SIRT1/FOXO Signaling Pathway in Breast Cancer Progression and Metastasis
Abstract
1. Introduction
2. Results
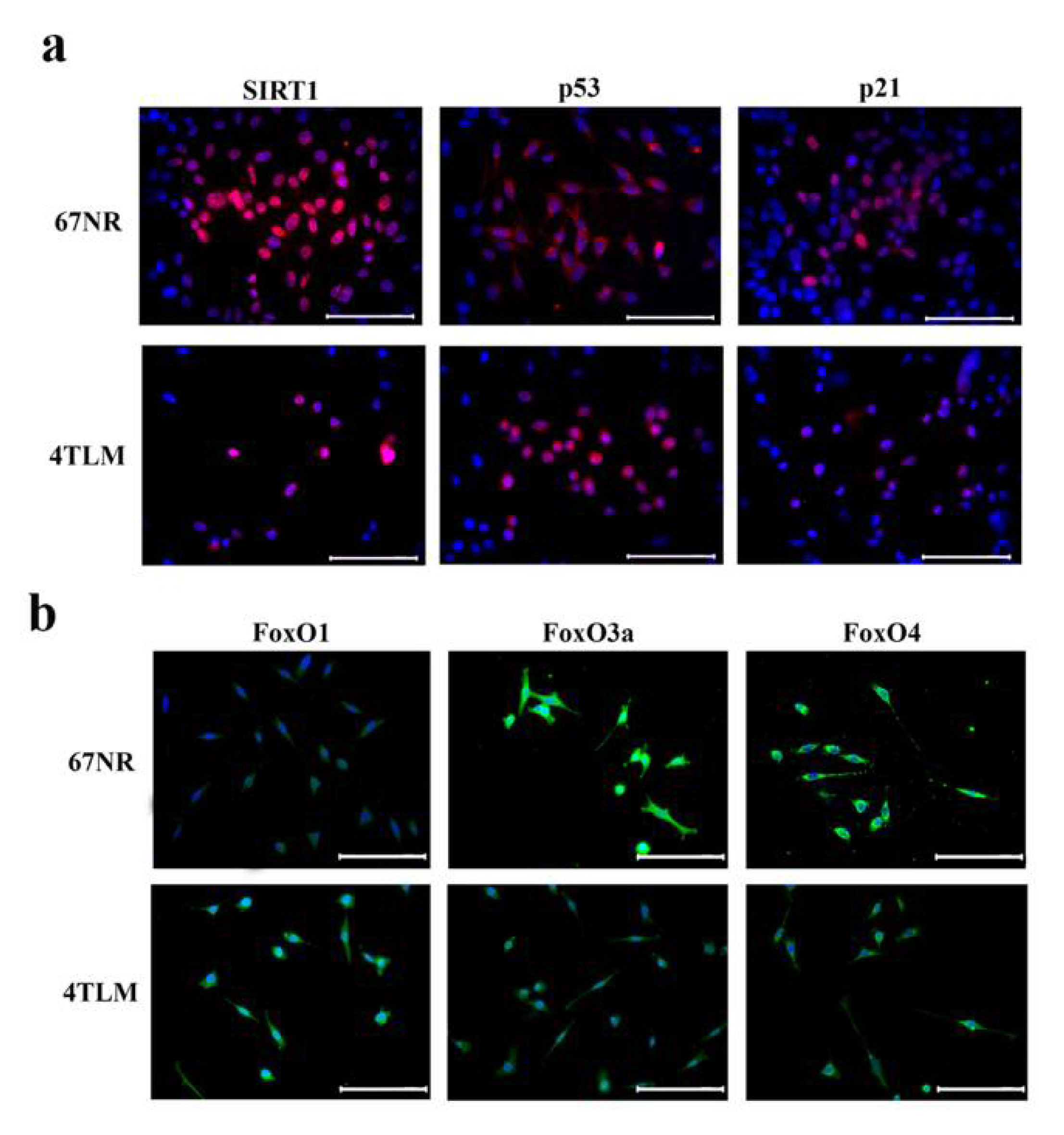
2.1. In Primary Tumors, Both 67NR and 4TLM Tumor Cells Exhibited Differential Expression Levels and Localizations of SIRT1 and FoxOs
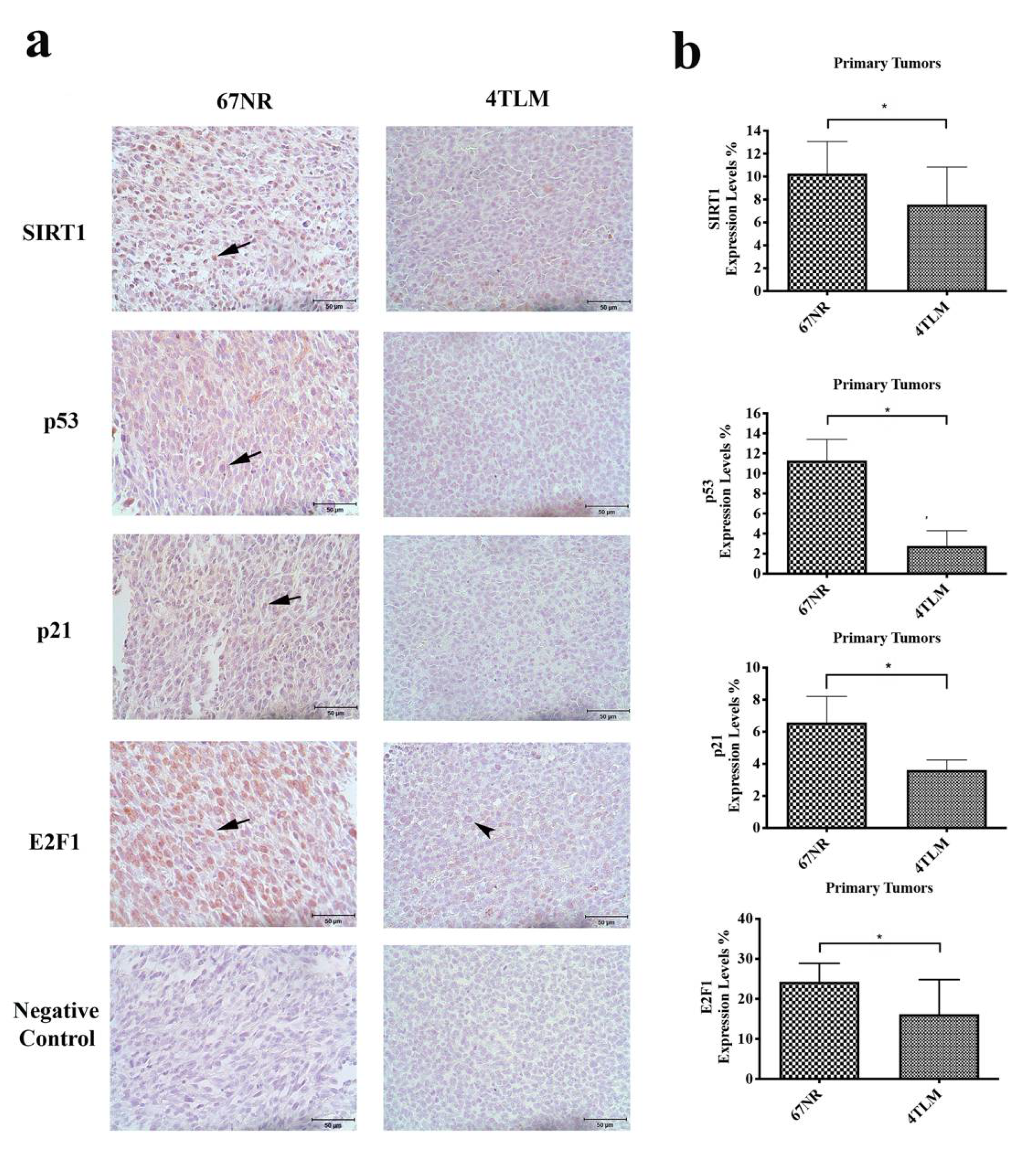
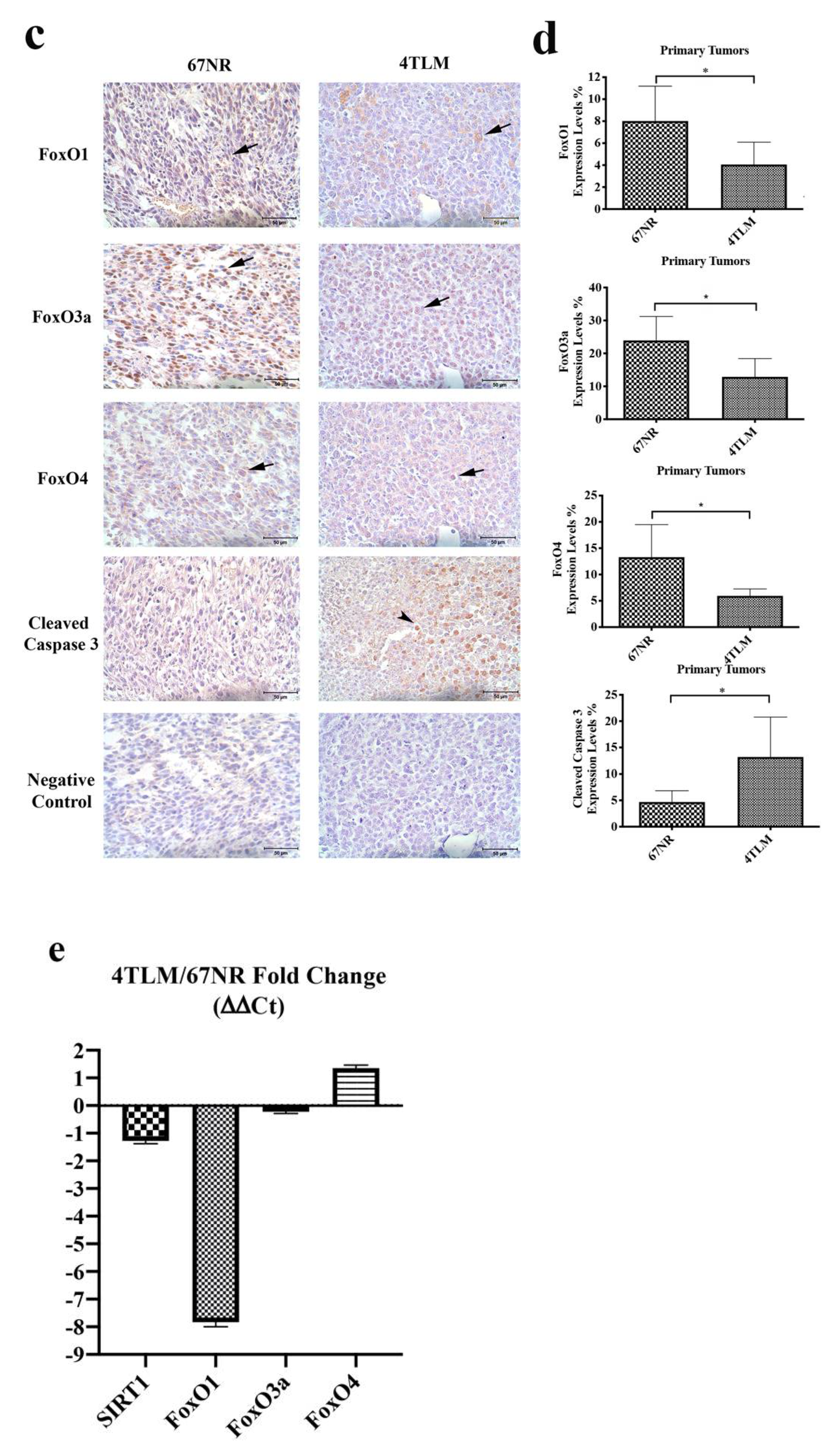
2.2. Expression of SIRT1 and FoxOs Differed between 67NR and 4TLM Primary Tumors in Mice
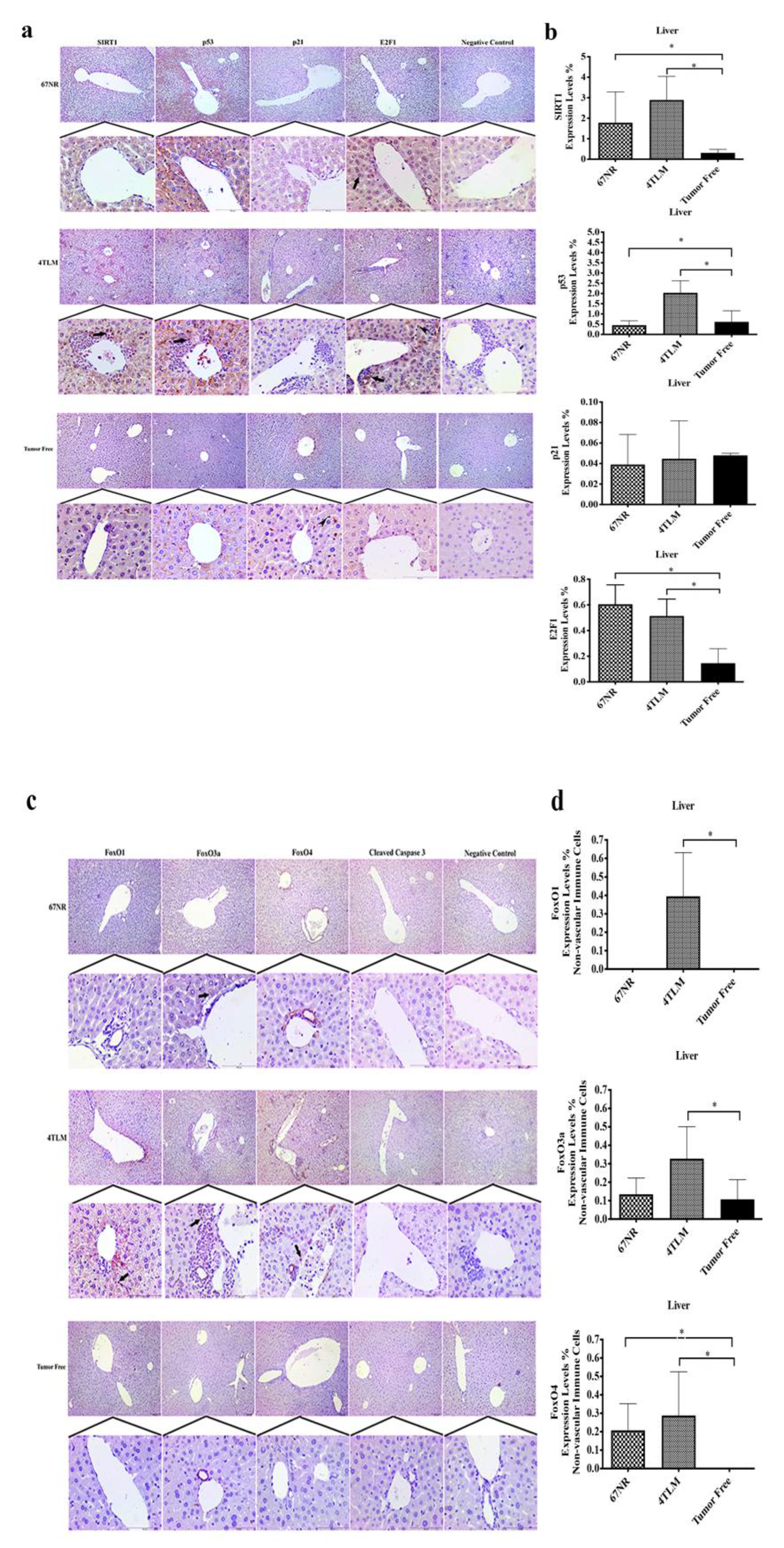
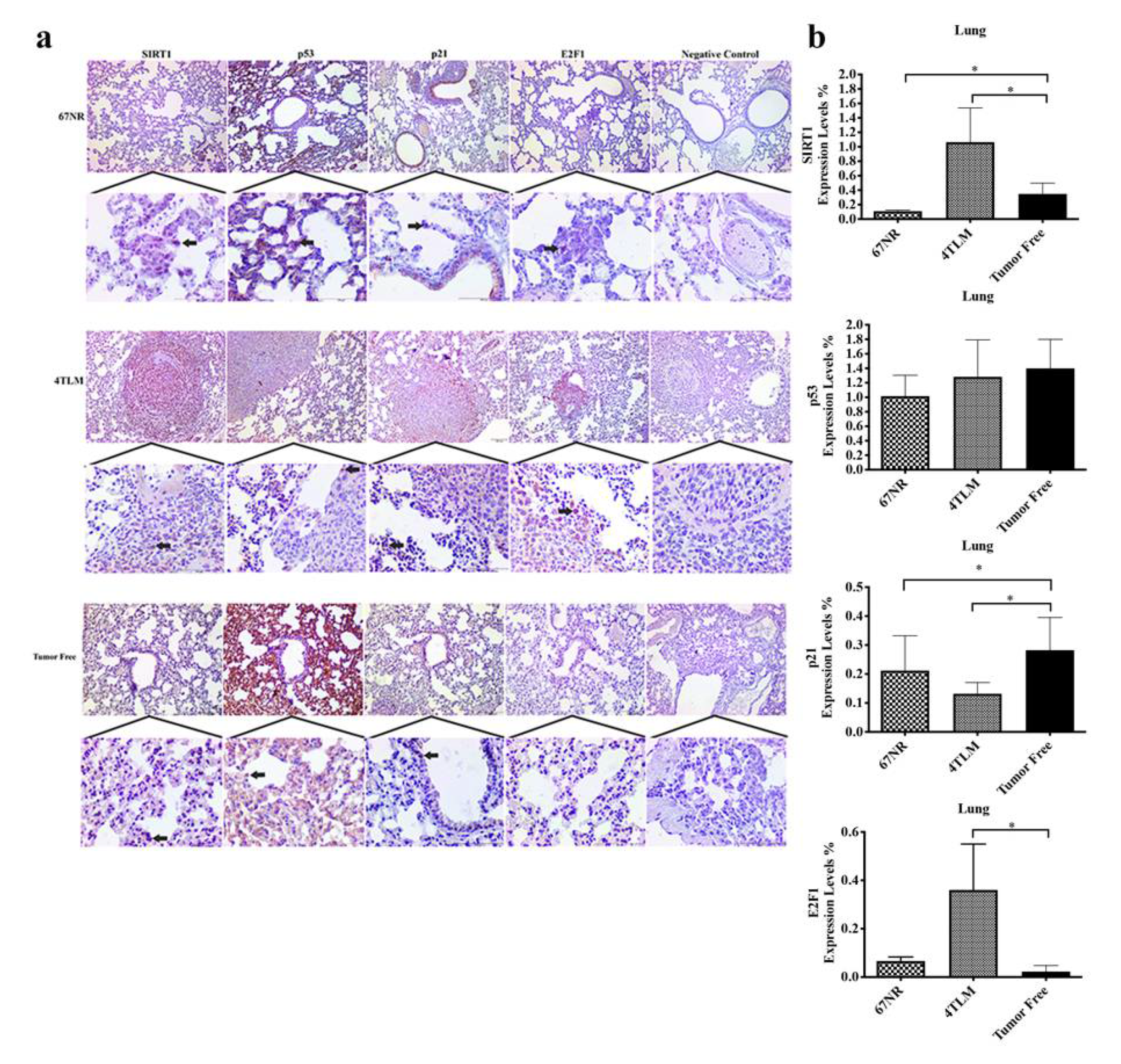
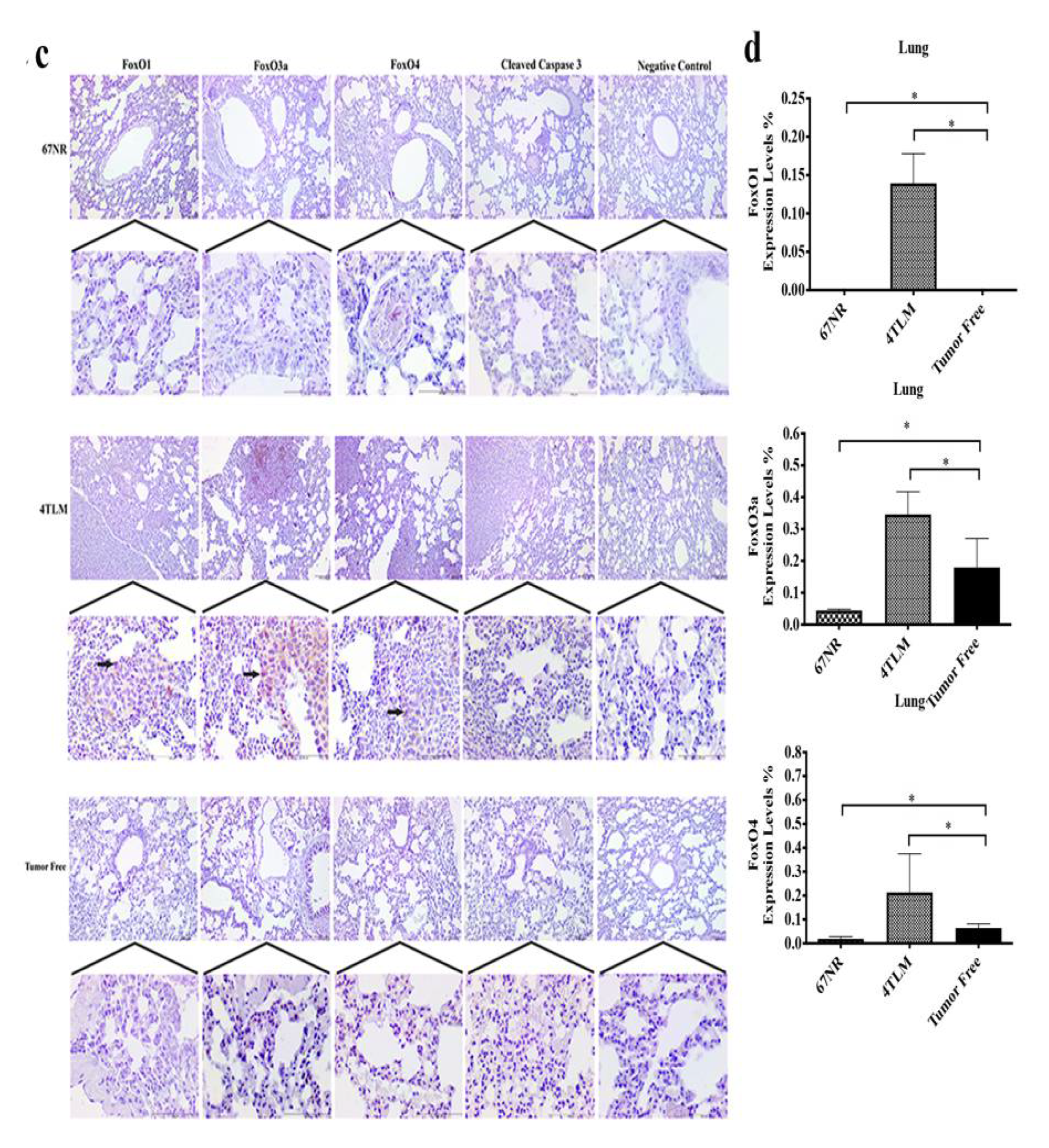
2.3. Expression Levels of SIRT1 and FoxOs in Metastatic Tissues
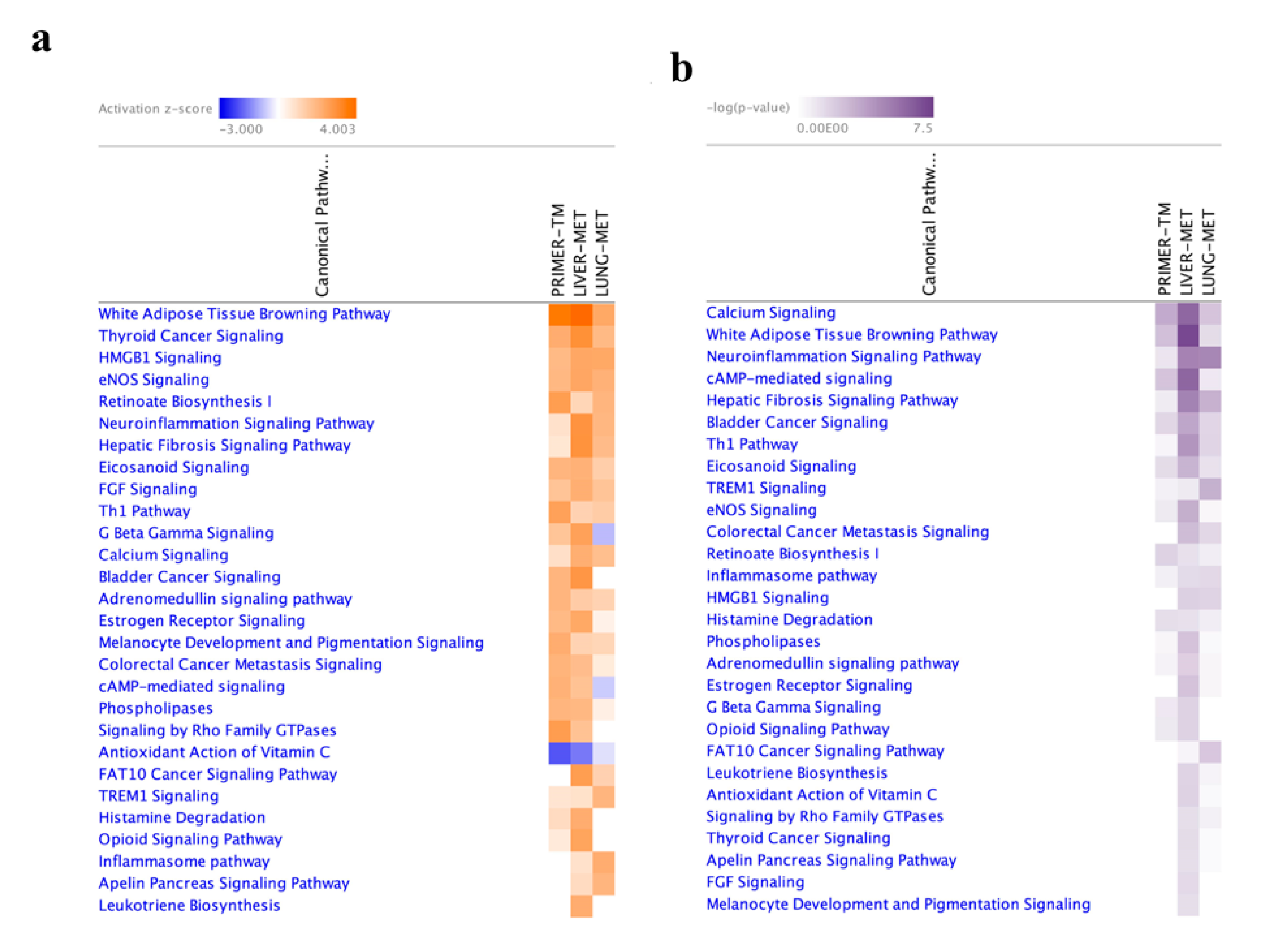
2.4. Functional Enrichment Analysis
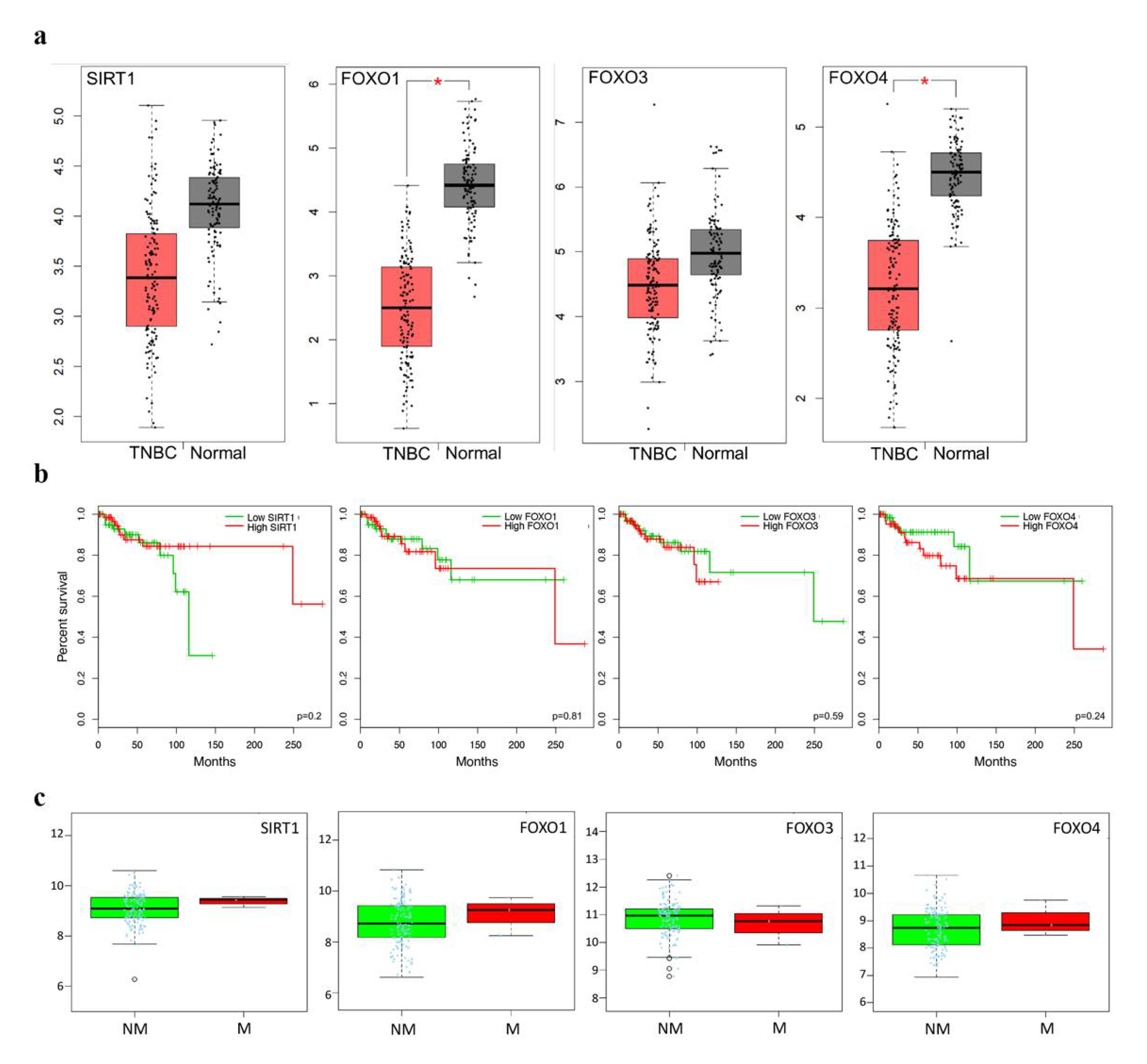
2.5. Clinicopathological Statistics of TNBC Patients
3. Discussion
4. Materials and Methods
4.1. In Vitro Experimental Procedures
4.1.1. Cell Culture
4.1.2. Immunocytochemistry
4.2. In Vivo Animal Studies
4.2.1. Animal Models
4.2.2. Immunohistochemistry
4.3. Image J Analysis of Immunohistochemical Staining
4.4. Reverse-Transcriptase qPCR
4.5. Bioinformatics Analysis
4.6. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Deshmukh, S.K.; Srivastava, S.K.; Poosarla, T.; Dyess, D.L.; Holliday, N.P.; Singh, A.P.; Singh, S. Inflammation, immunosuppressive microenvironment and breast cancer: Opportunities for cancer prevention and therapy. Ann. Transl. Med. 2019, 7, 593. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Tan, G.-Z.; Li, M.; Tan, X.; Shi, M.-L.; Mou, K. MiR-491 suppresses migration and invasion via directly targeting TPX2 in breast cancer. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 9996–10004. [Google Scholar] [PubMed]
- Lee, E.Y.; Muller, W.J. Oncogenes and tumor suppressor genes. Cold Spring Harb. Perspect. Biol. 2010, 2, a003236. [Google Scholar] [CrossRef] [PubMed]
- Osborne, C.; Wilson, P.; Tripathy, D. Oncogenes and Tumor Suppressor Genes in Breast Cancer: Potential Diagnostic and Therapeutic Applications. Oncologist 2004, 9, 361–377. [Google Scholar] [CrossRef]
