Direct Analysis of Mitochondrial Damage Caused by Misfolded/Destabilized Proteins
Abstract
:1. Introduction
2. Results
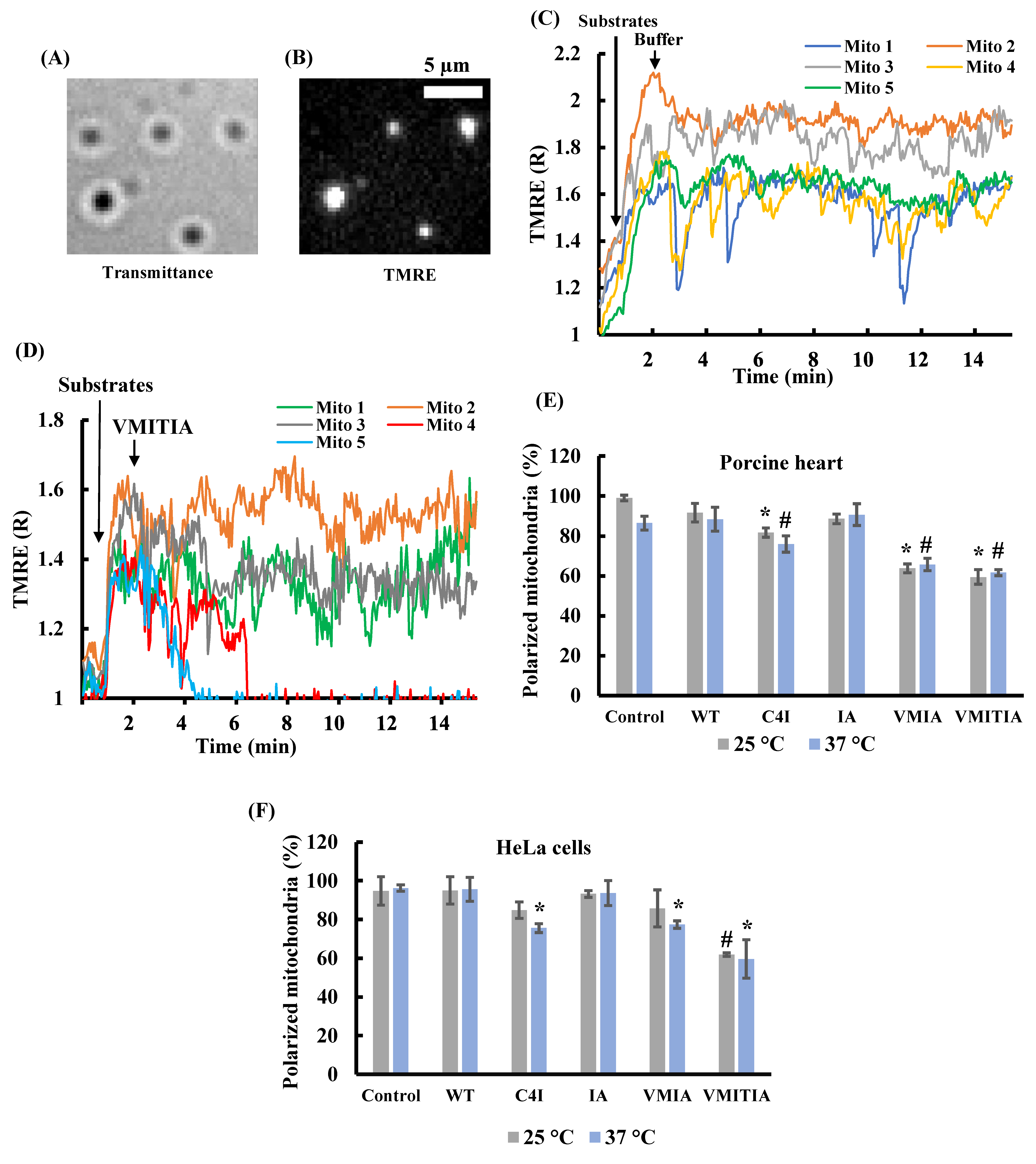
2.1. Effects of DENV3-ED3 Variants on Mitochondrial Membrane Potential
2.2. Effects of the DENV3-ED3 Variants on Mitochondrial Inner Membrane Integrity
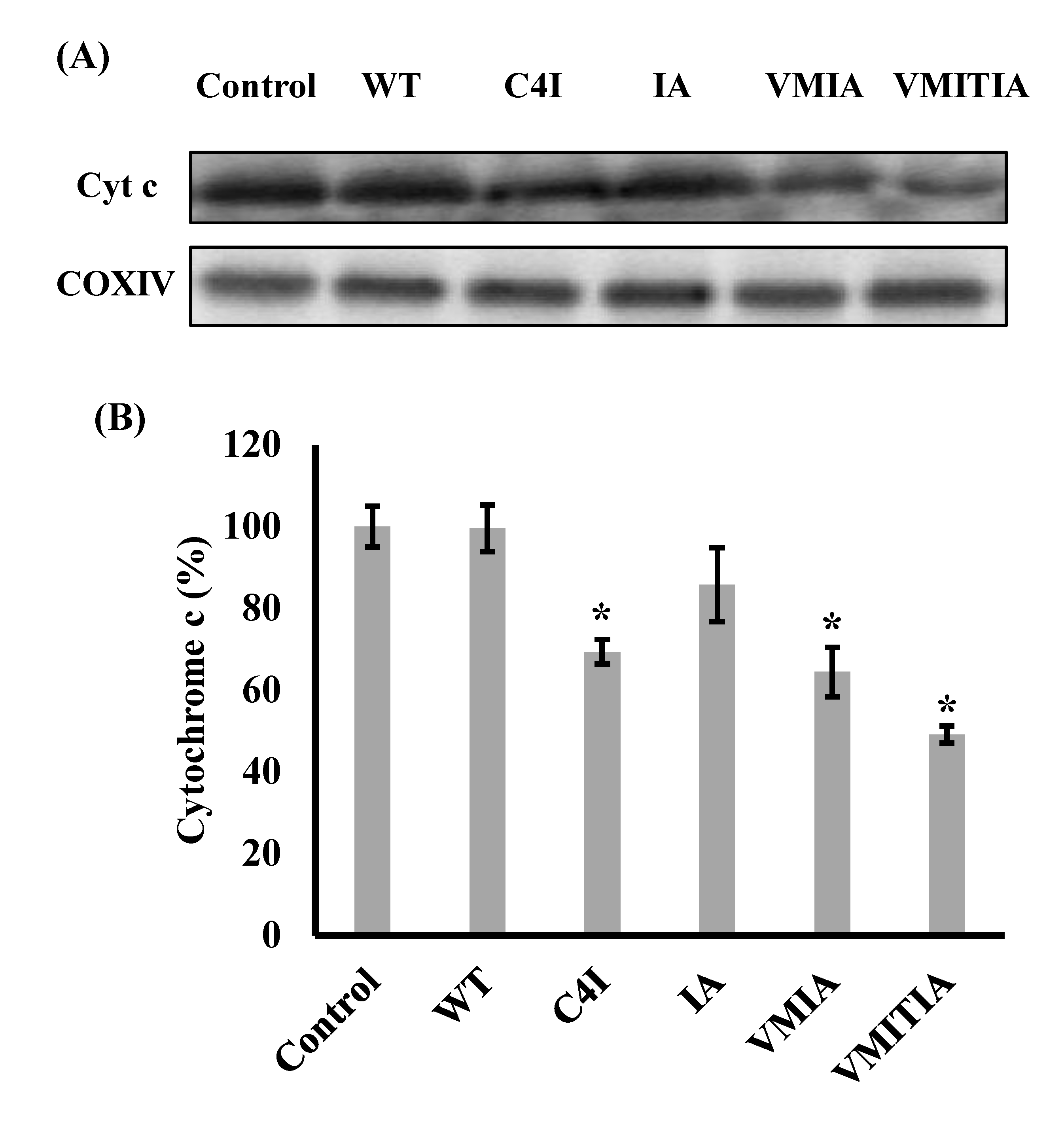
2.3. Effects of the DENV3-ED3 Variants on Cytochrome C Release from Mitochondria
2.4. Oligomer Size and Molten Globule-like Properties of the DENV3-ED3 Variants
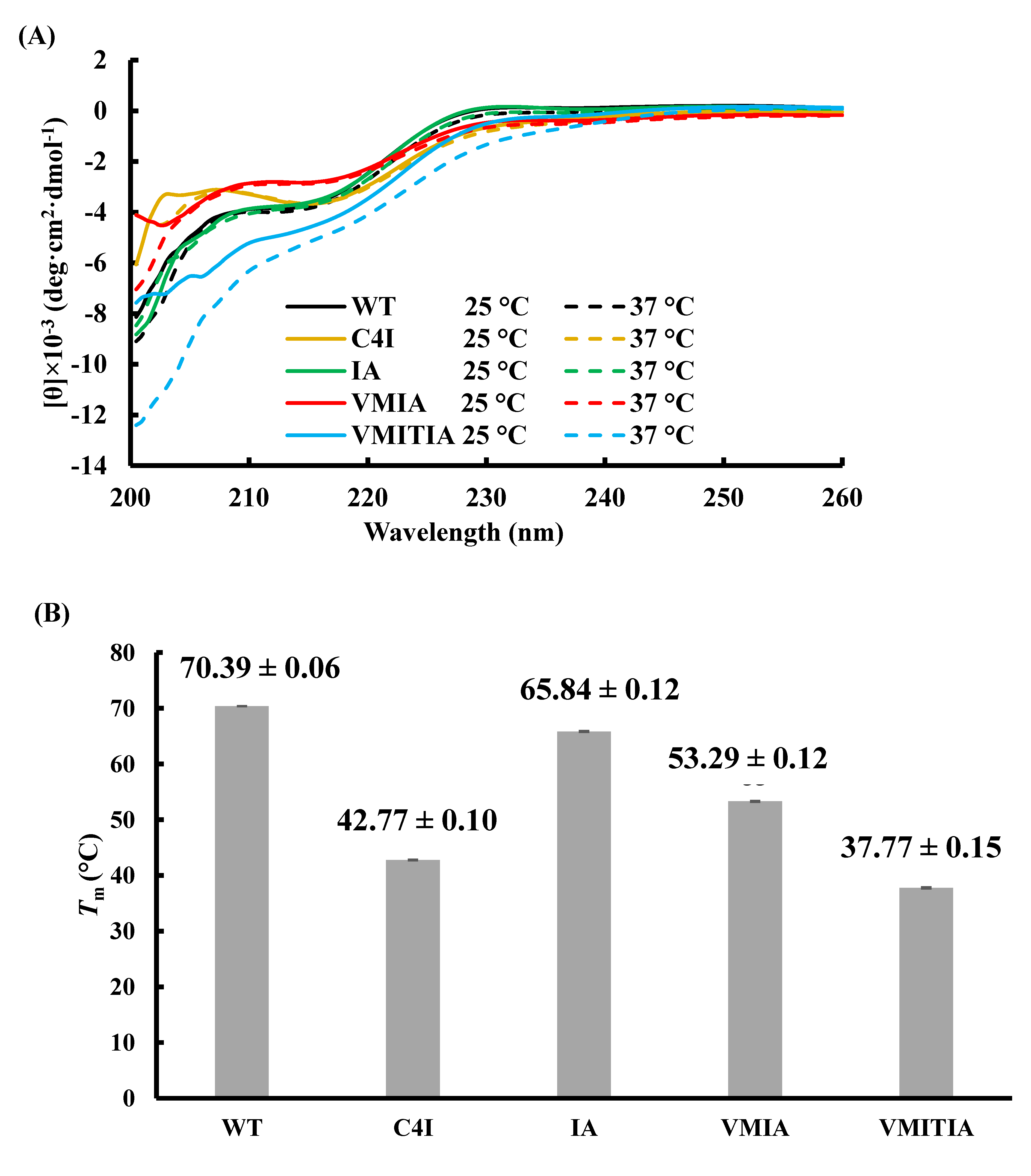
2.5. Secondary Structure Content and Thermal Stability of the DENV3-ED3 Variants
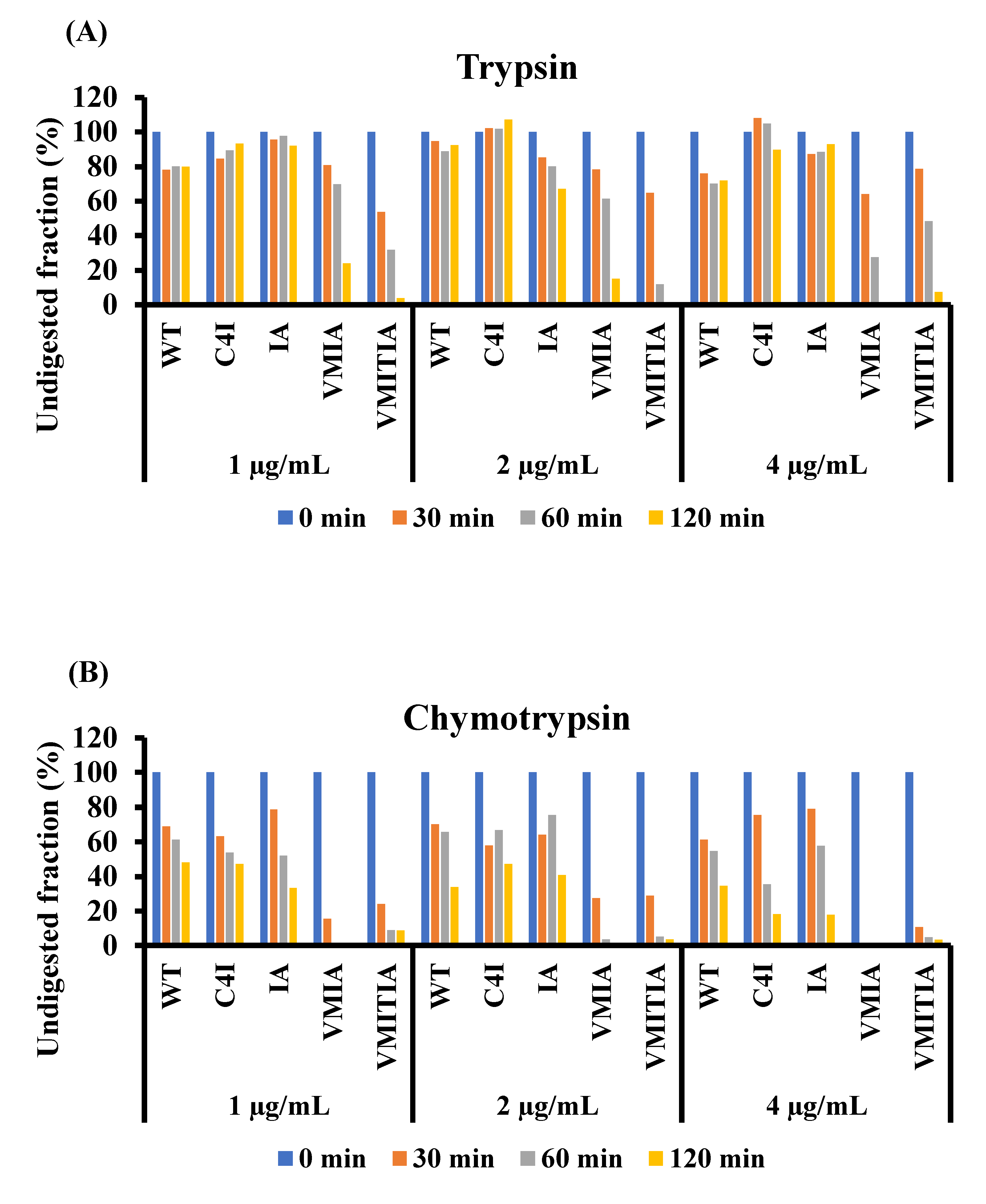
2.6. Resistance of the DENV3-ED3 Variants to Degradation by Proteolytic Enzymes
3. Discussion
4. Materials and Methods
4.1. Mutant DENV3-ED3 Design, Expression, and Purification
