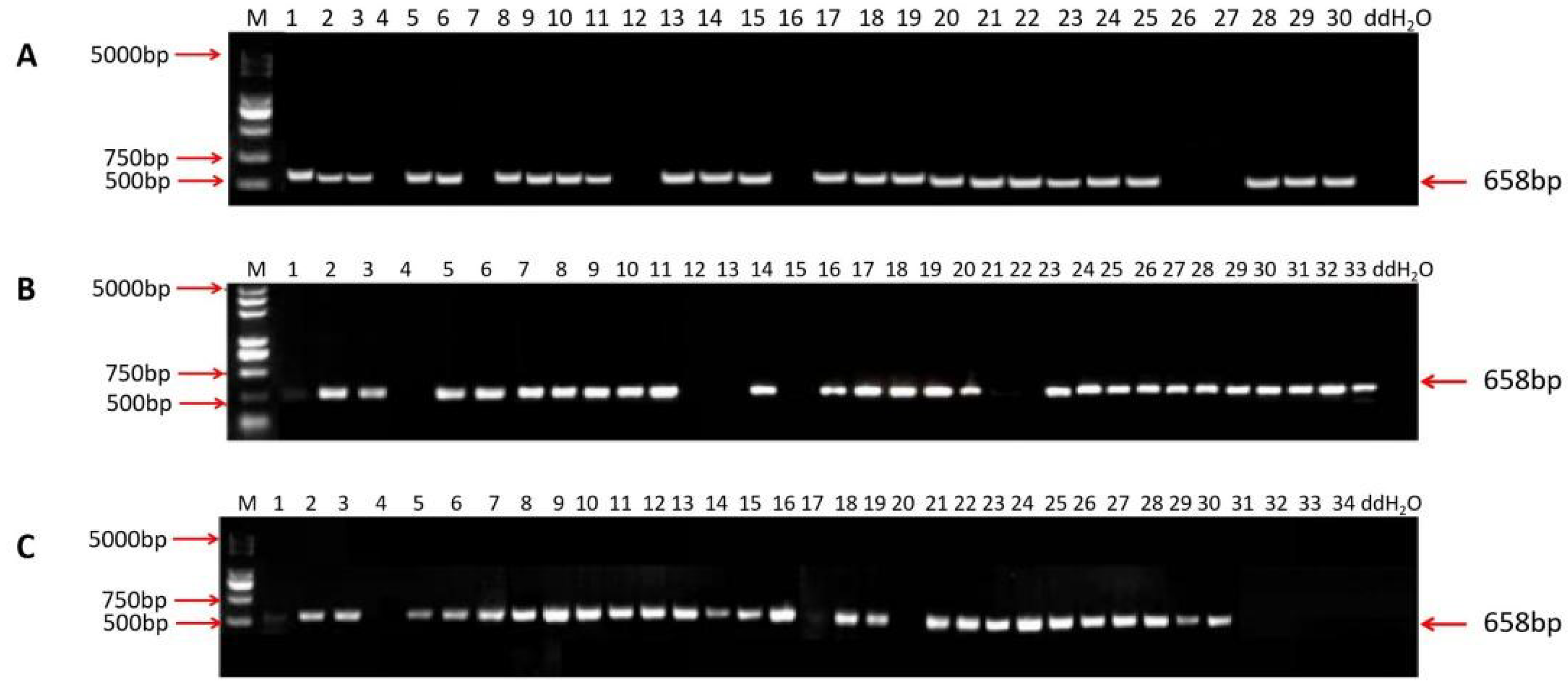
2.4. Screening of T-DNA-Free Plants in T1 Generations
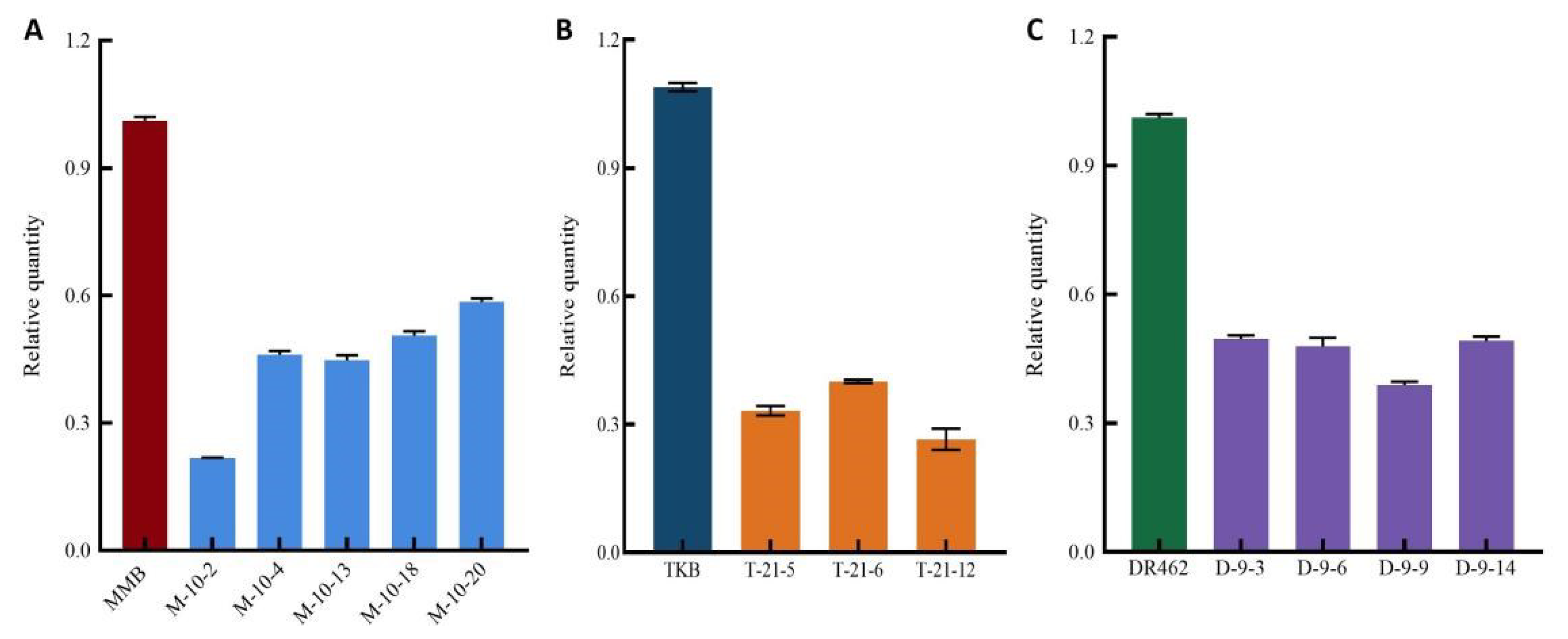
2.5. Quantitative Analysis of Wx Gene Expression in Mutant Lines in the T2 Generation
2.6. Determination of AC in Mutant Lines in the T2 Generation
2.7. Determination of Gel Consistency (GC) in Mutant Lines in the T2 Generation
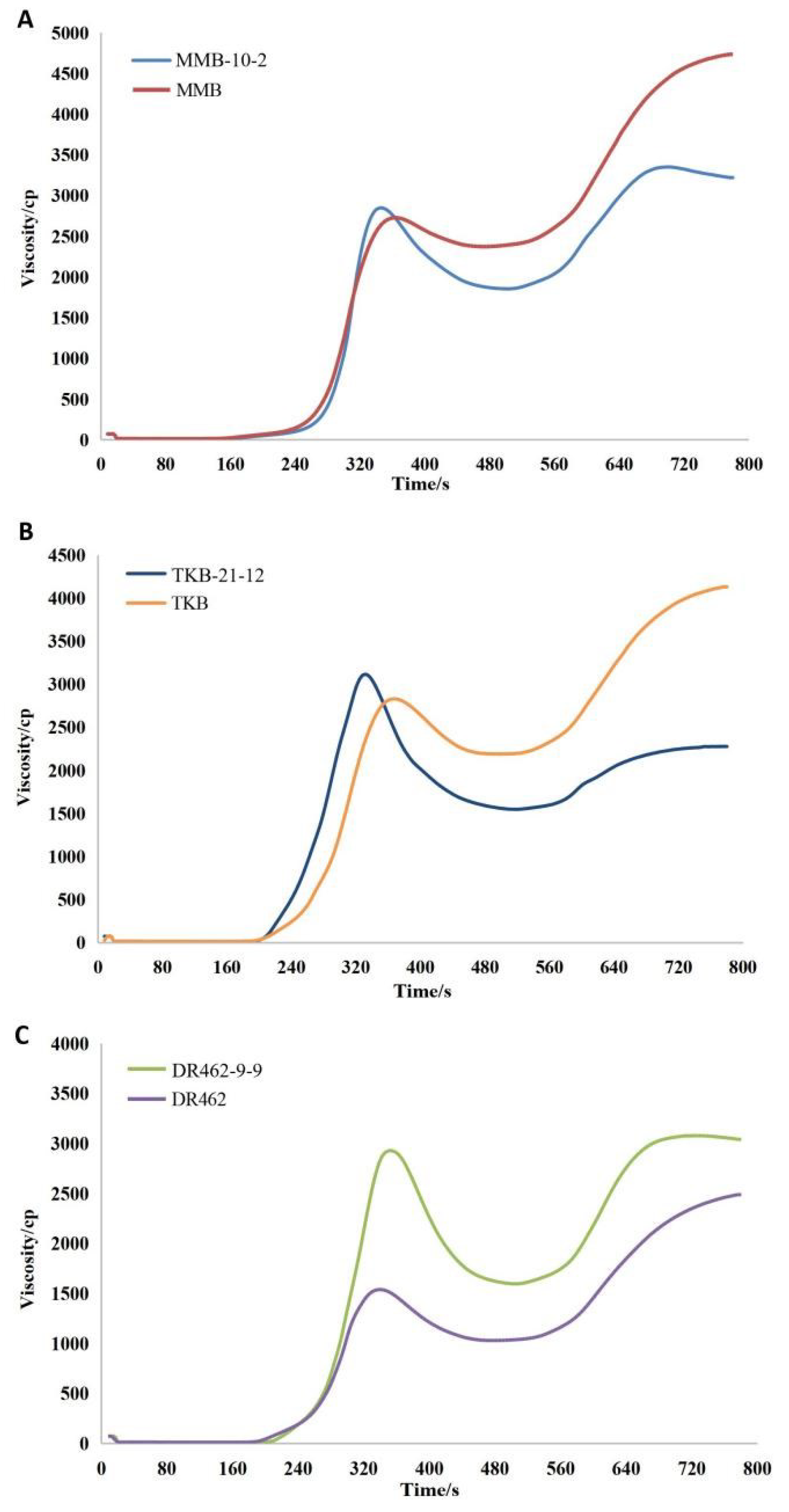
2.8. Analysis of RVA Profile in Mutant Lines in the T2 Generation
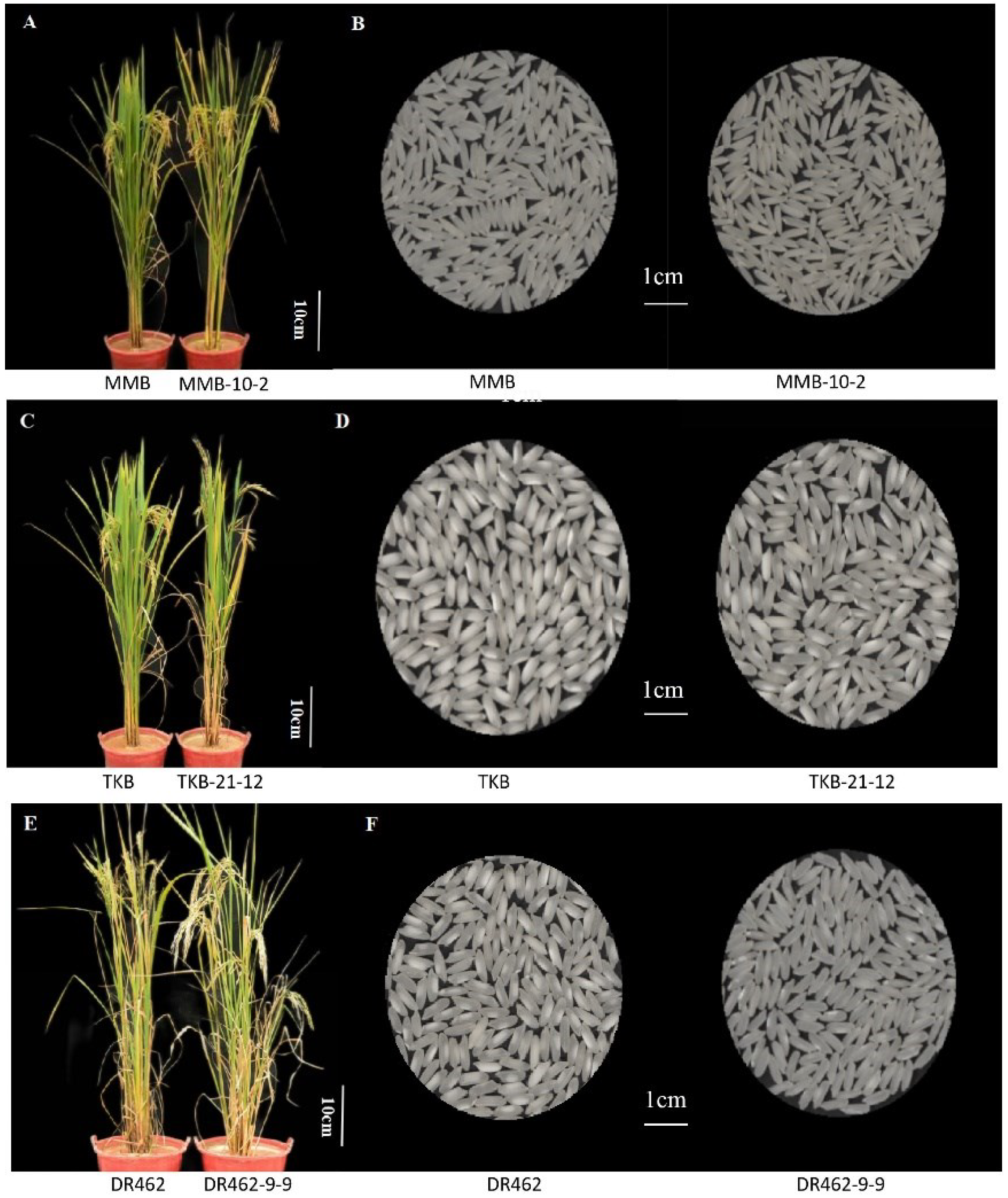
2.9. Analysis of the Main Agronomic Traits in Mutant Lines in the T2 Generation
3. Discussion