- Blander, G.; Guarente, L. The Sir2 Family of Protein Deacetylases. Annu. Rev. Biochem. 2004, 73, 417–435. [Google Scholar] [CrossRef]
- Cho, I.-R.; Koh, S.S.; Malilas, W.; Srisuttee, R.; Moon, J.; Choi, Y.-W.; Horio, Y.; Oh, S.; Chung, Y.-H. SIRT1 inhibits proliferation of pancreatic cancer cells expressing pancreatic adenocarcinoma up-regulated factor (PAUF), a novel oncogene, by suppression of β-catenin. Biochem. Biophys. Res. Commun. 2012, 423, 270–275. [Google Scholar] [CrossRef]
- Luo, J.; Su, F.; Chen, D.; Shiloh, A.; Gu, W. Deacetylation of p53 modulates its effect on cell growth and apoptosis. Nature 2000, 408, 377–381. [Google Scholar] [CrossRef]
- Peck, B.; Chen, C.-Y.; Ho, K.-K.; Di Fruscia, P.; Myatt, S.S.; Coombes, R.C.; Fuchter, M.J.; Hsiao, C.-D.; Lam, E.W.-F. SIRT Inhibitors Induce Cell Death and p53 Acetylation through Targeting Both SIRT1 and SIRT2. Mol. Cancer Ther. 2010, 9, 844–855. [Google Scholar] [CrossRef]
- Brunet, A.; Sweeney, L.B.; Sturgill, J.F.; Chua, K.F.; Greer, P.L.; Lin, Y.; Tran, H.; Ross, S.E.; Mostoslavsky, R.; Cohen, H.Y.; et al. Stress-Dependent Regulation of FOXO Transcription Factors by the SIRT1 Deacetylase. Science 2004, 303, 2011–2015. [Google Scholar] [CrossRef]
- Motta, M.C.; Divecha, N.; Lemieux, M.; Kamel, C.; Chen, D.; Gu, W.; Bultsma, Y.; McBurney, M.; Guarente, L. Mammalian SIRT1 Represses Forkhead Transcription Factors. Cell 2004, 116, 551–563. [Google Scholar] [CrossRef]
- Carter, M.E.; Brunet, A. FOXO transcription factors. Curr. Biol. 2007, 17, R113–R114. [Google Scholar] [CrossRef] [PubMed]
- Medema, R.H.; Jäättelä, M. Cytosolic FoxO1: Alive and killing. Nat. Cell Biol. 2010, 12, 642–643. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, Y.; Furukawa-Hibi, Y.; Chen, C.; Horio, Y.; Isobe, K.; Ikeda, K.; Motoyama, N. SIRT1 is critical regulator of FOXO-mediated transcription in response to oxidative stress. Int. J. Mol. Med. 2005, 16, 237–243. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, K.; Araki, K.Y.; Naka, K.; Arai, F.; Takubo, K.; Yamazaki, S.; Matsuoka, S.; Miyamoto, T.; Ito, K.; Ohmura, M.; et al. Foxo3a Is Essential for Maintenance of the Hematopoietic Stem Cell Pool. Cell Stem Cell 2007, 1, 101–112. [Google Scholar] [CrossRef]
