Molasses-Silver Nanoparticles: Synthesis, Optimization, Characterization, and Antibiofilm Activity
Abstract
:1. Introduction
2. Results and Discussion
2.1. Liquid Chromatography–Mass Spectroscopy (LCMS) Analysis
2.2. Mo-Capped AgNP Synthesis
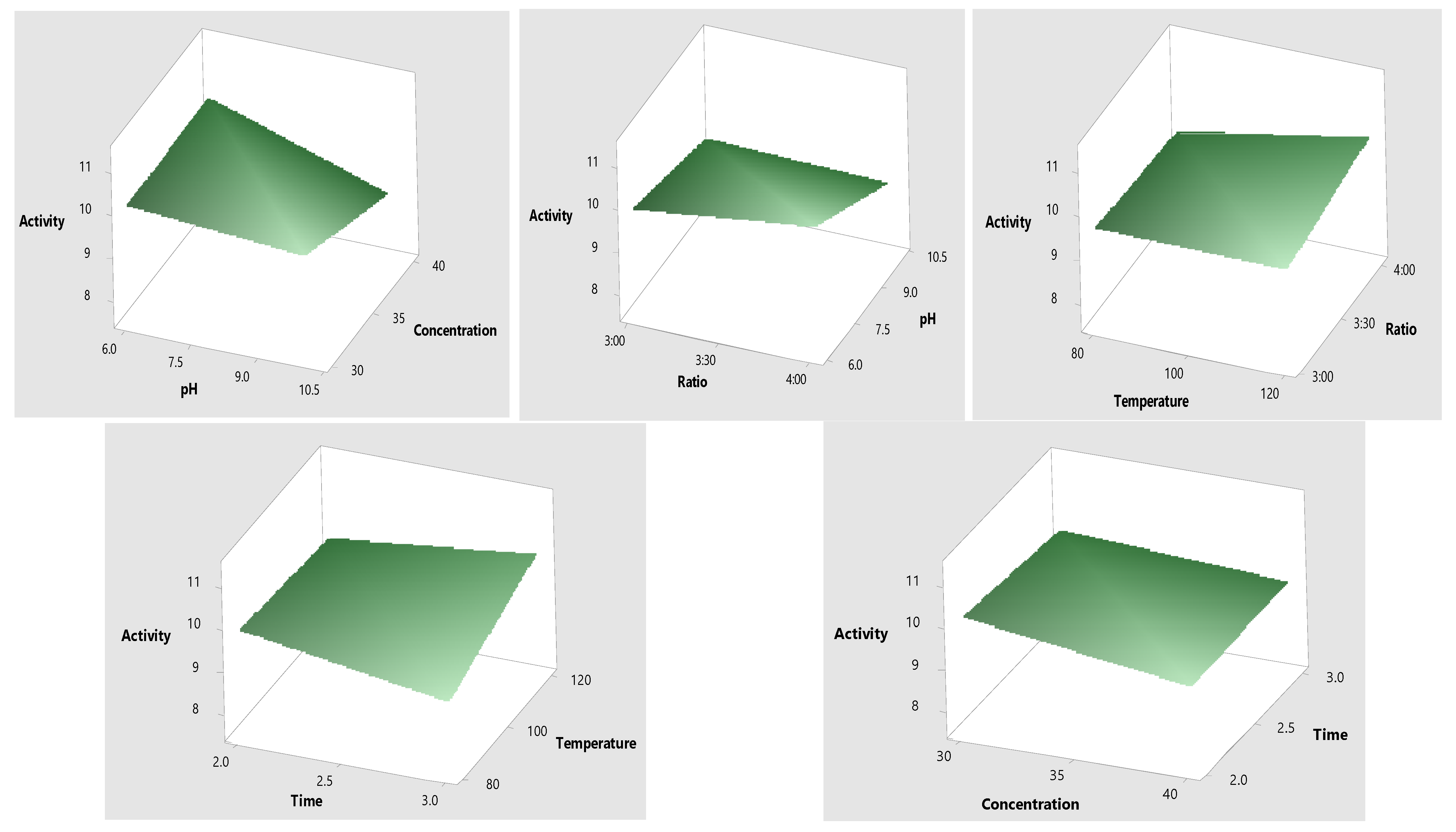
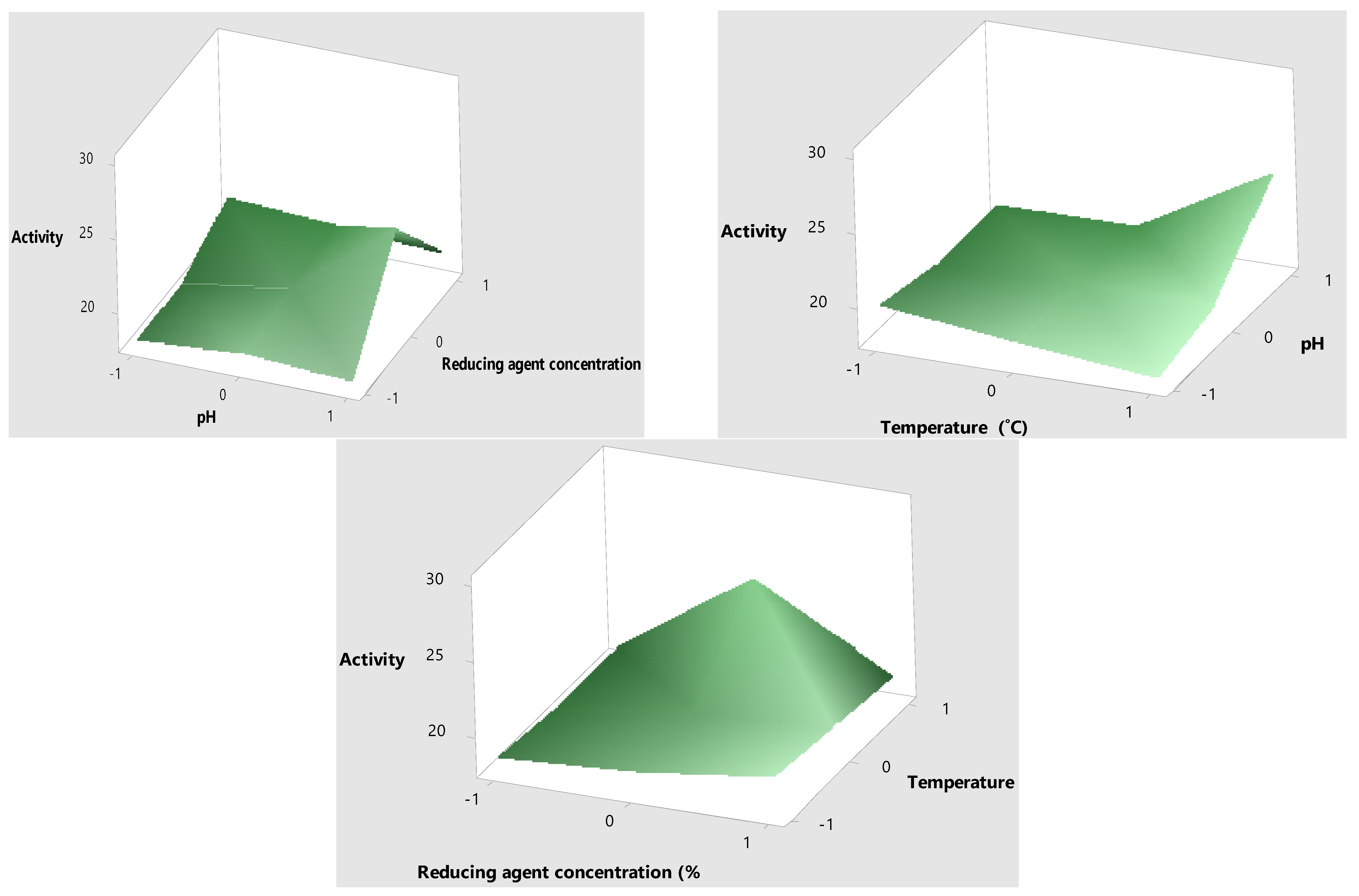
2.3. Optimization of Mo-Capped AgNPs
2.4. Antimicrobial Activity of Mo-Capped AgNPs
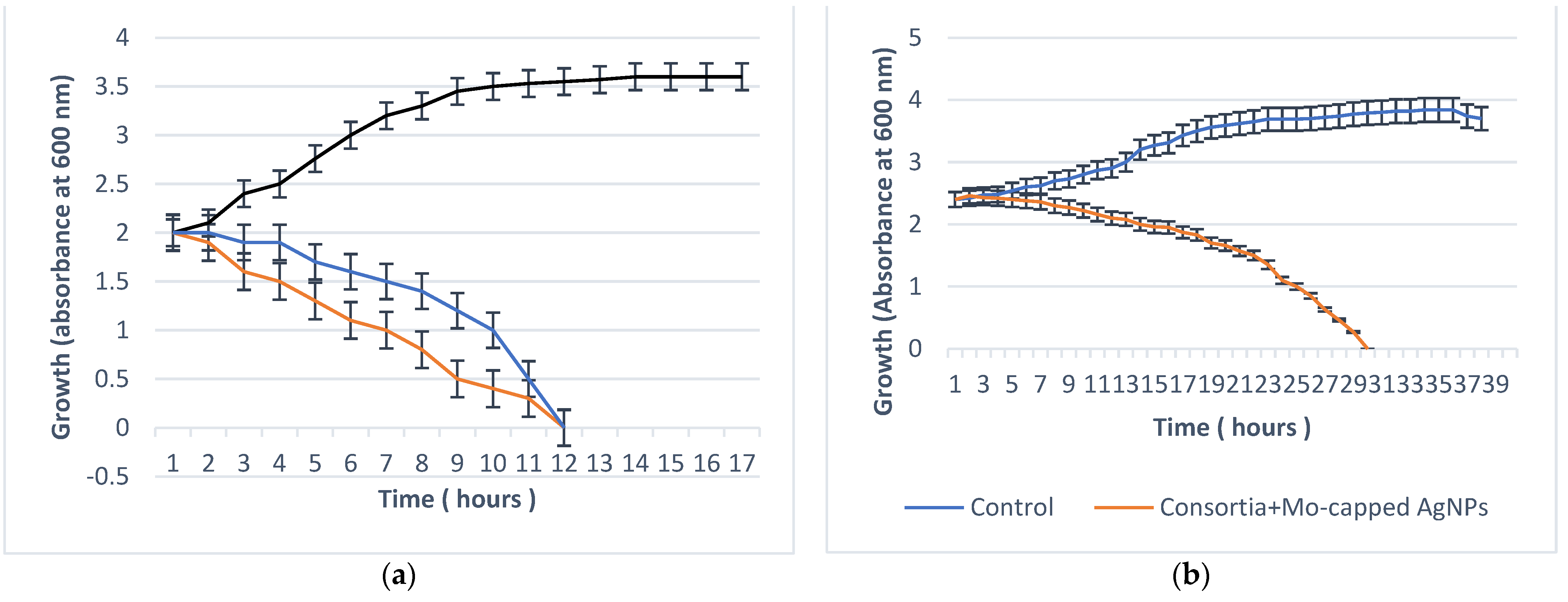
2.5. Antibiofilm Activity
2.5.1. Minimal Biofilm Eradication Concentration (MBEC)
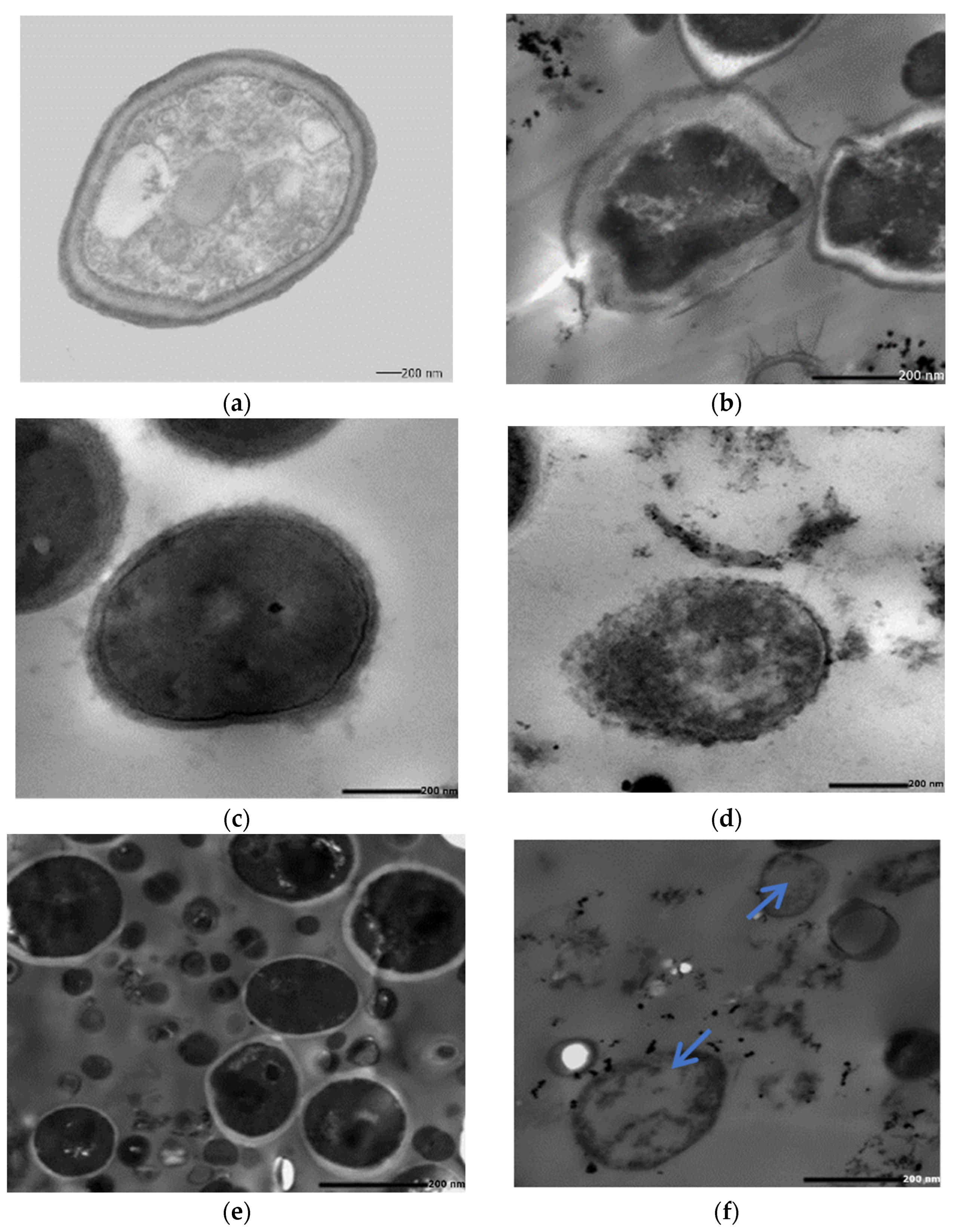
2.5.2. SEM of the Treated Biofilm
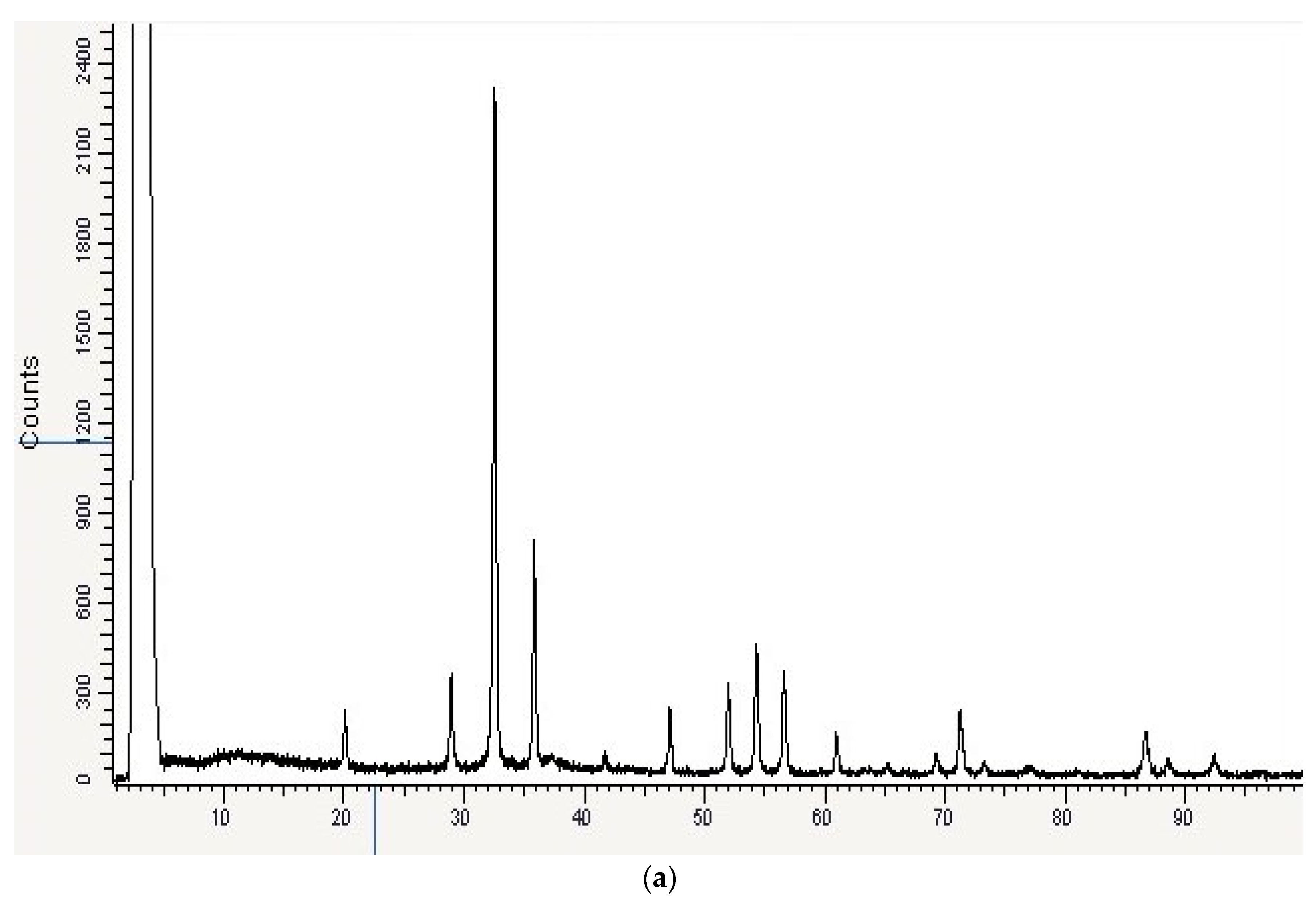
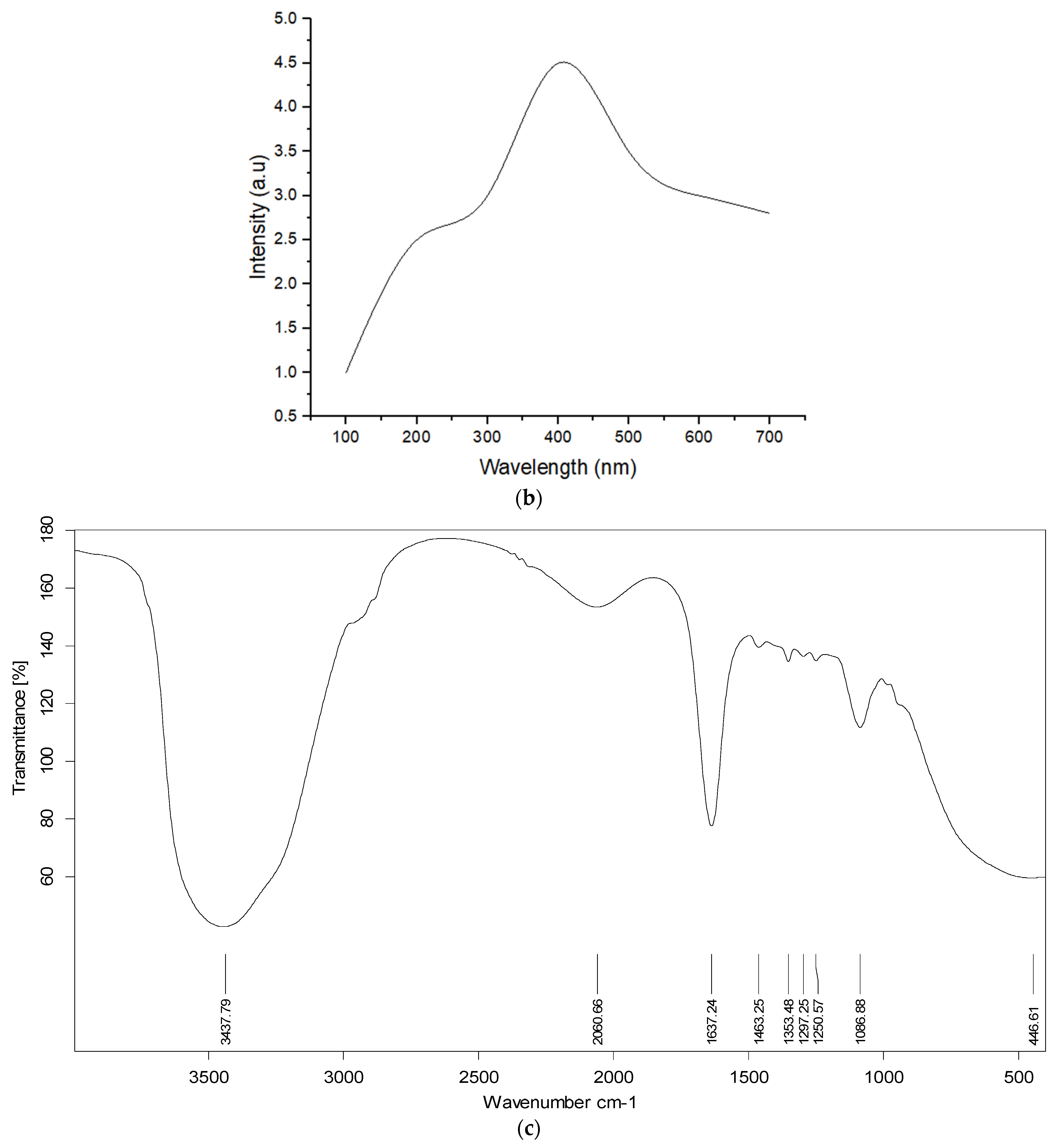
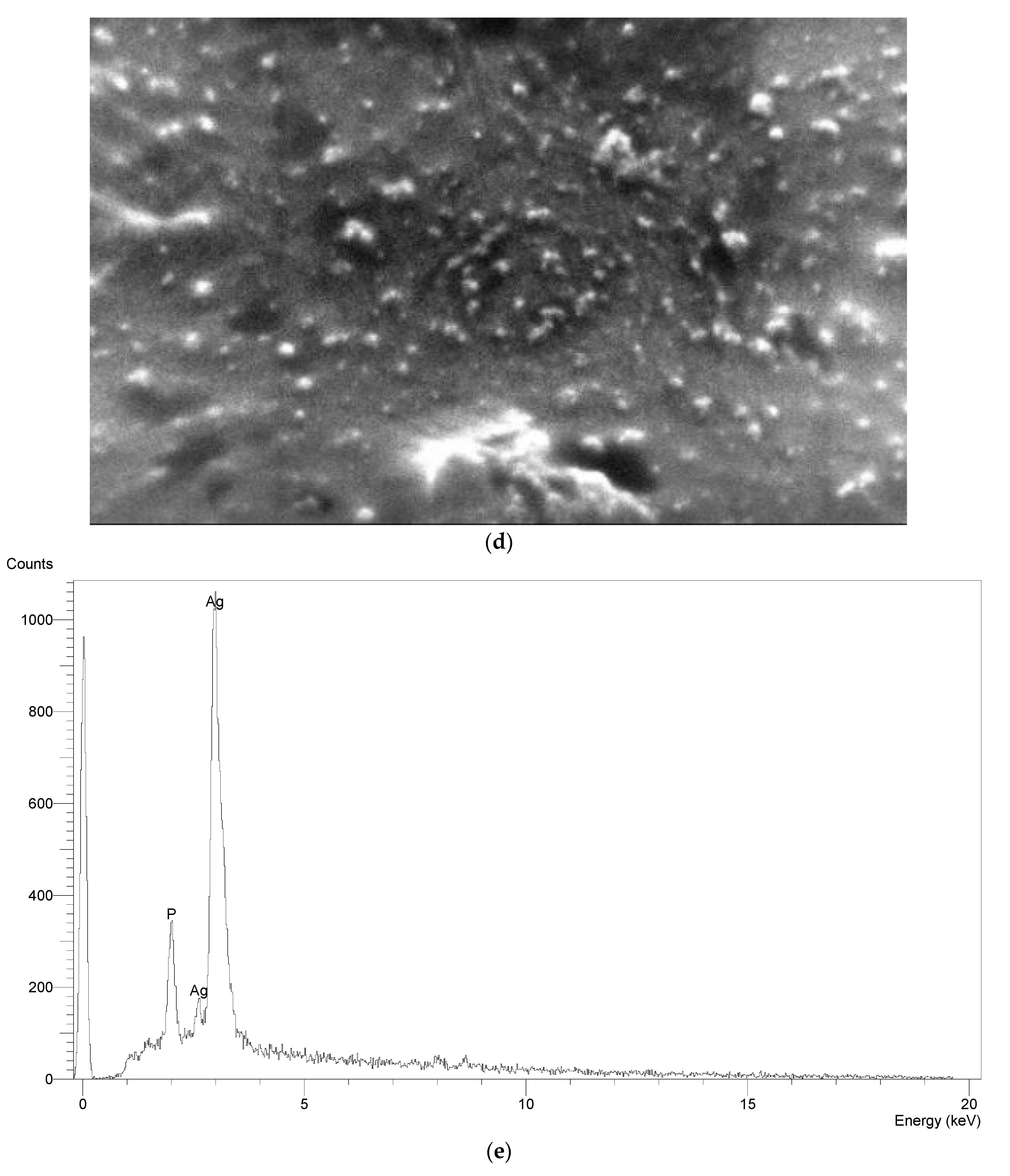
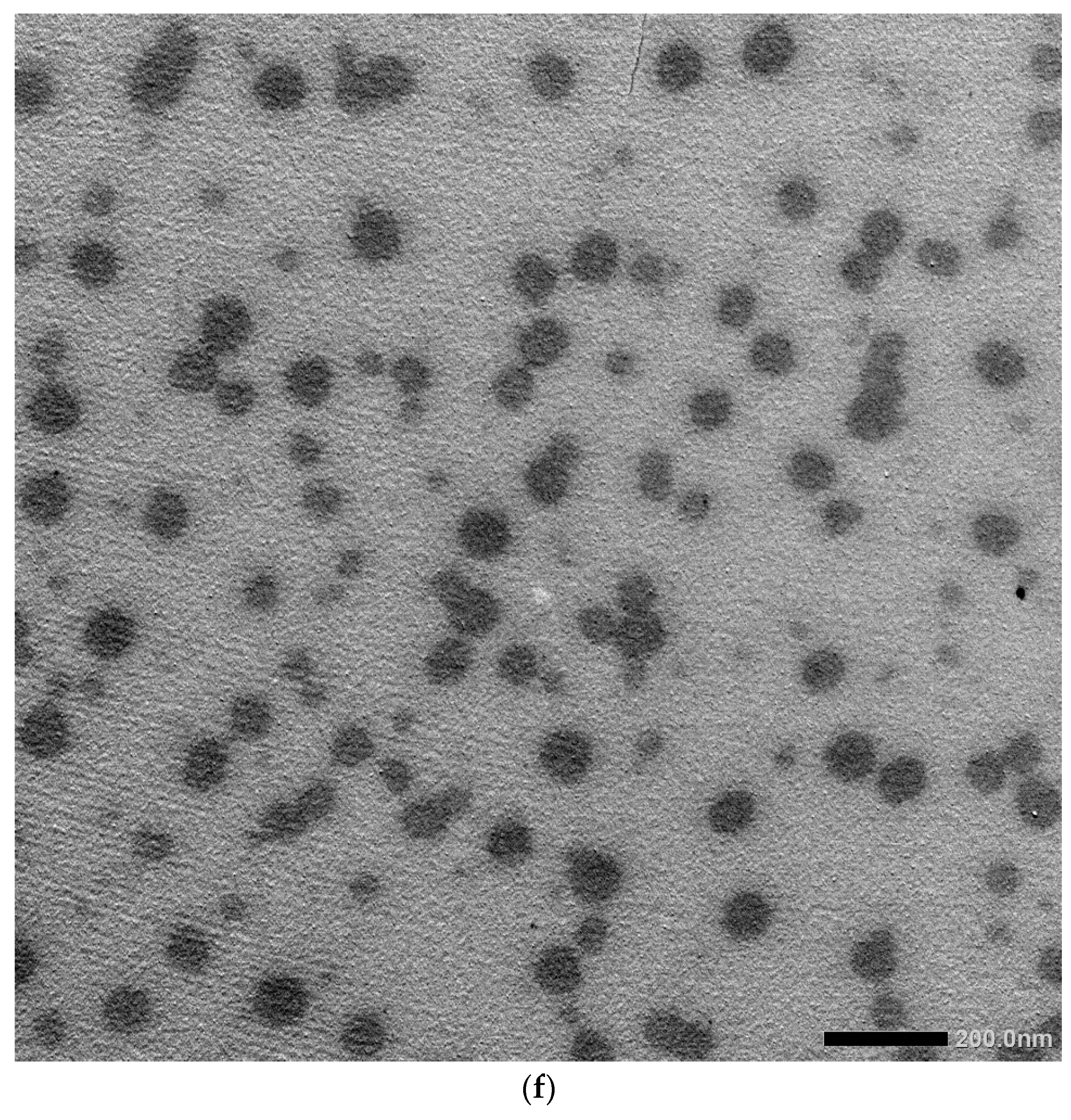
2.6. Characterization of AgNPs
3. Materials and Methods
3.1. Microorganisms
Raw Material
3.2. Liquid Chromatography–Mass Spectroscopy Analysis of the Obtained Sugarcane Molasses
3.3. Synthesis of AgNPs
3.3.1. Experimental Design
Box–Behnken Optimization
3.3.2. Antimicrobial Activity of the Optimized AgNPs
3.4. Antibiofilm Activity
3.4.1. Minimal Biofilm Eradication Concentration (MBEC)
3.4.2. Activity of the Prepared AgNPs against Preformed Biofilm
3.5. Characterization of the Optimized AgNPs
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rogers, S.A.; Bero, J.D.; Melander, C. Chemical synthesis and biological screening of 2-aminoimidazole-based bacterial and fungal antibiofilm agents. ChemBioChem 2010, 11, 396–410. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Meng, H.; Xing, G.; Chen, C.; Zhao, Y.; Jia, G.; Wang, T.; Yuan, H.; Ye, C.; Zhao, F.; et al. Acute toxicological effects of copper nanoparticles in vivo. Toxicol. Lett. 2006, 163, 109–120. [Google Scholar] [CrossRef] [PubMed]
- Sheetz, T.; Vidal, J.; Pearson, T.D.; Lozano, K. Nanotechnology: Awareness and societal concerns. Technol. Soc. 2005, 27, 329–345. [Google Scholar] [CrossRef]
- Kumar, P.; Singh, P.; Kumari, K.; Mozumdar, S.; Chandra, R. A green approach for the synthesis of gold nanotriangles using aqueous leaf extract of Callistemon viminalis. Mater. Lett. 2011, 65, 595–597. [Google Scholar] [CrossRef]