3.1. 5′UTR Region Regulates the Expression of Wx Gene
3.2. Application of CRISPR/Cas9 System in Wx Gene Editing
3.3. Effects on Agronomic Traits of Plants When Editing the 5′UTR of Wx Gene
3.4. Advantages of CRISPR/Cas9 Technology in Improving ECQ of Rice
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Construction of CRISPR/Cas9 Vectors and Screening of Homozygous Mutants
4.3. Quantitative Real-Time PCR (qRT-PCR) Expression Analysis
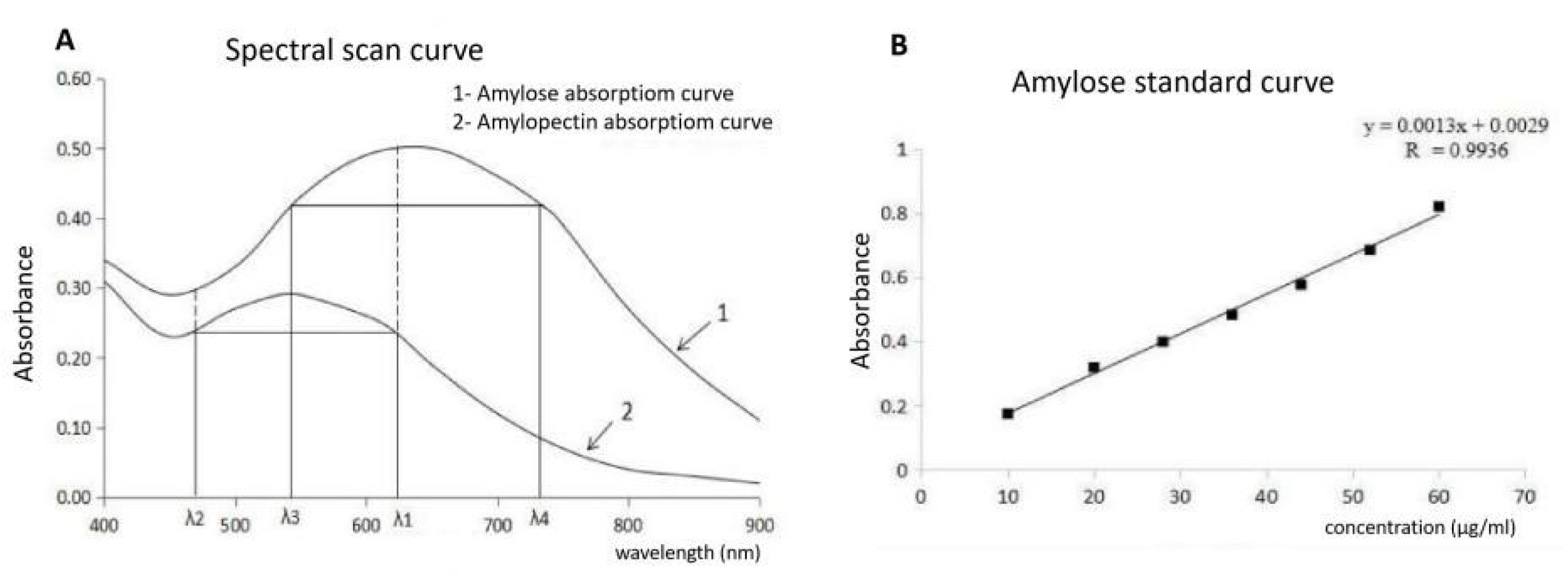
4.4. Measuring Rice Grain Physicochemical Properties
4.5. Agronomic Trait Investigation
4.6. Statistical Analysis
5. Conclusions
- 1.
- In the T0 generation of transgenic plants, the genetic transformation efficiency was more than 80%. The sequencing analysis showed that, among the four mutation types, homozygous mutation occupied a relatively large proportion, which not only demonstrated the extremely high editing efficiency of the CRISPR/cas9 gene editing technology, but also implied a great potential for application in designing targets in the 5′UTR region;
- 2.
- The expression analysis showed that the expression level of Wx gene in the edited lines was significantly lower than that of their wild type parents, indicating that editing 5′UISS changed the splicing mode and efficiency of introns;
- 3.
- The ECQ analysis of MMB-10-2, TKB-21-12, and DR462-9-9 showed that, compared with the wild type parents, the AC of the editing lines was significantly reduced, reaching the first-grade high-quality standard concerning AC. The GC of the edited lines was significantly increased, far exceeding the first-grade high-quality standard concerning GC. The RVA spectrum of the edited lines showed that the grain of the edited lines became softer and more elastic, and the eating quality was greatly improved.