- Hu, Q.; Wang, G.; Peng, J.; Qian, G.; Jiang, W.; Xie, C.; Xiao, Y.; Wang, X. Knockdown of SIRT1 Suppresses Bladder Cancer Cell Proliferation and Migration and Induces Cell Cycle Arrest and Antioxidant Response through FOXO3a-Mediated Pathways. BioMed Res. Int. 2017, 2017, 3781904. [Google Scholar] [CrossRef]
- Saretzki, G. Cellular Senescence in the Development and Treatment of Cancer. Curr. Pharm. Des. 2010, 16, 79–100. [Google Scholar] [CrossRef]
- Sarma, P.; Bag, I.; Ramaiah, J.; Kamal, A.; Bhadra, U.; Bhadra, M.P. Bisindole-PBD regulates breast cancer cell proliferation via SIRT-p53 axis. Cancer Biol. Ther. 2015, 16, 1486–1501. [Google Scholar] [CrossRef][Green Version]
- Li, K.; Luo, J.; Azabdaftari, G. The Role of SIRT1 in Tumorigenesis. Am. Chin. J. Med. Sci. 2011, 4, 104. [Google Scholar] [CrossRef]
- Abbas, T.; Dutta, A. p21 in cancer: Intricate networks and multiple activities. Nat. Cancer 2009, 9, 400–414. [Google Scholar] [CrossRef]
- Xiong, Y.; Zhang, H.; Beach, D. D type cyclins associate with multiple protein kinases and the DNA replication and repair factor PCNA. Cell 1992, 71, 505–514. [Google Scholar] [CrossRef]
- Bourgeois, B.; Madl, T. Regulation of cellular senescence via the FOXO4-p53 axis. FEBS Lett. 2018, 592, 2083–2097. [Google Scholar] [CrossRef] [PubMed]
- Biswas, A.K.; Johnson, D.G. Transcriptional and Nontranscriptional Functions of E2F1 in Response to DNA Damage. Cancer Res. 2012, 72, 13–17. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Padiernos, E.; Ding, F.; Lossos, I.S.; Lopez, C.D. Apoptosis-stimulating protein of p53-2 (ASPP2/53BP2L) is an E2F target gene. Cell Death Differ. 2005, 12, 358–368. [Google Scholar] [CrossRef]
- Fogal, V.; Kartasheva, N.N.; Trigiante, G.; Llanos, S.; Yap, D.; Vousden, K.H.; Lu, X. ASPP1 and ASPP2 are new transcriptional targets of E2F. Cell Death Differ. 2005, 12, 369–376. [Google Scholar] [CrossRef][Green Version]