- Geissel, F.J.; Platania, V.; Gogos, A.; Herrmann, I.K.; Belibasakis, G.N.; Chatzinikolaidou, M.; Sotiriou, G.A. Antibiofilm activity of nanosilver coatings against Staphylococcus aureus. J. Colloid Interface Sci. 2022, 608, 3141–3150. [Google Scholar] [CrossRef]
- Goderecci, S.S.; Kaiser, E.; Yanakas, M.; Norris, Z.; Scaturro, J.; Oszust, R.; Medina, C.D.; Waechter, F.; Heon, M.; Krchnavek, R.R.; et al. Silver oxide coatings with high silver-ion elution rates and characterization of bactericidal activity. Molecules 2017, 22, 1487. [Google Scholar] [CrossRef]
- Thukkaram, M.; Vaidulych, M.; Kylián, O.; Hanus, J.; Rigole, P.; Aliakbarshirazi, S.; Asadian, M.; Nikiforov, A.; Van Tongel, A.; Biederman, H.; et al. Investigation of Ag/aC: H nanocomposite coatings on titanium for orthopedic applications. ACS Appl. Mater. Interfaces 2020, 12, 23655–23666. [Google Scholar] [CrossRef]
- Taglietti, A.; Arciola, C.R.; D’Agostino, A.; Dacarro, G.; Montanaro, L.; Campoccia, D.; Cucca, L.; Vercellino, M.; Poggi, A.; Pallavicini, P.; et al. Antibiofilm activity of a monolayer of silver nanoparticles anchored to an amino-silanized glass surface. Biomaterials 2014, 35, 1779–1788. [Google Scholar] [CrossRef]
- Skandalis, N.; Dimopoulou, A.; Georgopoulou, A.; Gallios, N.; Papadopoulos, D.; Tsipas, D.; Theologidis, I.; Michailidis, N.; Chatzinikolaidou, M. The effect of silver nanoparticles size, produced using plant extract from Arbutus unedo, on their antibacterial efficacy. Nanomaterials 2017, 7, 178. [Google Scholar] [CrossRef]
- Jamir, L.; Kumar, V.; Kaur, J.; Kumar, S.; Singh, H. Composition, valorization and therapeutical potential of molasses: A critical review. Environ. Technol. Rev. 2021, 10, 131–142. [Google Scholar] [CrossRef]
- Othman, A.M.; Elsayed, M.A.; Elshafei, A.M.; Hassan, M.M. Application of response surface methodology to optimize the extracellular fungal mediated nanosilver green synthesis. J. Genet. Eng. Biotechnol. 2017, 15, 497–504. [Google Scholar] [CrossRef] [PubMed]