- 4.
- The main agronomic traits of MMB-10-2, TKB-21-12, and DR462-9-9 had no significant difference from their corresponding wild type, which implied that the trait changes caused by homozygous mutations could be stably inherited to self-bred offspring, reflecting the stability and reliability of CRISPR/cas9 gene editing technology in rice genetic improvement.
Author Contributions
Funding
Conflicts of Interest
References
- Zhong, Y.; Qu, J.; Li, Z.; Tian, Y.; Zhu, F.; Blennow, A.; Liu, X. Rice starch multi-level structure and functional relationships. Carbohydr. Polym. 2022, 275, 118777. [Google Scholar] [CrossRef] [PubMed]
- Wani, A.A.; Singh, P.; Shah, M.A.; Schweiggert-Weisz, U.; Gul, K.; Wani, I.A. Rice Starch Diversity: Effects on structural, morphological, thermal, and physicochemical properties—A review. Compr. Rev. Food Sci. Food Saf. 2012, 11, 417–436. [Google Scholar] [CrossRef]
- Huang, L.; Li, Q.-F.; Zhang, C.; Chu, R.; Gu, Z.; Tan, H.; Zhao, D.; Fan, X.; Liu, Q. Creating novel Wx alleles with fine-tuned amylose levels and improved grain quality in rice by promoter editing using CRISPR/Cas9 system. Plant Biotechnol. J. 2020, 18, 2164–2166. [Google Scholar] [CrossRef] [PubMed]
- Juliano, B.O. Structure, chemistry, and function of the rice grain and its fractions. Cereal Foods World 1992, 37, 772–779. [Google Scholar] [CrossRef]
- Zhu, D.W.; Zhang, H.C.; Guo, B.W.; Dai, Y.G.; Huo, Z.Y.; Xu, K.; Wei, H.Y.; Gao, H. Development and outlook of Chinese soft rice. J. Yangzhou Univ. (Agric. Life Sci. Ed.) 2015, 36, 47–52. (In Chinese) [Google Scholar] [CrossRef]
- Yang, J.; Wang, J.; Fan, F. Development of AS-PCR marker based on a key mutation confirmed by resequencing of Wx-mp in milky princess and its application in japonica soft rice (Oryza sativa L.) breeding. Plant Breed. 2013, 132, 595–603. [Google Scholar] [CrossRef]
- Zhu, D.-W.; Zhang, H.-C.; Guo, B.-W.; Xu, K.; Dai, Q.-G.; Wei, H.-Y.; Gao, H.; Hu, Y.-J.; Cui, P.-Y.; Huo, Z.-Y. Effects of nitrogen level on yield and quality of japonica soft super rice. J. Integr. Agric. 2017, 16, 1018–1027. [Google Scholar] [CrossRef]
- Xu, Z.; Yu, M.; Yin, Y.; Zhu, C.; Ji, W.; Zhang, C.; Li, Q.; Zhang, H.; Tang, S.; Yu, H.; et al. Generation of selectable marker-free soft transgenic rice with transparent kernels by down regulation of SSSII-2. Crop J. 2020, 8, 53–61. [Google Scholar] [CrossRef]
- Lu, J.G.; Gao, R.C.; Li, P.; Xu, M.L.; Li, J.J. Breeding of low-amylose content late japonica rice and its taste quality analysis. China Rice 2014, 20, 41–45. (In Chinese) [Google Scholar] [CrossRef]
- Lu, M.C. Rice variety Songxiangjing1018 and its direct seeding cultivation techniques. China Seed Ind. 2018, 36, 90–92. (In Chinese) [Google Scholar] [CrossRef]
- Zhao, C.F.; Yue, H.L.; Huang, S.J.; Zhou, L.H.; Zhao, L.; Zhang, Y.D.; Chen, T.; Zhu, Z.; Zhao, Q.Y.; Yao, S.; et al. Eating quality and physicochemical properties in Nanjing rice varieties. Sci. Agric. Sin. 2019, 52, 909–920. (In Chinese) [Google Scholar] [CrossRef]
- Wang, H.; La, R.M.; Qi, L.S. CRISPR/Cas9 in Genome Editing and Beyond. Annu. Rev. Biochem. 2016, 85, 227–264. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Han, Y.; Feng, X.; Gul, N.; Luo, L.; Liu, F.; Qin, B.X.; Liu, Y.G.; Li, R.B. Improvement of a traditional high-quality glutinous rice variety by Crispr-Cas9 gene editing system. Mol. Plant Breed. 2019, 17, 6332–6342. (In Chinese) [Google Scholar] [CrossRef]