- Hallstrom, T.C.; Mori, S.; Nevins, J.R. An E2F1-Dependent Gene Expression Program that Determines the Balance between Proliferation and Cell Death. Cancer Cell 2008, 13, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Yadav, R.K.; Chauhan, A.S.; Zhuang, L.; Gan, B. FoxO transcription factors in cancer metabolism. Semin. Cancer Biol. 2018, 50, 65–76. [Google Scholar] [CrossRef]
- Wilking, M.J.; Singh, C.; Nihal, M.; Zhong, W.; Ahmad, N. SIRT1 deacetylase is overexpressed in human melanoma and its small molecule inhibition imparts anti-proliferative response via p53 activation. Arch. Biochem. Biophys. 2014, 563, 94–100. [Google Scholar] [CrossRef]
- Ford, J.; Jiang, M.; Milner, J. Cancer-specific functions of SIRT1 enable human epithelial cancer cell growth and survival. Cancer Res. 2005, 65, 10457–10463. [Google Scholar] [CrossRef]
- Yi, J.; Luo, J. SIRT1 and p53, effect on cancer, senescence and beyond. Biochim. Biophys. Acta (BBA) Proteins Proteom. 2010, 1804, 1684–1689. [Google Scholar] [CrossRef]
- Green, D.R.; Kroemer, G. Cytoplasmic functions of the tumour suppressor p53. Nature 2009, 458, 1127–1130. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Fang, D. The Roles of SIRT1 in Cancer. Genes Cancer 2013, 4, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Guttilla, I.K.; White, B.A. Coordinate Regulation of FOXO1 by miR-27a, miR-96, and miR-182 in Breast Cancer Cells. J. Biol. Chem. 2009, 284, 23204–23216. [Google Scholar] [CrossRef] [PubMed]
- Ertosun, M.G.; Hapil, F.Z.; Nidai, O.O. E2F1 transcription factor and its impact on growth factor and cytokine signaling. Cytokine Growth Factor Rev. 2016, 31, 17–25. [Google Scholar] [CrossRef]
- Mori, K.; Uchida, T.; Fukumura, M.; Tamiya, S.; Higurashi, M.; Sakai, H.; Ishikawa, F.; Shibanuma, M. Linkage of E2F1 transcriptional network and cell proliferation with respiratory chain activity in breast cancer cells. Cancer Sci. 2016, 107, 963–971. [Google Scholar] [CrossRef]
- Jin, X.; Wei, Y.; Xu, F.; Zhao, M.; Dai, K.; Shen, R.; Yang, S.; Zhang, N. SIRT1 promotes formation of breast cancer through modulating Akt activity. J. Cancer 2018, 9, 2012–2023. [Google Scholar] [CrossRef]