- Daâssi, D.; Frikha, F.; Zouari-Mechichi, H.; Belbahri, L.; Woodward, S.; Mechichi, T. Application of response surface methodology to optimize decolourization of dyes by the laccase-mediator system. J. Environ. Manag. 2012, 108, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Poonkuzhali, K.; Palvannan, T. Thermostabilization of laccase by polysaccharide additives: Enhancement using central composite design of RSM. Carbohydr. Polym. 2011, 86, 860–864. [Google Scholar] [CrossRef]
- Lu, S.Y.; Qian, J.Q.; Wu, Z.G.; Ye, W.D.; Wu, G.F.; Pan, Y.B.; Zhang, K.Y. Application of statistical method to evaluate immobilization variables of trypsin entrapped with sol-gel method. J. Biochem. Technol. 2009, 1, 79–84. [Google Scholar]
- Deseo, M.A.; Elkins, A.; Rochfort, S.; Kitchen, B. Antioxidant activity and polyphenol composition of sugarcane molasses extract. Food Chem. 2020, 314, 126180. [Google Scholar] [CrossRef] [PubMed]
- Vilchis-Nestor, A.R.; Sánchez-Mendieta, V.; Camacho-López, M.A.; Gómez-Espinosa, R.M.; Camacho-López, M.A.; Arenas-Alatorre, J.A. Solventless synthesis and optical properties of Au and Ag nanoparticles using Camellia sinensis extract. Mater. Lett. 2008, 62, 3103–3105. [Google Scholar] [CrossRef]
- Banerjee, P.; Satapathy, M.; Mukhopahayay, A.; Das, P. Leaf extract mediated green synthesis of silver nanoparticles from widely available Indian plants: Synthesis, characterization, antimicrobial property and toxicity analysis. Bioresour. Bioprocess. 2014, 1, 3. [Google Scholar] [CrossRef]
- Pourmortazavi, S.M.; Taghdiri, M.; Makari, V.; Rahimi-Nasrabadi, M. Procedure optimization for green synthesis of silver nanoparticles by aqueous extract of Eucalyptus oleosa. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2015, 136, 1249–1254. [Google Scholar] [CrossRef]
- Ebrahimzadeh, H.; Behbahani, M.; Yamini, Y.; Adlnasab, L.; Asgharinezhad, A.A. Optimization of Cu (II)-ion imprinted nanoparticles for trace monitoring of copper in water and fish samples using a Box–Behnken design. React. Funct. Polym. 2013, 73, 23–29. [Google Scholar] [CrossRef]
- Nishio, E.K.; Ribeiro, J.M.; Oliveira, A.G.; Andrade, C.G.; Proni, E.A.; Kobayashi, R.K.; Nakazato, G. Antibacterial synergic effect of honey from two stingless bees: Scaptotrigona bipunctata Lepeletier, 1836, and S. postica Latreille, 1807. Sci. Rep. 2016, 6, 21641. [Google Scholar] [CrossRef]