- Huang, L.C.; Gu, Z.W.; Tan, H.Y.; Zhao, W.; Xiao, Y.; Chu, R.; Fan, X.L.; Zhang, C.Q.; Li, Q.F.; Liu, Q.Q. Creating novel glutinous rice germplasms by editing Wx gene via CRISPR/Cas9 technology. J. Plant Genet. Resour. 2021, 22, 789–799. (In Chinese) [Google Scholar] [CrossRef]
- Liu, X.; Ding, Q.; Wang, W.; Pan, Y.; Tan, C.; Qiu, Y.; Chen, Y.; Li, H.; Li, Y.; Ye, N.; et al. Targeted deletion of the first intron of the Wxb allele via CRISPR/Cas9 significantly increases grain amylose content in rice. Rice 2022, 15, 1. [Google Scholar] [CrossRef] [PubMed]
- Teng, K.; Wang, X.; Guo, X. Generation of a new glutinous photothermosensitive genic-male-sterile (PTGMS) line by CRISPR/Cas9-directed mutagenesis of Wx in rice (Oryza sativa L.). Agriculture 2021, 11, 1044. [Google Scholar] [CrossRef]
- Fu, Y.; Luo, T.; Hua, Y.; Yan, X.; Liu, X.; Liu, Y.; Liu, Y.; Zhang, B.; Liu, R.; Zhu, Z.; et al. Assessment of the Characteristics of Waxy Rice Mutants Generated by CRISPR/Cas9. Front. Plant Sci. 2022, 13, 881964. [Google Scholar] [CrossRef]
- Dou, L.L.; Xiao, L.; Li, J.Y. Improving tasting quality of the main parent line “93-11” of Indica hybrid rice. J. Shanghai Norm. Univ. (Nat. Sci.) 2014, 43, 245–250. (In Chinese) [Google Scholar] [CrossRef]
- NY/T593-2013; Cooking Rice Variety Quality. Standards Press of China: Beijing, China, 2013.
- Patron, N.J.; Smith, A.M.; Fahy, B.F.; Hylton, C.M.; Naldrett, M.J.; Rossnagel, B.G.; Denyer, K. The altered pattern of amylose accumulation in the endosperm of low-amylose barley cultivars is attributable to a single mutant allele of granule-bound starch synthase I with a deletion in the 5′-non-coding region. Plant Physiol. 2002, 130, 190–198. [Google Scholar] [CrossRef]
- Yao, Y.; Thompson, D.B.; Guiltinan, M.J. Maize starch-branching enzyme isoforms and amylopectin structure. In the absence of starch-branching enzyme IIB, the further absence of starch-branching enzyme la leads to increased branching. Plant Physiol. 2004, 136, 3515–3523. [Google Scholar] [CrossRef] [Green Version]
- Cai, X.; Wang, Z.; Zheng, F.; Hong, M. A regulation-related intron in 5′ untranslated region of rice Waxy gene. Acta Phytophysiol. Sin. 1997, 23, 257–261. (In Chinese) [Google Scholar]
- Cai, X.; Wang, Z.; Xing, Y.; Zhang, J.; Hong, M. Alteration of RNA secondary structure of rice Waxy intron 1 caused by naturally ocurred mutations. Acta Phytophysiol. Sin. 2000, 26, 59–63. (In Chinese) [Google Scholar] [CrossRef]
- Cheng, S.; Ge, H.; Wang, Z.; Hong, M. Analysis of influence of Wx intron 1 on gene expression in transgenic rice plant. Acta Phytophysiol. Sin. 2001, 27, 381–386. (In Chinese) [Google Scholar] [CrossRef]
- Li, Q.; Pan, Z.F.; Deng, G.B. Effect of Waxy gene polymorphism on starch properties of highland barley in Qinghai Tibet plateau. Conf. Chin. Genet. Soc. 2015, 64–65. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, Q.; Zhu, Q. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 2015, 8, 1274–1284. [Google Scholar] [CrossRef]
- Fan, M.Y.; Mei, F.T.; Zhu, Y.W.; Lin, Y.R.; Zhao, L.L.; Tian, G.L.; Wang, F. Greating new glutinous rice by CRISPR/Cas9-targeted mutagenesis in rice. Fujian J. Agric. Sci. 2019, 34, 503–508. (In Chinese) [Google Scholar] [CrossRef]
