Bioconjugated Thymol-Zinc Oxide Nanocomposite as a Selective and Biocompatible Antibacterial Agent against Staphylococcus Species
Abstract
:1. Introduction
2. Results and Discussion
2.1. Material Properties
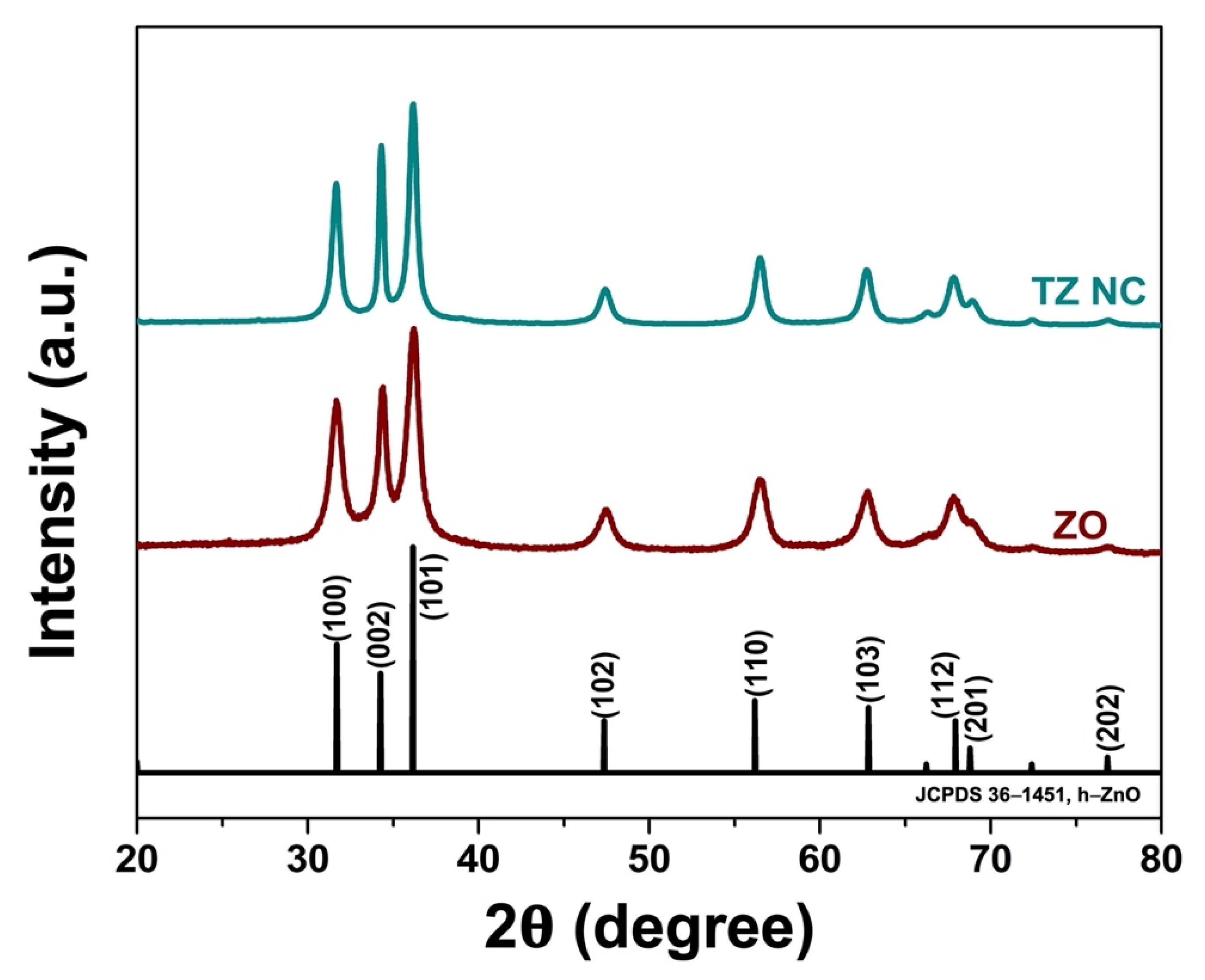
2.1.1. Phase Composition
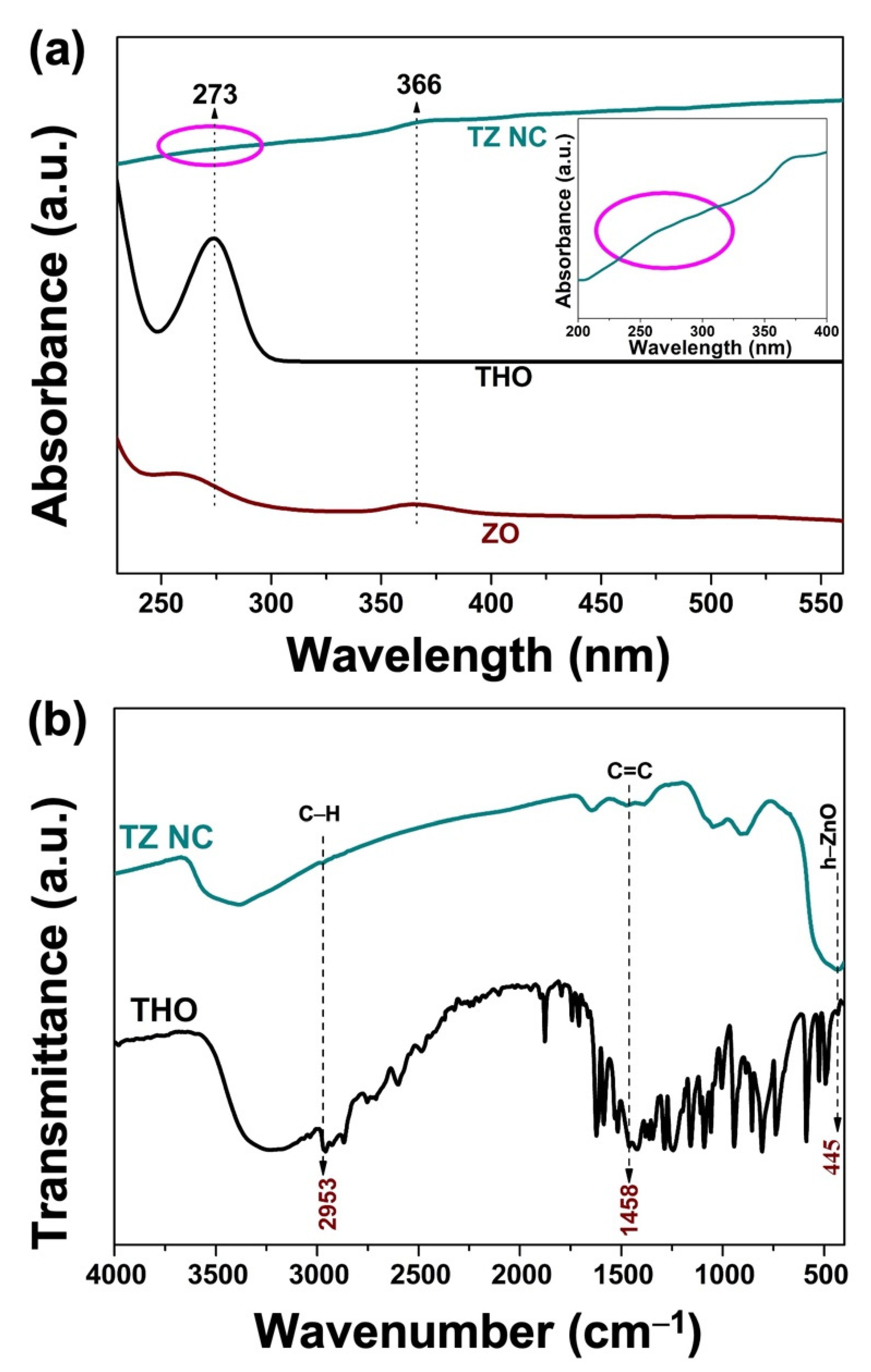
2.1.2. UV–Vis Analysis
2.1.3. FT-IR Analysis
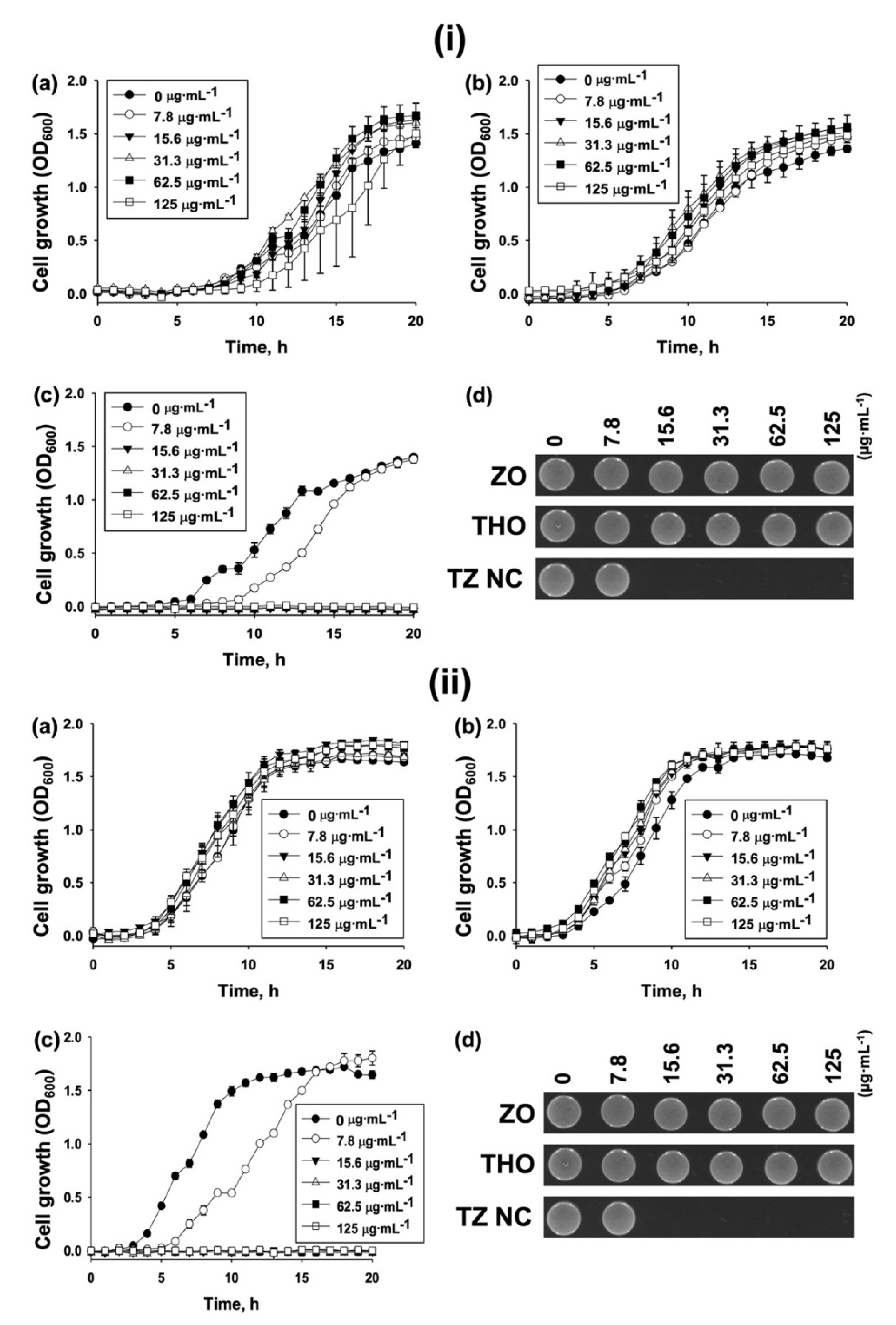
2.2. Biological Activity
Antibacterial Activity
2.3. Plausible Antibacterial Mechanism
2.3.1. Inhibition of Bacterial Biofilm Formation by TZ NC
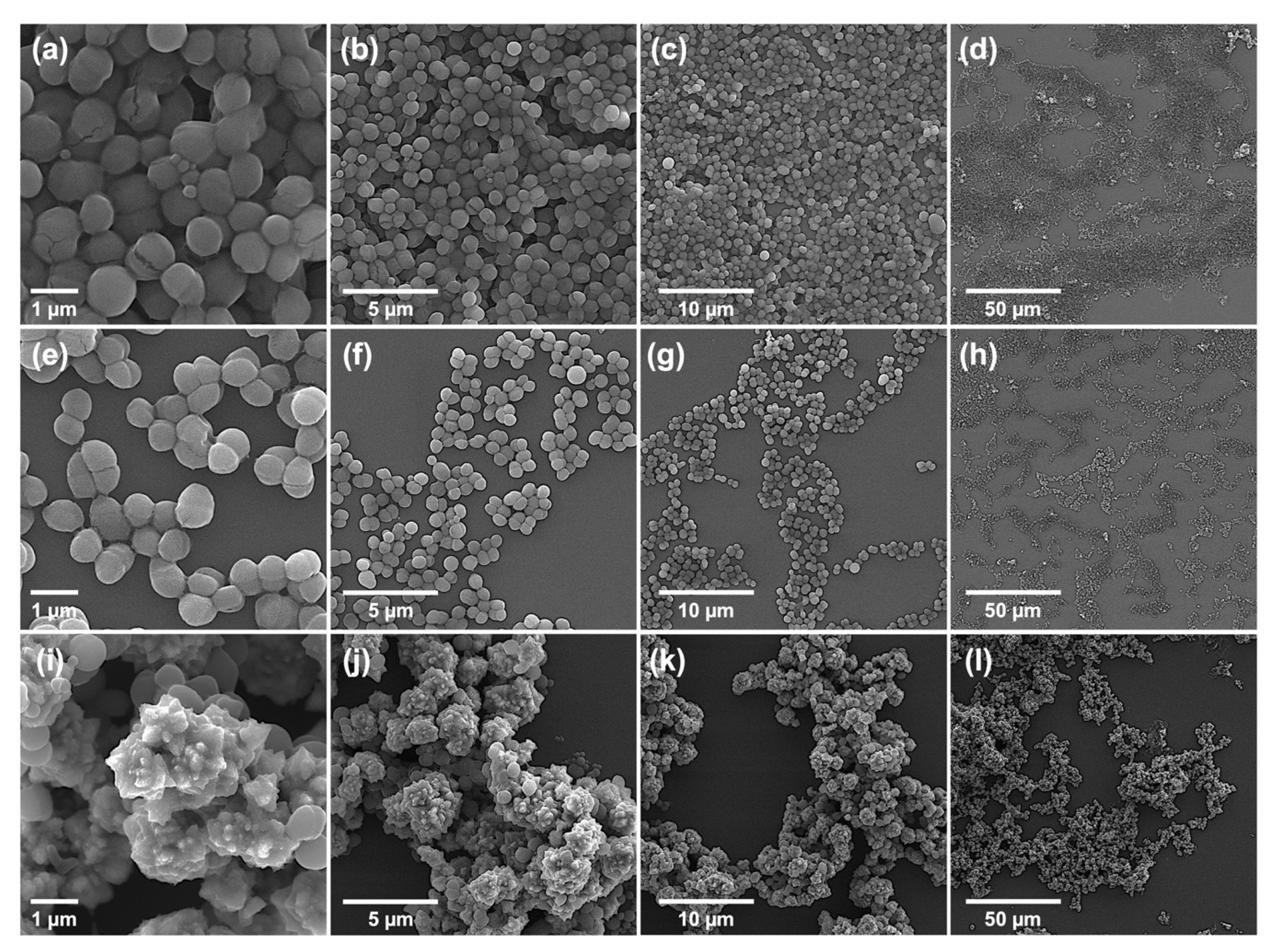
2.3.2. Morphological Characterization of Bacteria: Bursting from Within
2.3.3. Synergistic Action of TZ NC with Antibiotics
2.4. Biocompatibility of TZ NC
3. Materials and Methods
3.1. Biosynthesis of TZ NC
3.2. Characterization
3.2.1. Properties of the Materials
3.2.2. Preparation of Bacterial Cells
3.2.3. Determination of MICs
3.2.4. Growth Curve Analysis
3.2.5. Crystal Violet Assays
3.2.6. Morphological Characterization of Bacterial Cells and Nanocomposite
3.2.7. Screening of Synergistic Antibiotics with TZ NC
3.2.8. Biocompatibility Assays
3.2.9. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brown, M.M.; Horswill, A.R. Staphylococcus epidermidis—Skin friend or foe? PLoS Pathog. 2020, 16, e1009026. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, R.; Priyadarshini, A.; Pandey, R.P.; Raj, V.S. Antimicrobial Resistance in Staphylococcus aureus. In Insights into Drug Resistance in Staphylococcus aureus; Aqib, A., Ed.; IntechOpen: London, UK, 2021. [Google Scholar] [CrossRef]
- Diallo, O.O.; Baron, S.A.; Abat, C.; Colson, P.; Chaudet, H.; Rolain, J.M. Antibiotic resistance surveillance systems: A review. J. Glob. Antimicrob. Resist. 2020, 23, 430–438. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Han, R.; Xu, Y.; Li, N.; Wang, J.; Dan, W. Recent progress of antibacterial natural products: Future antibiotics candidates. Bioorg. Chem. 2020, 101, 103922. [Google Scholar] [CrossRef] [PubMed]