- Haque, M.A.; Imamura, R.; Brown, G.A.; Krishnamurthi, V.R.; Niyonshuti, I.I.; Marcelle, T.; Mathurin, L.E.; Chen, J.; Wang, Y. An experiment-based model quantifying antimicrobial activity of silver nanoparticles on Escherichia coli. RSC Adv. 2017, 7, 56173–56182. [Google Scholar] [CrossRef] [Green Version]
- Morones, J.R.; Elechiguerra, J.L.; Camacho, A.; Holt, K.; Kouri, J.B.; Ramírez, J.T.; Yacaman, M.J. The bactericidal effect of silver nanoparticles. Nanotechnology 2005, 16, 2346. [Google Scholar] [CrossRef] [PubMed]
- Cao, C.; Zhang, T.; Yang, N.; Niu, X.; Zhou, Z.; Wang, J.; Yang, D.; Chen, P.; Zhong, L.; Dong, X.; et al. POD Nanozyme optimized by charge separation engineering for light/pH activated bacteria catalytic/photodynamic therapy. Signal Transduct. Target. Ther. 2022, 7, 86. [Google Scholar] [CrossRef] [PubMed]
- Palanisamy, N.K.; Ferina, N.; Amirulhusni, A.N.; Mohd-Zain, Z.; Hussaini, J.; Ping, L.J.; Durairaj, R. Antibiofilm properties of chemically synthesized silver nanoparticles found against Pseudomonas aeruginosa. J. Nanobiotechnol. 2014, 12, 2. [Google Scholar] [CrossRef] [PubMed]
- Ansari, M.A.; Khan, H.M.; Khan, A.A.; Cameotra, S.S.; Alzohairy, M.A. Anti-biofilm efficacy of silver nanoparticles against MRSA and MRSE isolated from wounds in a tertiary care hospital. Indian J. Med. Microbiol. 2015, 33, 101. [Google Scholar] [CrossRef]
- Hosny, A.M.; Kashef, M.T.; Rasmy, S.A.; Aboul-Magd, D.S.; El-Bazza, Z.E. Antimicrobial activity of silver nanoparticles synthesized using honey and gamma radiation against silver-resistant bacteria from wounds and burns. Adv. Nat. Sci. Nanosci. Nanotechnol. 2017, 8, 045009. [Google Scholar] [CrossRef]
- Shervani, Z.; Ikushima, Y.; Sato, M.; Kawanami, H.; Hakuta, Y.; Yokoyama, T.; Nagase, T.; Kuneida, H.; Aramaki, K. Morphology and size-controlled synthesis of silver nanoparticles in aqueous surfactant polymer solutions. Colloid Polym. Sci. 2008, 286, 403–410. [Google Scholar] [CrossRef]