- Wang, X.; Han, Y.; Feng, X.; Li, Y.-Z.; Qin, B.-X.; Luo, J.-J.; Wei, Z.; Qiu, Y.-F.; Liu, F.; Li, R.-B. Breeding of indica glutinous cytoplasmic male sterile line Wx 209A via CRISPR/Cas9 mediated genomic editing. Czech J. Genet. Plant Breed. 2019, 55, 93–100. [Google Scholar] [CrossRef]
- Zeng, D.; Liu, T.; Ma, X. Quantitative regulation of Waxy expression by CRISPR/Cas9-based promoter and 5′UTR-intron editing improves grain quality in rice. Plant Biotechnol. J. 2020, 18, 2385–2387. [Google Scholar] [CrossRef]
- Larkin, P.D.; Park, W.D. Association of waxy gene single nucleotide polymorphisms with starch characteristics in rice (Oryza sativa L.). Mol. Breed. 2003, 12, 335–339. [Google Scholar] [CrossRef]
- Li, X.; Xie, Y.; Zhu, Q.; Liu, Y.-G. Targeted genome editing in genes and cis-regulatory regions improves qualitative and quantitative traits in crops. Mol. Plant 2017, 10, 1368–1370. [Google Scholar] [CrossRef]
- Rodríguez-Leal, D.; Lemmon, Z.H.; Man, J.; Bartlett, M.E.; Lippman, Z.B. Engineering quantitative trait variation for crop improvement by genome editing. Cell 2017, 171, 470–480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.W. Creation of Waxy Allelic Mutants in Rice (Oryza sativa L.) by CRISPR/Cas9 System and Analysis of Starch Quality; Shanghai Normal University: Shanghai, China, 2021. [Google Scholar] [CrossRef]
- Li, Z.; Xiong, X.; Wang, F. Gene disruption through base editing-induced messenger RNA missplicing in plants. New Phytol. 2019, 222, 1139–1148. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.J. Research on Starch Quality Using Different Transgenic Constructs in the Endosperm of Rice (Oryza sativa L.); Yangzhou University: Yangzhou, China, 2007. [Google Scholar]
- Terada, R.; Nakajima, M.; Isshiki, M. Antisense waxy genes with highly active promoters effectively suppress Waxy gene expression in transgenic rice. Plant Cell Physiol. 2000, 41, 881–888. [Google Scholar] [CrossRef]
- Mao, Y.; Zhang, H.; Xu, N. Application of the CRISPR/Cas system for efficient genome engineering in plants. Mol. Plant 2013, 6, 2008–2011. [Google Scholar] [CrossRef] [PubMed]
- Yarra, R.; Sahoo, L. Base editing in rice: Current progress, advances, limitations, and future perspectives. Plant Cell Rep. 2021, 40, 595–604. [Google Scholar] [CrossRef]
- Gupta, D.; Bhattacharjee, O.; Mandal, D. CRISPR-Cas9 system: A new-fangled dawn in gene editing. Life Sci. 2019, 232, 116636. [Google Scholar] [CrossRef]
- The China Rice Data Center. Available online: https://www.ricedata.cn/ (accessed on 20 March 2021).
- The CRISPR-GE. Available online: http://skl.scau.edu.cn/ (accessed on 20 March 2021).
- Murray, M.G.; Thompson, W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 1980, 8, 4321–4325. [Google Scholar] [CrossRef]
- Sanger, F.; Coulson, A.R. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J. Mol. Biol. 1975, 94, 441–448. [Google Scholar] [CrossRef]
- NY/T83-2017; Determination of Rice Quality. Standards Press of China: Beijing, China, 2017.


| Parent | Mutation Type | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Homozygous | Bi-Allelic | Heterozygous | Wild | Total | ||||||
| No. of Plants | Rate (%) | No. of Plants | Rate (%) | No. of Plants | Rate (%) | No. of Plants | Rate (%) | No. of Plants | Rate (%) | |
| MMB | 8 | 33.3 | 7 | 29.2 | 4 | 16.7 | 5 | 20.8 | 24 | 100 |
| TKB | 9 | 34.6 | 7 | 26.9 | 5 | 19.2 | 6 | 23.1 | 26 | 100 |
| DR462 | 9 | 34.6 | 8 | 30.8 | 5 | 19.2 | 4 | 15.4 | 26 | 100 |
| Wild Type | GC (mm) | Mutant Lines | GC (mm) |
|---|---|---|---|