- Farhan, M.; Wang, H.; Gaur, U.; Little, P.; Xu, J.; Zheng, W. FOXO Signaling Pathways as Therapeutic Targets in Cancer. Int. J. Biol. Sci. 2017, 13, 815–827. [Google Scholar] [CrossRef]
- Storz, P.; Döppler, H.; Copland, J.A.; Simpson, K.J.; Toker, A. FOXO3a Promotes Tumor Cell Invasion through the Induction of Matrix Metalloproteinases. Mol. Cell. Biol. 2009, 29, 4906–4917. [Google Scholar] [CrossRef]
- Hornsveld, M.; Dansen, T.B.; Derksen, P.W.; Burgering, B.M.T. Re-evaluating the role of FOXOs in cancer. In Seminars in Cancer Biology; Elsevier: Amsterdam, The Netherlands, 2018; pp. 90–100. [Google Scholar]
- Feng, X.; Wu, Z.; Wu, Y.; Hankey, W.; Prior, T.W.; Li, L.; Ganju, R.K.; Shen, R.; Zou, X. Cdc25A Regulates Matrix Metalloprotease 1 through Foxo1 and Mediates Metastasis of Breast Cancer Cells. Mol. Cell. Biol. 2011, 31, 3457–3471. [Google Scholar] [CrossRef]
- Liu, H.; Song, Y.; Qiu, H.; Liu, Y.; Luo, K.; Yi, Y.; Jiang, G.; Lu, M.; Zhang, Z.; Yin, J.; et al. Downregulation of FOXO3a by DNMT1 promotes breast cancer stem cell properties and tumorigenesis. Cell Death Differ. 2019, 27, 966–983. [Google Scholar] [CrossRef]
- Shukla, S.; Shukla, M.; MacLennan, G.T.; Fu, P.; Gupta, S. Deregulation of FOXO3A during prostate cancer progression. Int. J. Oncol. 2009, 34, 1613–1620. [Google Scholar] [CrossRef][Green Version]
- Pardoll, D. Metastasis-Promoting Immunity: When T Cells Turn to the Dark Side. Cancer Cell 2009, 16, 81–82. [Google Scholar] [CrossRef] [PubMed]
- DeNardo, D.G.; Barreto, J.B.; Andreu, P.; Vasquez, L.; Tawfik, D.; Kolhatkar, N.; Coussens, L.M. CD4+ T Cells Regulate Pulmonary Metastasis of Mammary Carcinomas by Enhancing Protumor Properties of Macrophages. Cancer Cell 2009, 16, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Bong, A.H.; Monteith, G.R. Calcium signaling and the therapeutic targeting of cancer cells. Biochim. Biophys. Acta 2018, 1865, 1786–1794. [Google Scholar] [CrossRef] [PubMed]