- Marchese, A.; Orhan, I.E.; Daglia, M.; Barbieri, R.; Lorenzo, A.D.; Nabavi, S.F.; Gortzi, O.; Izadi, M.; Nabavi, S.M. Antibacterial and antifungal activities of thymol: A brief review of the literature. Food Chem. 2016, 210, 402–414. [Google Scholar] [CrossRef]
- Olasupo, N.A.; Fitzgerald, D.J.; Gasson, M.J.; Narbad, A. Activity of natural antimicrobial compounds against Escherichia coli and Salmonella enterica serovar Typhimurium. Lett. Appl. Microbiol. 2003, 37, 448–451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trombetta, D.; Castelli, F.; Sarpietro, M.G.; Venuti, V.; Cristani, M.; Daniele, C.; Saija, A.; Mazzanti, G.; Bisignano, G. Mechanisms of antibacterial action of three monoterpenes. Antimicrob. Agents Chemother. 2005, 49, 2474–2478. [Google Scholar] [CrossRef] [Green Version]
- Tian, L.; Wang, X.Y.; Liu, R.J.; Zhang, D.; Wang, X.; Sun, R.C.; Guo, W.Y.; Yang, S.Q.; Li, H.; Gong, G.L. Antibacterial mechanism of thymol against Enterobacter sakazakii. Food Control 2021, 123, 107716. [Google Scholar] [CrossRef]
- Liang, C.; Huang, S.H.; Geng, Y.; Huang, X.L.; Chen, D.F.; Lai, W.M.; Guo, H.R.; Deng, H.D.; Fang, J.; Yin, L.Z.; et al. A Study on the antibacterial mechanism of thymol against Aeromonas hydrophila in vitro. Aquac. Int. 2022, 30, 115–129. [Google Scholar] [CrossRef]
- Pivetta, T.P.; Simoes, S.; Araujo, M.M.; Carvalho, T.; Arruda, C.; Marcato, P.D. Development of nanoparticles from natural lipids for topical delivery of thymol: Investigation of its anti-inflammatory properties. Colloids Surf. B 2018, 164, 281–290. [Google Scholar] [CrossRef]
- Pires, F.Q.; Pinho, L.A.; Freire, D.O.; Silva, I.C.R.; Sa-Barreto, L.L.; Cardozo, L.; Gratieri, T.; Gelfuso, G.M.; Cunha, M. Thermal analysis used to guide the production of thymol and Lippia origanoides essential oil inclusion complexes with cyclodextrin. Therm. Anal. Calorim. 2019, 137, 543–553. [Google Scholar] [CrossRef]
- Carofiglio, M.; Barui, S.; Cauda, V.; Laurenti, M. Doped Zinc Oxide Nanoparticles: Synthesis, Characterization and Potential Use in Nanomedicine. Appl. Sci. 2020, 10, 5194. [Google Scholar] [CrossRef]