- Logeswari, P.; Silambarasan, S.; Abraham, J. Synthesis of silver nanoparticles using plants extract and analysis of their antimicrobial property. J. Saudi Chem. Soc. 2015, 19, 311–317. [Google Scholar] [CrossRef]
- Ali, Z.A.; Yahya, R.; Sekaran, S.D.; Puteh, R. Green synthesis of silver nanoparticles using apple extract and its antibacterial properties. Adv. Mater. Sci. Eng. 2016, 2016, 4102196. [Google Scholar] [CrossRef]
- El-Attar, A.A.; El-Wakil, H.B.; Hassanin, A.H.; Bakr, B.A.; Almutairi, T.M.; Hagar, M.; Elwakil, B.H.; Olama, Z.A. Silver/Snail Mucous PVA nanofibers: Electrospun synthesis and antibacterial and wound healing activities. Membranes 2022, 12, 536. [Google Scholar] [CrossRef]
- Muthukumar, M.; Mohan, D.; Rajendran, M. Optimization of mix proportions of mineral aggregates using Box Behnken design of experiments. Cem. Concr. Compos. 2003, 25, 751–758. [Google Scholar] [CrossRef]
- Elnaggar, Y.S.; Elwakil, B.H.; Elshewemi, S.S.; El-Naggar, M.Y.; A Bekhit, A.; A Olama, Z. Novel Siwa propolis and colistin-integrated chitosan nanoparticles: Elaboration, in vitro and in vivo appraisal. Nanomedicine 2020, 15, 1269–1284. [Google Scholar] [CrossRef] [PubMed]
- Elnaggar, Y.S.; Elwakil, B.H.; Elshewemi, S.S.; El-Naggar, M.Y.; A Bekhit, A.; A Olama, Z. In vivo bio-distribution and acute toxicity evaluation of greenly synthesized ultra-small gold nanoparticles with different biological activities. Sci. Rep. 2022, 12, 6269. [Google Scholar]
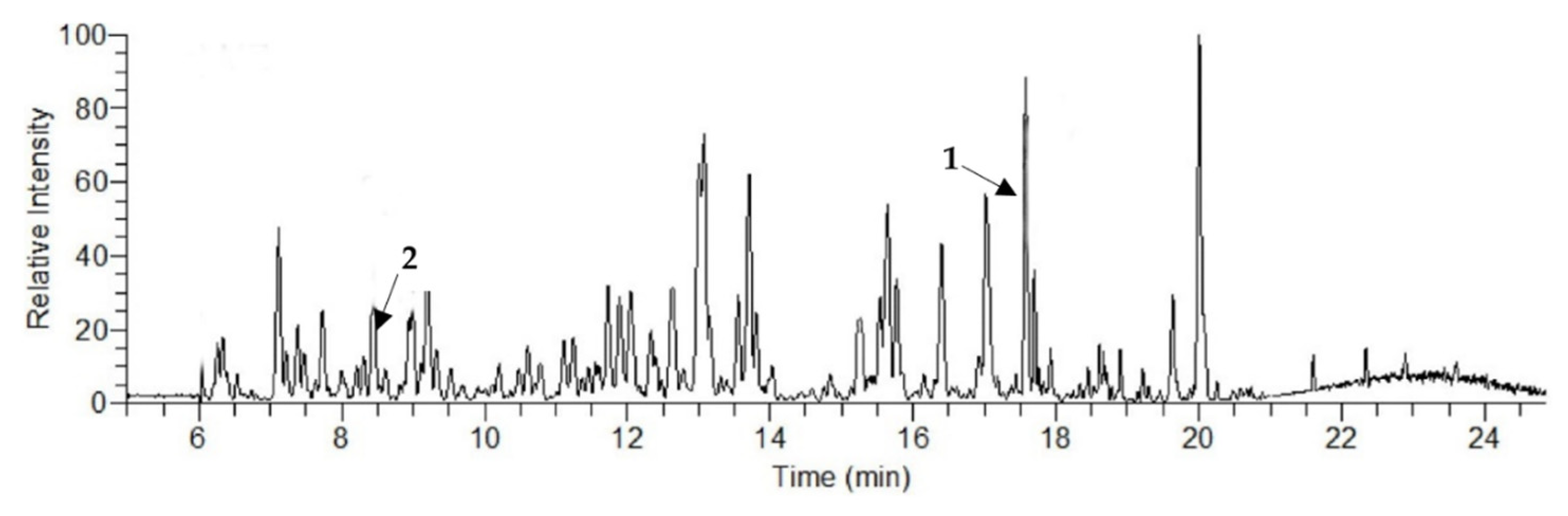


| Analyte | RT (min) | Concentration (μg/g) |
|---|---|---|
| Chlorogenic acid | 8.9 | 5.91 |
| Caffeic acid | 9.7 | 0.46 |
| Vanillin | 12.4 | 0.21 |
| Orientin | 13.5 | 0.37 |
| Sinapic acid | 14.5 | 0.14 |
| Diosmin | 17.1 | 13.1 |
| Apigenin | 18.3 | 0.01 |
| Diosmetin | 20.0 | 0.24 |
| Tested Strains | AgNO3 | Molasses | Mo-Capped AgNPs | |||
|---|---|---|---|---|---|---|
| IZ (mm) | MIC (µg/mL) | IZ (mm) | MIC (µg/mL) | IZ (mm) | MIC (µg/mL) | |
| C. albicans DAY185 | R | 256.0 | R | 256.0 | 13.0 ± 0.79 | 64.0 |
| S. aureus ATCC 6538 | 15.0 ± 0.58 | 128.0 | R | 256.0 | 19.5 ± 0.91 | 32.0 |
| Trails | Antimicrobial Activity (Inhibition Zone Diameter, mm) | Physical Characteristics | ||
|---|---|---|---|---|