- Journé, F.; Dumon, J.-C.; Kheddoumi, N.; Fox, J.; Laïos, I.; Leclercq, G.; Body, J.-J. Extracellular calcium downregulates estrogen receptor alpha and increases its transcriptional activity through calcium-sensing receptor in breast cancer cells. Bone 2004, 35, 479–488. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S.; Sharma, S.; Vora, J.; Shrivastava, N. Emerging role of sirtuins in breast cancer metastasis and multidrug resistance: Implication for novel therapeutic strategies targeting sirtuins. Pharmacol. Res. 2020, 158, 104880. [Google Scholar] [CrossRef]
- Aslakson, C.J.; Miller, F.R. Selective events in the metastatic process defined by analysis of the sequential dissemination of subpopulations of a mouse mammary tumor. Cancer Res. 1992, 52, 1399–1405. [Google Scholar]
- Erin, N.; Boyer, P.J.; Bonneau, R.H.; Clawson, G.A.; Welch, D.R. Capsaicin-mediated denervation of sensory neurons promotes mammary tumor metastasis to lung and heart. Anticancer. Res. 2004, 24, 1003–1010. [Google Scholar]
- Erin, N.; Wang, N.; Xin, P.; Bui, V.; Weisz, J.; Barkan, G.A.; Zhao, W.; Shearer, D.; Clawson, G.A. Altered gene expression in breast cancer liver metastases. Int. J. Cancer 2009, 124, 1503–1516. [Google Scholar] [CrossRef]
- Kennecke, H.; Yerushalmi, R.; Woods, R.; Cheang, M.C.U.; Voduc, D.; Speers, C.H.; Nielsen, T.O.; Gelmon, K. Metastatic Behavior of Breast Cancer Subtypes. J. Clin. Oncol. 2010, 28, 3271–3277. [Google Scholar] [CrossRef]
- Liang, Y.; Zhang, H.; Song, X.; Yang, Q. Metastatic heterogeneity of breast cancer: Molecular mechanism and potential therapeutic targets. Semin. Cancer Biol. 2020, 60, 14–27. [Google Scholar] [CrossRef] [PubMed]
- Cuesta, S.; Kireev, R.; García, C.; Rancan, L.; Vara, E.; Tresguerres, J.A.F. Melatonin can improve insulin resistance and aging-induced pancreas alterations in senescence-accelerated prone male mice (SAMP8). AGE 2013, 35, 659–671. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Jiang, X.; Chehab, F.F. FoxO4 interacts with the sterol regulatory factor SREBP2 and the hypoxia inducible factor HIF2α at the CYP51 promoter. J. Lipid Res. 2014, 55, 431–442. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Belinda, P.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef] [PubMed]
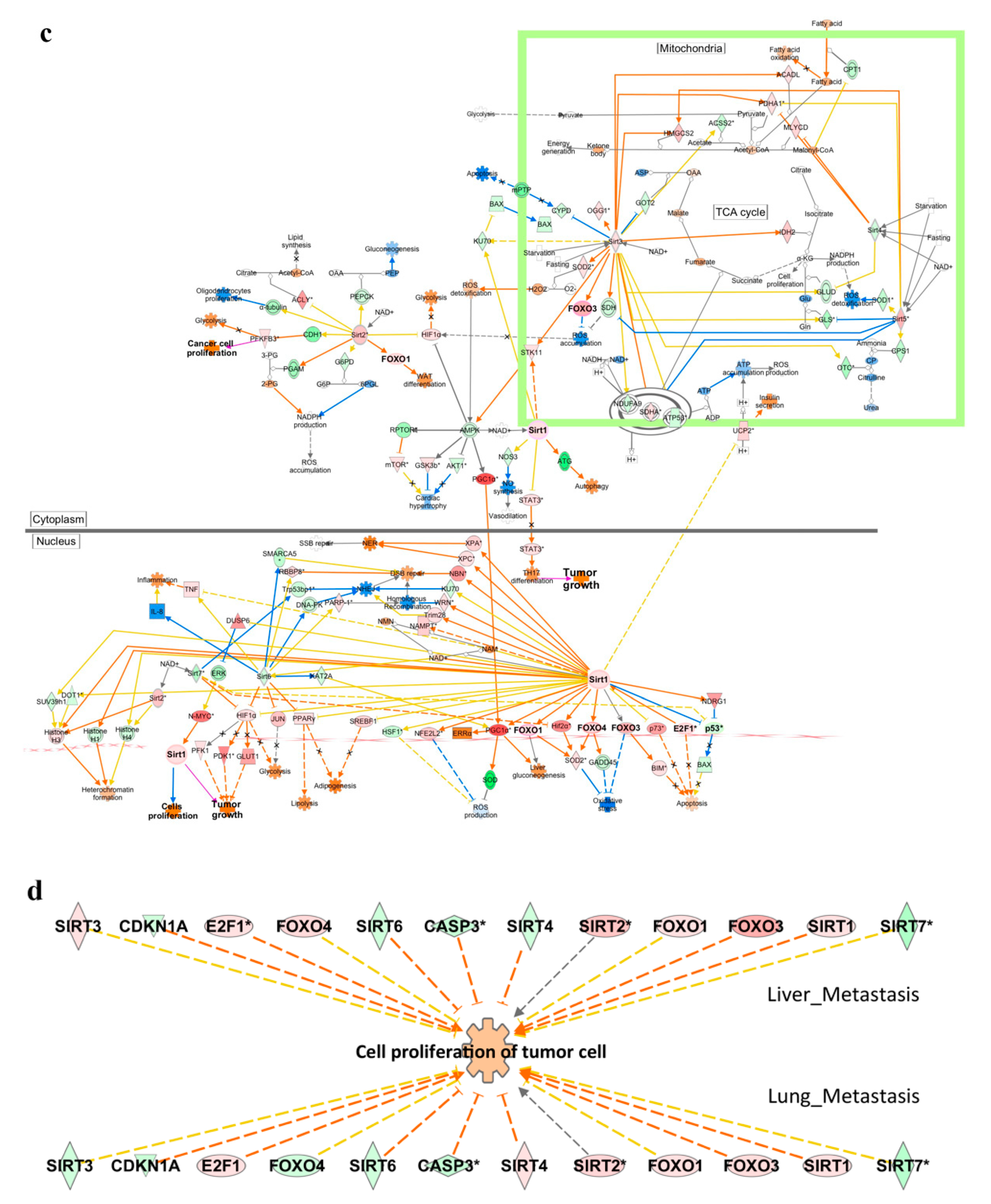