- Ma, L.; Kohli, M.; Smith, A. Nanoparticles for combination drug therapy. ACS Nano 2013, 7, 9518–9525. [Google Scholar] [CrossRef] [Green Version]
- Alavi, M.; Rai, M. Recent advances in antibacterial applications of metal nanoparticles (MNPs) and metal nanocomposites (MNCs) against multidrug-resistant (MDR) bacteria. Expert Rev. Anti-Infect. Ther. 2019, 17, 419–428. [Google Scholar] [CrossRef]
- Naskar, A.; Kim, K.S. Nanomaterials as Delivery Vehicles and Components of New Strategies to Combat Bacterial Infections: Advantages and Limitations. Microorganisms 2019, 7, 356. [Google Scholar] [CrossRef] [Green Version]
- Pozdnyakov, A.; Emel’yanov, A.; Ivanova, A.; Kuznetsova, N.; Semenova, T.; Bolgova, Y.; Korzhova, S.; Trofimova, O.; Fadeeva, T.; Prozorova, G. Strong Antimicrobial Activity of Highly Stable Nanocomposite Containing AgNPs Based on Water-Soluble Triazole-Sulfonate Copolymer. Pharmaceutics 2022, 14, 206. [Google Scholar] [CrossRef]
- Naskar, A.; Lee, S.; Lee, Y.; Kim, S.; Kim, K.S. A New Nano-Platform of Erythromycin Combined with Ag Nano-Particle ZnO Nano-Structure against Methicillin-Resistant Staphylococcus aureus. Pharmaceutics 2020, 12, 841. [Google Scholar] [CrossRef]
- Slavin, Y.N.; Asnis, J.; Hafeli, U.O.; Bach, H. Metal nanoparticles: Understanding the mechanisms behind antibacterial activity. J. Nanobiotechnol. 2017, 15, 65. [Google Scholar] [CrossRef]
- Gudkov, S.V.; Burmistrov, D.E.; Serov, D.A.; Rebezov, M.B.; Semenova, A.A.; Lisitsyn, A.B. A Mini Review of Antibacterial Properties of ZnO Nanoparticles. Front. Phys. 2021, 9, 641481. [Google Scholar] [CrossRef]
- Premanathan, M.; Karthikeyan, K.; Jeyasubramanian, K.; Manivannan, G. Selective toxicity of ZnO nanoparticles toward Gram-positive bacteria and cancer cells by apoptosis through lipid peroxidation. Nanomedicine 2011, 7, 184–192. [Google Scholar] [CrossRef]