| S. aureus ATCC 6538 | C. albicans DAY185 | Size (nm) | PDI | |
| 1 | 16.0 | 9.8 | 75.0 | 0.785 |
| 2 | 9.2 | 8.2 | 85.0 | 0.836 |
| 3 | 17.7 | 11.8 | 58.2 | 0.537 |
| 4 | 18.7 | 10.8 | 63.7 | 0.627 |
| 5 | 21.5 | 8.7 | 80.0 | 0.819 |
| 6 | 23.5 | 10.5 | 75.0 | 0.785 |
| 7 | 12.7 | 13.3 | 37.4 | 0.397 |
| 8 | 21.5 | 10.2 | 67.7 | 0.635 |
| 9 | 21.2 | 11.0 | 65.2 | 0.684 |
| 10 | 20.2 | 12.3 | 49.3 | 0.496 |
| 11 | 22.5 | 9.8 | 67.7 | 0.627 |
| 12 | 17.0 | 9.7 | 67.7 | 0.627 |
| 13 | 15.7 | 9.5 | 65.2 | 0.616 |
| 14 | 20.7 | 8.7 | 81.2 | 0.836 |
| 15 | 17.5 | 12.0 | 43.1 | 0.425 |
| 16 | 19.7 | 10.7 | 73.1 | 0.741 |
| 17 | 22.0 | 7.7 | 88.0 | 0.875 |
| 18 | 21.0 | 8.5 | 80.0 | 0.819 |
| 19 | 17.5 | 8.7 | 82.0 | 0.846 |
| 20 | 21.5 | 8.2 | 84.0 | 0.860 |
| 21 | 20.0 | 11.8 | 58.2 | 0.553 |
| 22 | 17.0 | 10.7 | 60.1 | 0.601 |
| 23 | 15.2 | 12.7 | 50.1 | 0.512 |
| 24 | 20.2 | 12.5 | 57.3 | 0.573 |
| 25 | 27.2 | 16.0 | 29.0 | 0.354 |
| 26 | 10.0 | 12.7 | 53.5 | 0.517 |
| 27 | 16.7 | 11.0 | 64.3 | 0.611 |
| 28 | 17.2 | 8.7 | 78.0 | 0.763 |
| 29 | 12.7 | 10.7 | 62.4 | 0.608 |
| 30 | 12.5 | 9.3 | 78.0 | 0.763 |
| 31 | 18.0 | 11.0 | 65.4 | 0.627 |
| 32 | 10.2 | 11.0 | 62.4 | 0.618 |
| Trails | Antimicrobial Activity (Inhibition Zone Diameter, mm) | Physical Characteristics | ||
|---|---|---|---|---|
| S. aureus ATCC 6538 | C. albicans DAY185 | Size (nm) | PDI | |
| 1 | 28.0 | 26.0 | 52.7 | 0.467 |
| 2 | 26.0 | 28.0 | 81.4 | 0.724 |
| 3 | 27.0 | 26.5 | 75.4 | 0.634 |
| 4 | 28.0 | 27.0 | 80.3 | 0.712 |
| 5 | 23.0 | 24.0 | 38.2 | 0.375 |
| 6 | 18.0 | 18.5 | 88.0 | 0.776 |
| 7 | 24.5 | 26.0 | 35.5 | 0.362 |
| 8 | 29.0 | 28.0 | 39.7 | 0.383 |
| 9 | 28.0 | 26.0 | 47.2 | 0.434 |
| 10 | 30.0 | 29.0 | 29.0 | 0.356 |
| 11 | 23.6 | 24.2 | 50.2 | 0.467 |
| 12 | 23.7 | 25.4 | 42.0 | 0.410 |
| 13 | 24.5 | 25.6 | 61.4 | 0.567 |
| 14 | 20.1 | 19.0 | 52.0 | 0.534 |
| 15 | 24.0 | 25.0 | 46.0 | 0.434 |
| 16 | 18.3 | 19.0 | 72.5 | 0.612 |
| 17 | 20.0 | 18.7 | 65.4 | 0.578 |
| Tested Microorganism | Mo-Capped AgNPs (µg/mL) | ||
|---|---|---|---|
| MIC | MBC | MIC Index | |
| S. aureus ATCC 6538 | 4.0 | 16.0 | 4.0 |
| C. albicans DAY185 | 16.0 | 64.0 | 4.0 |