| Upstream Regulator | Z Score | p-Value of Overlap | Target Genes |
|---|---|---|---|
| FOXO1 Liver-met Lung-met | 0.92 1.427 | 1.19 × 10−4 5.16 × 10−7 | Acan, AFF3, ANGPT1, ANGPT2, APLN, APOA5, BLNK, BNIP3, CCL20, CCN2, CCN3, CCNG2, CD40, CERS4, CTSK, CXCL10, DAPK1, EDN1, EOMES, FABP4, FBXO32, FLT4, GADD45A, HSPD1, ICAM1, IFNG, IGF1R, IKZF2, IKZF5, ITGB2, ITGB6, JAG1, MMP9, OVOL1, PPARGC1A, PSMB8, PTGS1, RIPOR2, SELL, SEMA6D, SERPINB2, SERPINB5, SERPINE1, SESN3, SFN, SLC2A4, SOX5, TNFRSF9, TNFSF10, UCP1,V CAM, ACLY, ADIPOQ, AICDA, ATP6V0D2, BIRC3, CA2, CAMK4, CASP14, CCND1, CTSV, EDNRB, EFHD1, ERG, FASLG, FGF21, FOXC1, GABARAPL1, GPX3, IGLL1/IGLL5, IKZF1, IL1B, IL22, IL23R, IL7R, KLK3, LAMP2, MLXIPL, MYCN, PIK3C3, PNPLA2, POMC, PRKAA2, RUNX2, SCD, SOX9, TIE, A2M, AQP9, BCL2, COL4A, EBF1, FLT4, GSTK, IKZF5, IL18, IL6, ITGAM, ITGAV, KIF11, MAP1LC3B, MRPL57, MYOG, PLA2G2D, PTEN, RICTOR, RPS6KA3, SREBF2, STAT5B, TBX21, TIMM8A, TKT, TNNC1 |
| FOXO3 Liver-met | 0.82 | 2.94 × 10−5 | Acan, ACLY, Acot1, ACTA2, ANGPT1, ANGPT2, APLN, AQP4, ATP6V0D2, BNIP3, CCN2, CCND1, CCND2, CCNG2, CDH1, CERS4, CLDN1, CTSV, CXCL10, EDNRB, ERG, FABP4, FASLG, FBXO32, FOXC1, GABARAPL1, GABRR2, GADD45A, GPX3, ICAM1, Ifna4, IFNG, IGF1R, IKZF2, IL12A, INHBA, ITGB2, JAG1, MAP1LC3A, MMP13, MMP9, Mt1, Mt2, NDRG1, NOS2, NUPR1, OVOL1, PLAU, POMC, PPARGC1A, PRKAA2, PTGS1, RTN3, RUNX2, SBSN, SELL, SERPINE1, SESN3, SLC40A1, SOX5, SOX9, TCIM, TIE1, TNFSF10, TP53INP1, TP63, VCAM1, VEGFA, VIM |
| FOXO4 Liver-met Lung-met | 0.292 0.151 | 2.39 × 10−8 1.0 × 10−6 | Acan, ANGPT1, APLN, BNIP3, CCN2, CCNG2, CERS4, EDNRB, ERG, FABP4, FASLG, FOXC1, GADD45A, ITGB2, JAG1, MMP9, OVOL1, PTGS1, SELL, SERPINE1, SESN3, SOX5, TIE1, VCAM1, ACLY, CCND1, CCND2, GABARAPL1, GPX3, PRKAA2, PSMD11, RUNX2, SCD, SLC2A1, SOX9, VEGFA, COL4A1, FLT4, IDI1, ITGAM, MAP1LC3B, PLA2G2D, RICTOR, SREBF2 |