- Sanchez-Lopez, E.; Gomes, D.; Esteruelas, G.; Bonilla, L.; Lopez-Machado, A.L.; Galindo, R.; Cano, A.; Espina, M.; Ettcheto, M.; Camins, A.; et al. Metal-Based Nanoparticles as Antimicrobial Agents: An Overview. Nanomaterials 2020, 10, 292. [Google Scholar] [CrossRef] [Green Version]
- Singh, P.; Kim, Y.J.; Zhang, D.B.; Yang, D.C. Biological Synthesis of Nanoparticles from Plants and Microorganisms. Trends Biotechnol. 2016, 34, 588–599. [Google Scholar] [CrossRef]
- Vigneshwaran, N.; Ashtaputre, N.M.; Varadarajan, P.V.; Nachane, R.P.; Paralikar, K.M.; Balasubramanya, R.H. Biological synthesis of silver nanoparticles using the fungus Aspergillus flavus. Mater. Lett. 2007, 61, 1413–1418. [Google Scholar] [CrossRef]
- Yusof, H.M.; Rahman, N.A.; Mohamad, R.; Zaidan, U.H.; Samsudin, A.A. Biosynthesis of zinc oxide nanoparticles by cell-biomass and supernatant of Lactobacillus plantarum TA4 and its antibacterial and biocompatibility properties. Sci. Rep. 2020, 10, 19996. [Google Scholar] [CrossRef]
- Yusof, H.M.; Mohamad, R.; Zaidan, U.H.; Rahman, N.A.A. Microbial synthesis of zinc oxide nanoparticles and their potential application as an antimicrobial agent and a feed supplement in animal industry: A review. J. Anim. Sci. Biotechnol. 2019, 10, 57. [Google Scholar] [CrossRef] [PubMed]
- Kitching, M.; Ramani, M.; Marsili, E. Fungal biosynthesis of gold nanoparticles: Mechanism and scale up. Microb. Biotechnol. 2015, 8, 904–917. [Google Scholar] [CrossRef] [PubMed]
- Singh, J.; Kaur, S.; Kaur, G.; Basu, S.; Rawat, M. Biogenic ZnO nanoparticles: A study of blueshift of optical band gap and photocatalytic degradation of reactive yellow 186 dye under direct sunlight. Green Process. Synth. 2019, 8, 272–280. [Google Scholar] [CrossRef] [Green Version]
- Lin, L.; Zhu, Y.; Thangaraj, B.; Abdel-Samie, M.A.S.; Cui, H. Improving the stability of thyme essential oil solid liposome by using β-cyclodextrin as a cryoprotectant. Carbohydr. Polym. 2018, 188, 243–251. [Google Scholar] [CrossRef]
- Pleva, P.; Bartošová, L.; Máčalová, D.; Zálešáková, L.; Sedlaříková, J.; Janalíková, M. Biofilm Formation Reduction by Eugenol and Thymol on Biodegradable Food Packaging Material. Foods 2022, 11, 2. [Google Scholar] [CrossRef]