| Microorganism | MBEC (µg/mL) |
|---|---|
| S. aureus ATCC 6538 | 16.0 |
| C. albicans DAY185 | 32.0 |
| Consortia (S. aureus ATCC 6538 and C. albicans DAY185) | 32.0 |
| Trail/Variable | Reducing Agent Concentration (%) | pH | Ratio (Reducing Agent: AgNO3) | Temperature (°C) | Time (h) |
|---|---|---|---|---|---|
| 1 | −1 (30.0) | +1 (10.0) | −1 (3:1) | +1 (120.0) | +1 (3.0) |
| 2 | +1 (40.0) | +1 (10.0) | +1 (4:1) | −1 (80.0) | +1 (3.0) |
| 3 | −1 (30.0) | −1 (6.0) | +1 (4:1) | +1 (120.0) | +1 (3.0) |
| 4 | −1 (30.0) | −1 (6.0) | +1 (4:1) | +1 (120.0) | −1 (2.0) |
| 5 | −1 (30.0) | −1 (6.0) | +1 (4:1) | −1 (80.0) | +1 (3.0) |
| 6 | −1 (30.0) | +1 (10.0) | +1 (4:1) | +1 (120.0) | −1 (2.0) |
| 7 | −1 (30.0) | +1 (10.0) | +1 (4:1) | −1 (80.0) | +1 (3.0) |
| 8 | +1 (40.0) | −1 (6.0) | −1 (3:1) | +1 (120.0) | −1 (2.0) |
| 9 | +1 (40.0) | +1 (10.0) | −1 (3:1) | +1 (120.0) | +1 (3.0) |
| 10 | −1 (30.0) | −1 6.0) | −1 (3:1) | +1 (120.0) | +1 (3.0) |
| 11 | −1 (30.0) | −1 (6.0) | −1 (3:1) | +1 (120.0) | −1 (2.0) |
| 12 | +1 (40.0) | −1 (6.0) | −1 (3:1) | −1 (80.0) | +1 (3.0) |
| 13 | +1 (40.0) | −1 (6.0) | −1 (3:1) | +1 (120.0) | +1 (3.0) |
| 14 | −1 (30.0) | +1 (10.0) | −1 (3:1) | −1 (80.0) | +1 (3.0) |
| 15 | −1 (30.0) | +1 (10.0) | +1 (4:1) | −1 (80.0) | −1 (2.0) |
| 16 | −1 (30.0) | −1 (6.0) | −1 (3:1) | −1 (80.0) | +1 (3.0) |
| 17 | +1 (40.0) | +1 (10.0) | +1 (4:1) | −1 (80.0) | −1 (2.0) |
| 18 | +1 (40.0) | +1 (10.0) | −1 (3:1) | −1 (80.0) | +1 (3.0) |
| 19 | +1 (40.0) | +1 (10.0) | +1 (4:1) | +1 (120.0) | −1 (2.0) |
| 20 | +1 (40.0) | +1 (10.0) | −1 (3:1) | +1 (120.0) | −1 (2.0) |
| 21 | +1 (40.0) | −1 (6.0) | +1 (4:1) | −1 (80.0) | −1 (2.0) |
| 22 | +1 (40.0) | −1 (6.0) | +1 (4:1) | +1 (120.0) | −1 (2.0) |
| 23 | −1 (30.0) | −1 (6.0) | +1 (4:1) | −1 (80.0) | −1 (2.0) |
| 24 | −1 (30.0) | −1 (6.0) | −1 (3:1) | −1 (80.0) | −1 (2.0) |
| 25 | −1 (30.0) | +1 (10.0) | +1 (4:1) | +1 (120.0) | +1 (3.0) |
| 26 | +1 (40.0) | −1 (6.0) | +1 (4:1) | −1 (80.0) | +1 (3.0) |
| 27 | +1 (40.0) | +1 (10.0) | −1 (3:1) | +1 (120.0) | −1 (2.0) |
| 28 | +1 (40.0) | +1 (10.0) | +1 (4:1) | +1 (120.0) | +1 (3.0) |
| 29 | +1 (40.0) | −1 (6.0) | +1 (4:1) | +1 (120.0) | +1 (3.0) |