| SIRT1 Liver-met Lung-met | 1.011 −0.595 | 3.4 × 10−4 3.27 × 10−9 | ABCB1, APLN, BDNF, BNIP3, CDH1, DDAH2, FABP4, FBXO32, GADD45A, GBP3, GBP6, HLAA, HLADQB1, ICAM1, IFNG, IGF1R, Iigp1, LAMA4, LGALS3BP, MMP13, MMP9, PDGFRA, PNPLA3, PPARGC1A, PRDM16, RIMS2, RTP4, RUNX2, Sectm1b, SERPINE1, SLC7A11, SP110, SYNPO, TAC, STD2, TMPRSS4, ZEB1, ABCA1, ABCG1, ACAP1, ADIPOQ, AGT, BIRC3, Ccl2, CCND1, CCND2, CCNG2, CEBPB, CLEC10A, CTNNB1, EPAS1, FGF21, FGFR1, GABARAPL1, GLI2, GRIP1, GSTM3, Ifi47, IGHM, IL1B, KALRN, KMT2B, LAMA2, LRCH1, NANOG, NCAM2, NECTIN4, NKG7, NOS2, PCSK2, RBPJL, SLC27A6, TNFSF11, TP73, TRIM31, ZNF296, AFP, BCL2, CCNG2, CD74, CMPK2, CNTN6, CPB, Csprs, CYP2B6, DDX60, DHX58, EDEMEP3, ERMP1, HIVEP3, IFI44, IFIT1B, IFIT3, Igtp, IL12B, IL6, IRGM, Irgm1, KDM5B, MYOG, NAIP, NF1, NLRC5, NTRK2, Oasl2, P3H3, PRDM1, PSMB9, TAP1, TFPI, Tgtp1/Tgtp2, TIMP2, Trim30a/Trim30d, UBA7 |
| FORWARD | REVERSE | |
|---|---|---|
| FoxO1 | CTTCAAGGATAAGGGCGACA | GACAGATTGTGGCGAATTGA |
| FoxO3a | GCTAAGCAGGCCTCATCTCA | TTCCGTCAGTTTGAGGGTCT |
| FoxO4 | TCATCAAGGTTCACAACGAGGC | AGGACAGACGGCTTCTTCTTGG |
| SIRT 1 | TCGTGGAGACATTTTTAATCAGG | GCTTCATGATGGCAAGTGG |
|
r18S (Housekeeping gene) | GGTGCATGGCCGTTCTTA | TCGTTCGTTATCGGAATTAACC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dilmac, S.; Kuscu, N.; Caner, A.; Yildirim, S.; Yoldas, B.; Farooqi, A.A.; Tanriover, G. SIRT1/FOXO Signaling Pathway in Breast Cancer Progression and Metastasis. Int. J. Mol. Sci. 2022, 23, 10227. https://doi.org/10.3390/ijms231810227
Dilmac S, Kuscu N, Caner A, Yildirim S, Yoldas B, Farooqi AA, Tanriover G. SIRT1/FOXO Signaling Pathway in Breast Cancer Progression and Metastasis. International Journal of Molecular Sciences. 2022; 23(18):10227. https://doi.org/10.3390/ijms231810227
Chicago/Turabian StyleDilmac, Sayra, Nilay Kuscu, Ayse Caner, Sendegul Yildirim, Burcak Yoldas, Ammad Ahmad Farooqi, and Gamze Tanriover. 2022. "SIRT1/FOXO Signaling Pathway in Breast Cancer Progression and Metastasis" International Journal of Molecular Sciences 23, no. 18: 10227. https://doi.org/10.3390/ijms231810227
APA StyleDilmac, S., Kuscu, N., Caner, A., Yildirim, S., Yoldas, B., Farooqi, A. A., & Tanriover, G. (2022). SIRT1/FOXO Signaling Pathway in Breast Cancer Progression and Metastasis. International Journal of Molecular Sciences, 23(18), 10227. https://doi.org/10.3390/ijms231810227