- Trivedi, S.P.M.K.; Mishra, R.; Jana, S. Structural and Physical Properties of Biofield Treated Thymol and Menthol. J. Mol. Pharm. Org. Process Res. 2015, 3, 1000127. [Google Scholar] [CrossRef]
- Murray, C.J.L.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Aguilar, G.R.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Otto, M. Staphylococcus epidermidis—The ‘accidental’ pathogen. Nat. Rev. Microbiol. 2009, 7, 555–567. [Google Scholar] [CrossRef] [Green Version]
- Guo, Y.L.; Song, G.H.; Sun, M.L.; Wang, J.; Wang, Y. Prevalence and Therapies of Antibiotic-Resistance in Staphylococcus aureus. Front. Cell Infect. Microbiol. 2020, 10, 107. [Google Scholar] [CrossRef] [Green Version]
- Shin, J.; Magar, K.B.S.; Lee, J.; Kim, K.S.; Lee, Y.R. Design, synthesis, and discovery of novel oxindoles bearing 3-heterocycles as species-specific and combinatorial agents in eradicating Staphylococcus species. Sci. Rep. 2019, 9, 8012. [Google Scholar] [CrossRef] [Green Version]
- Costa, S.S.; Viveiros, M.; Pomba, C.; Couto, I. Active antimicrobial efflux in Staphylococcus epidermidis: Building up of resistance to fluoroquinolones and biocides in a major opportunistic pathogen. J. Antimicrob. Chemother. 2018, 73, 320–324. [Google Scholar] [CrossRef] [Green Version]
- Dopcea, G.N.; Dopcea, I.; Nanu, A.E.; Diguţă, C.F.; Matei, F. Resistance and cross-resistance in Staphylococcus spp. strains following prolonged exposure to different antiseptics. J. Glob. Antimicrob. 2020, 21, 399–404. [Google Scholar] [CrossRef]
- Águila-Arcos, S.; Álvarez-Rodríguez, I.; Garaiyurrebaso, O.; Garbisu, C.; Grohmann, E.; Alkorta, I. Biofilm-Forming Clinical Staphylococcus Isolates Harbor Horizontal Transfer and Antibiotic Resistance Genes. Front. Microbiol. 2017, 8, 2018. [Google Scholar] [CrossRef]