| MMB | 60.2 ± 0.2 | MMB-10-2 | 78.6 ** ± 0.1 |
| TKB | 61.2 ± 0.2 | TKB-21-6 | 77.4 ** ± 0.4 |
| DR462 | 61.4 ± 0.6 | DR462-9-9 | 79.6 ** ± 0.5 |
| Lines | PKV | BDV | SBV | CSV | HPV | CPV |
|---|---|---|---|---|---|---|
| MMB | 2729.6 ± 43.0 | 355.0 ± 6.5 | 2353.6 ± 21.5 | 2708.6 ± 26.8 | 2374.7 ± 40.8 | 5083.3 ± 37.2 |
| MMB-10-2 | 2850.3 ± 39.0 | 994.0 ± 9.0 | 1366.3 ± 11.5 | 2360.3 ± 29.1 | 1856.3 ± 24.0 | 4216.7 ± 50.0 |
| TKB | 2832.3 ± 10.1 | 641.3 ± 22.5 | 1978.0 ± 14.7 | 2619.3 ± 36.1 | 2191.0 ± 28.1 | 4810.3 ± 39.7 |
| TKB-21-12 | 3116.0 ± 6.03 | 1533.6 ± 15.1 | 729.6 ± 4.5 | 2263.3 ± 14.4 | 1539.0 ± 11.7 | 3802.0 ± 39.0 |
| DR462 | 1539.6 ± 38.1 | 508.7 ± 18.0 | 1457.6 ± 19.8 | 1966.3 ± 24.5 | 1031.0 ± 25.0 | 2997.3 ± 49.2 |
| DR462-9-9 | 2875.3 ± 34.6 | 1293.3 ± 21.0 | 1444.6 ± 3.5 | 2738.0 ± 21.1 | 1582.0 ± 14.1 | 4320.0 ± 35.0 |
| Lines | Plant Height (cm) | 1000-Grain Weight (g) | Panicle Length (cm) | Grain Number per Panicle | Grain Set Rate (%) | Effective Spike Number |
|---|---|---|---|---|---|---|
| MMB | 83.9 ± 1.3 a | 19.4 ± 3.1 a | 27.8 ± 0.1 a | 186 ± 0.1 a | 87.4 ± 2.0 a | 8.5 ± 2.5 a |
| MMB-10-2 | 85.1 ± 0.9 a | 19.1 ± 2.8 a | 27.7 ± 0.3 a | 196 ± 0.1 a | 87.2 ± 2.5 a | 8.7 ± 2.7 a |
| TKB | 77.8 ± 1.2 b | 27.4 ± 2.5 b | 27.6 ± 0.3 b | 190 ± 0.1 b | 86.8 ± 1.8 b | 8.2 ± 1.8 b |
| TKB-21-12 | 76.8 ± 2.1 b | 27.0 ± 3.0 b | 27.6 ± 0.3 b | 186 ± 0.1 b | 87.0 ± 2.0 b | 8.3 ± 1.4 b |
| DR462 | 94.7 ± 1.6 c | 24.1 ± 3.1 c | 28.0 ± 0.2 c | 188 ± 0.1 c | 86.6 ± 1.9 c | 7.9 ± 2.0 c |
| DR462-9-9 | 93.7 ± 1.4 c | 23.8 ± 2.9 c | 27.9 ± 0.2 c | 180 ± 0.1 c | 85.9 ± 1.8 c | 7.7 ± 2.5 c |
| Primer Name | Primer Sequence 5′-3′ |
|---|---|
| Wx-text-F | TCCGCCACGGGTTCCAG |
| Wx-text-R | CTCCTACCTCAGCCACAACG |
| U-F | CTCCGTTTTACCTGTGGAATCG |
| gR-R | CGGAGGAAAATTCCATCCAC |
| Wx-U6a-1F | TGTGTGCTTACAGCCATGGCGTTTTAGAGCTAGAAAT |
| Wx-U6a-1R | GCCATGGCTGTAAGCACACACGGCAGCCAAGCCAGCA |
| Pps-GGL | TTCAGAGGTCTCTCTCGACTAGTATGGAATCGGCAGCAAAGG |
| Pgs-GGR | AGCGTGGGTCTCGACCGACGCGTATCCATCCACTCCAAGCTC |
| PB-R | GCGCGCGGTCTCTACCGACGCGTATCC |
| PB-L | GCGCGCgGTCTCGCTCGACTAGTATGG |
| HPT-F | ATTTGTGTACGCCCGACAGT |
| HPT-R | GTGCTTGACATTGGGGAGTT |
| CAS9-F | CTGACGCTAACCTCGACAAG |
| CAS9-R | CCGATCTAGTAACATAGATGACACC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, J.; Guo, X.; Wang, X.; Fang, Y.; Liu, F.; Qin, B.; Li, R. Development of Soft Rice Lines by Regulating Amylose Content via Editing the 5′UTR of the Wx Gene. Int. J. Mol. Sci. 2022, 23, 10517. https://doi.org/10.3390/ijms231810517
Yang J, Guo X, Wang X, Fang Y, Liu F, Qin B, Li R. Development of Soft Rice Lines by Regulating Amylose Content via Editing the 5′UTR of the Wx Gene. International Journal of Molecular Sciences. 2022; 23(18):10517. https://doi.org/10.3390/ijms231810517
Chicago/Turabian StyleYang, Jinlian, Xinying Guo, Xuan Wang, Yaoyu Fang, Fang Liu, Baoxiang Qin, and Rongbai Li. 2022. "Development of Soft Rice Lines by Regulating Amylose Content via Editing the 5′UTR of the Wx Gene" International Journal of Molecular Sciences 23, no. 18: 10517. https://doi.org/10.3390/ijms231810517
APA StyleYang, J., Guo, X., Wang, X., Fang, Y., Liu, F., Qin, B., & Li, R. (2022). Development of Soft Rice Lines by Regulating Amylose Content via Editing the 5′UTR of the Wx Gene. International Journal of Molecular Sciences, 23(18), 10517. https://doi.org/10.3390/ijms231810517