| 30 | +1 (40.0) | +1 (10.0) | −1 (3:1) | −1 (80.0) | −1 (2.0) |
| 31 | −1 (30.0) | +1 (10.0) | −1 (3:1) | −1 (80.0) | −1 (2.0) |
| 32 | +1 (40.0) | −1 (6.0) | −1 (3:1) | −1 (80.0) | −1 (2.0) |
| Trial Number | Reducing Agent Concentration (%) | pH | Temperature (°C) |
|---|---|---|---|
| 1 | 0 (30.0) | 0 (8.0) | 0 (100.0) |
| 2 | 0 (30.0) | −1 (6.0) | +1 (120.0) |
| 3 | +1 (40.0) | 0 (8.0) | −1 (80.0) |
| 4 | −1 (20.0) | −1 (6.0) | 0 (100.0) |
| 5 | 0 (30.0) | +1 (10.0) | −1 (80.0) |
| 6 | +1 (40.0) | −1(6.0) | −1 (80.0) |
| 7 | −1 (20.0) | 0 (8.0) | −1 (80.0) |
| 8 | +1 (40.0) | +1 (10.0) | 0 (100.0) |
| 9 | +1 (40.0) | −1 (6.0) | 0 (100.0) |
| 10 | 0 (30.0) | +1 (10.0) | +1 (120.0) |
| 11 | +1 (40.0) | 0 (8.0) | +1 (120.0) |
| 12 | −1 (20.0) | +1 (10.0) | +1 (120.0) |
| 13 | −1 (20.0) | 0 (8.0) | +1 (120.0) |
| 14 | 1 (40.0) | −1 (6.0) | +1 (120.0) |
| 15 | 0 (30.0) | −1 (6.0) | 0 (100.0) |
| 16 | −1 (20.0) | 1 (8.0) | −1 (80.0) |
| 17 | 1 (40.0) | 0 (7.0) | 0 (100.0) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dorgham, R.A.; Abd Al Moaty, M.N.; Chong, K.P.; Elwakil, B.H. Molasses-Silver Nanoparticles: Synthesis, Optimization, Characterization, and Antibiofilm Activity. Int. J. Mol. Sci. 2022, 23, 10243. https://doi.org/10.3390/ijms231810243
Dorgham RA, Abd Al Moaty MN, Chong KP, Elwakil BH. Molasses-Silver Nanoparticles: Synthesis, Optimization, Characterization, and Antibiofilm Activity. International Journal of Molecular Sciences. 2022; 23(18):10243. https://doi.org/10.3390/ijms231810243
Chicago/Turabian StyleDorgham, Rabab A., Mohamed N. Abd Al Moaty, Khim Phin Chong, and Bassma H. Elwakil. 2022. "Molasses-Silver Nanoparticles: Synthesis, Optimization, Characterization, and Antibiofilm Activity" International Journal of Molecular Sciences 23, no. 18: 10243. https://doi.org/10.3390/ijms231810243
APA StyleDorgham, R. A., Abd Al Moaty, M. N., Chong, K. P., & Elwakil, B. H. (2022). Molasses-Silver Nanoparticles: Synthesis, Optimization, Characterization, and Antibiofilm Activity. International Journal of Molecular Sciences, 23(18), 10243. https://doi.org/10.3390/ijms231810243