- Haunreiter, V.D.; Boumasmoud, M.; Häffner, N.; Wipfli, D.; Leimer, N.; Rachmühl, C.; Kühnert, D.; Achermann, Y.; Zbinden, R.; Benussi, S.; et al. In-host evolution of Staphylococcus epidermidis in a pacemaker-associated endocarditis resulting in increased antibiotic tolerance. Nat. Commun. 2019, 10, 1149. [Google Scholar] [CrossRef]
- Choo, P.; Liu, T.T.; Odom, T.W. Nanoparticle Shape Determines Dynamics of Targeting Nanoconstructs on Cell Membranes. J. Am. Chem. Soc. 2021, 143, 4550–4555. [Google Scholar] [CrossRef]
- Hari, T.K.T.N.; Nair, A.J. Comparative Study on the Synergistic Action of Differentially Synthesized Silver Nanoparticles with β-Cephem Antibiotics and Chloramphenicol. J. Nanosci. 2014, 2014, 8. [Google Scholar] [CrossRef] [Green Version]
- Leach, K.L.; Brickner, S.J.; Noe, M.C.; Miller, P.F. Linezolid, the first oxazolidinone antibacterial agent, Pharmaceutical Science to Improve the Human Condition. Ann. N. Y. Acad. Sci. 2011, 1222, 49–54. [Google Scholar] [CrossRef]
- Soares, G.M.; Figueiredo, L.C.; Faveri, M.; Cortelli, S.C.; Duarte, P.M.; Feres, M. Mechanisms of action of systemic antibiotics used in periodontal treatment and mechanisms of bacterial resistance to these drugs. J. Appl. Oral Sci. 2012, 20, 295–304. [Google Scholar] [CrossRef] [Green Version]
- Schedlbauer, A.; Kaminishi, T.; Ochoa-Lizarralde, B.; Dhimole, N.; Zhou, S.; Lopez-Alonso, J.P.; Connell, S.R.; Fucini, P. Structural characterization of an alternative mode of tigecycline binding to the bacterial ribosome. Antimicrob. Agents Chemother. 2015, 59, 2849–2854. [Google Scholar] [CrossRef] [Green Version]
- Cho, H.; Naskar, A.; Lee, S.; Kim, S.; Kim, K.S. A New Surface Charge Neutralizing Nano-Adjuvant to Potentiate Polymyxins in Killing Mcr-1 Mediated Drug-Resistant Escherichia coli. Pharmaceutics 2021, 13, 250. [Google Scholar] [CrossRef]
- Naskar, A.; Shin, J.; Kim, K.S. A MoS2 based silver-doped ZnO nanocomposite and its antibacterial activity against β-lactamase expressing Escherichia coli. RSC Adv. 2022, 12, 7268–7275. [Google Scholar] [CrossRef]
- Kim, T.; Lee, J.; Kim, K.S. Escherichia coli YmdB regulates biofilm formation independently of its role as an RNase III modulator. BMC Microbiol. 2013, 13, 266. [Google Scholar] [CrossRef] [Green Version]
- Ansari, M.A.; Albetran, H.M.; Alheshibri, M.H.; Timoumi, A.; Algarou, N.A.; Akhtar, S.; Slimani, Y.; Almessiere, M.A.; Alahmari, F.S.; Baykal, A.; et al. Synthesis of Electrospun TiO2 Nanofibers and Characterization of Their Antibacterial and Antibiofilm Potential against Gram-Positive and Gram-Negative Bacteria. Antibiotics 2020, 9, 572. [Google Scholar] [CrossRef]
- Wiegand, I.; Hilpert, K.; Hancock, R.E.W. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008, 3, 163–175. [Google Scholar] [CrossRef]


| Species | Strain Type | Strain Identification Number 1 | Resistant Antibiotics 2 | MIC (µg·mL−1) | ||
|---|---|---|---|---|---|---|
| ZO | THO | TZ | ||||
| Escherichia coli | Type | ATCC 25922 | None | >500 | 500 | >500 |
| MDR | ATCC BAA-2452 | AMI, AMP, AZT, CEF, CTX, ETP, GEN, IMP, MER, PIP, TIC, TOB | >500 | >500 | >500 | |
| ATCC BAA-2469 | AMI, AMP, AZT, CEF, CIP, CTX, ETP, GEN, IMP, MER, NAL, NOR, PIP, TIC, TOB | >500 | >500 | >500 | ||
| ATCC BAA-2471 | AMP, AZT, CEF, CIP, CTX, ETP, GEN, IMP, MER, NAL, NOR, PIP, TIC, TOB | >500 | >500 | >500 | ||
| Staphylococcus aureus | Type | ATCC 25923 | None | >500 | >500 | 62.5 |
| MDR 3 | MRSA1 | MET, OXA | >500 | >500 | 250 | |
| MRSA2 | MET, OXA | >500 | >500 | 500 | ||
| MRSA3 | MET, OXA | >500 | >500 | 125 | ||
| MRSA4 | MET, OXA | >500 | >500 | >500 | ||
| S. epidermidis | Type | ATCC 14990 | None | >500 | >500 | 15.6 |
| KCTC 13171 | None | >500 | >500 | 31.3 | ||
| MDR 4 | ATCC 12228 | STR, AMP, PEN | >500 | >500 | 15.6 | |
| S. warneri | Type | ATCC 27836 | None | >500 | >500 | 250 |
| Antibiotics | Acronym | Subclass | MIC (μg·mL−1) 1 | |
|---|---|---|---|---|
| −TZ | +TZ | |||
| Ampicillin | AMP | β-lactam | 0.125 | 0.0625 |
| Chloramphenicol | CHL | Amphenicol | >4 | 4 |
| Ciprofloxacin | CIP | Fluoroquinolone | <0.25 | <0.25 |
| Clindamycin | CLI | Lincosamide | <0.125 | <0.125 |
| Daptomycin | DAP | Cyclic lipopeptide | >1 | 1 |
| Erythromycin | ERY | Macrolide | 0.5 | 0.25 |
| Gentamicin | GEN | Aminoglycoside | <0.5 | <0.5 |
| Levofloxacin | LEVO | Fluoroquinolone | 0.25 | 0.125 |
| Linezolid | LZD | Oxazolidinone | >2 | 1 |
| Moxifloxacin | MXF | Fluoroquinolone | 0.125 | 0.125 |
| Nitrofurantoin | NIT | Nitrofuran | 16 | <8 |
| Oxacillin +2%NaCl | OXA+ | β-lactam | 0.125 | 0.125 |
| Penicillin | PEN | β-lactam | 0.25 | 0.0625 |
| Quinupristin/ Dalfopristin | SYN | Lincosamide | <0.125 | <0.125 |
| Rifampin | RIF | Rifampicin | <0.125 | <0.125 |
| Streptomycin | STR | Aminoglycoside | <250 | <250 |
| Tetracycline | TET | Tetracycline | >4 | >4 |
| Tigecycline | TGC | Tetracycline | 0.125 | 0.03 |
| Trimethoprim/sulfamethoxazole | SXT | Co-trimoxazole | 0.25/4.75 | 0.5/9.5 |
| Vancomycin | VAN | Glycopeptide | 2 | 2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shin, J.; Naskar, A.; Ko, D.; Kim, S.; Kim, K.-s. Bioconjugated Thymol-Zinc Oxide Nanocomposite as a Selective and Biocompatible Antibacterial Agent against Staphylococcus Species. Int. J. Mol. Sci. 2022, 23, 6770. https://doi.org/10.3390/ijms23126770
Shin J, Naskar A, Ko D, Kim S, Kim K-s. Bioconjugated Thymol-Zinc Oxide Nanocomposite as a Selective and Biocompatible Antibacterial Agent against Staphylococcus Species. International Journal of Molecular Sciences. 2022; 23(12):6770. https://doi.org/10.3390/ijms23126770
Chicago/Turabian StyleShin, Joonho, Atanu Naskar, Dongjoon Ko, Semi Kim, and Kwang-sun Kim. 2022. "Bioconjugated Thymol-Zinc Oxide Nanocomposite as a Selective and Biocompatible Antibacterial Agent against Staphylococcus Species" International Journal of Molecular Sciences 23, no. 12: 6770. https://doi.org/10.3390/ijms23126770
APA StyleShin, J., Naskar, A., Ko, D., Kim, S., & Kim, K.-s. (2022). Bioconjugated Thymol-Zinc Oxide Nanocomposite as a Selective and Biocompatible Antibacterial Agent against Staphylococcus Species. International Journal of Molecular Sciences, 23(12), 6770. https://doi.org/10.3390/ijms23126770