4.2. Preparation of Porcine Heart Mitochondria
4.3. HeLa Cell Culture and Mitochondria Isolation
4.4. Mitochondrial TMRE Fluorescence
4.5. Measurement of Intramitochondrial Calcein Release
4.6. Western Blot Analysis
4.7. DLS Assays
4.8. ANS and ThT Assays
4.9. Far-UV CD Assays
4.10. Proteolysis Assay
4.11. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Nussbaum, R.L.; Polymeropoulos, M.H. Genetics of Parkinson’s disease. Hum. Mol. Genet. 1997, 6, 1687–1691. [Google Scholar] [CrossRef] [PubMed]
- Bosco, D.A.; Morfini, G.; Karabacak, N.M.; Song, Y.; Gros-Louis, F.; Pasinelli, P.; Goolsby, H.; Fontaine, B.A.; Lemay, N.; McKenna-Yasek, D.; et al. Wildtype and mutant SOD1 share an aberrant conformation and a common pathogenic pathway in ALS. Nat. Neurosci. 2010, 13, 1396–1403. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Vicente, M.; Talloczy, Z.; Wong, E.; Tang, G.; Koga, H.; Kaushik, S.; de Vries, R.; Arias, E.; Harris, S.; Sulzer, D.; et al. Cargo recognition failure is responsible for inefficient autophagy in Huntington’s disease. Nat. Neurosci. 2010, 13, 567–576. [Google Scholar] [CrossRef] [PubMed]
- Deriziotis, P.; Andre, R.; Smith, D.M.; Goold, R.; Kinghorn, K.J.; Kristiansen, M.; Nathan, J.A.; Rosenzweig, R.; Krutauz, D.; Glickman, M.H.; et al. Misfolded PrP impairs the UPS by interaction with the 20S proteasome and inhibition of substrate entry. EMBO J. 2011, 30, 3065–3077. [Google Scholar] [CrossRef]
- Tai, H.C.; Serrano-Pozo, A.; Hashimoto, T.; Frosch, M.P.; Spires-Jones, T.L.; Hyman, B.T. The synaptic accumulation of hyperphosphorylated tau oligomers in Alzheimer disease is associated with dysfunction of the ubiquitin–proteasome system. Am. J. Pathol. 2012, 181, 1426–1435. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.H.; Wu, P.; Pan, Y.Y.; Sun, X.L.; Yang, H.Y.; Difiglia, M.; Lu, B.X. A toxic mutant huntingtin species is resistant to selective autophagy. Nat. Chem. Biol. 2017, 13, 1152–1154. [Google Scholar] [CrossRef]
- Sweeney, P.; Park, H.; Baumann, M.; Dunlop, J.; Frydman, J.; Kopito, R.; McCampbell, A.; Leblanc, G.; Venkateswaran, A.; Nurmi, A.; et al. Protein misfolding in neurodegenerative diseases: Implications and strategies. Transl. Neurodegener. 2017, 6, 6. [Google Scholar] [CrossRef]
- Pedersen, J.T.; Heegaard, N.H. Analysis of protein aggregation in neurodegenerative disease. Anal. Chem. 2013, 85, 4215–4227. [Google Scholar] [CrossRef]
- Briston, T.; Hicks, A.R. Mitochondrial dysfunction and neurodegenerative proteinopathies: Mechanisms and prospects for therapeutic intervention. Biochem. Soc. Trans. 2018, 46, 829–842. [Google Scholar] [CrossRef]
- Noji, M.; Samejima, T.; Yamaguchi, K.; So, M.; Yuzu, K.; Chatani, E.; Akazawa-Ogawa, Y.; Hagihara, Y.; Kawata, Y.; Ikenaka, K.; et al. Breakdown of supersaturation barrier links protein folding to amyloid formation. Commun. Biol. 2021, 4, 120. [Google Scholar] [CrossRef]
- Nagai, Y.; Inui, T.; Popiel, H.A.; Fujikake, N.; Hasegawa, K.; Urade, Y.; Goto, Y.; Naiki, H.; Toda, T. A toxic monomeric conformer of the polyglutamine protein. Nat. Struct. Mol. Biol. 2007, 14, 332–340. [Google Scholar] [CrossRef] [PubMed]
- Sugiyama, M.; Fujii, N.; Morimoto, Y.; Itoh, K.; Mori, K.; Fukunaga, T.; Fuji, N. SAXS and SANS Observations of Abnormal Aggregation of Humana-Crystallin. Chem. Biodivers. 2010, 7, 1380–1388. [Google Scholar] [CrossRef] [PubMed]
- Constantinescu, P.; Brown, R.A.; Wyatt, A.R.; Ranson, M.; Wilson, M.R. Amorphous protein aggregates stimulate plasminogen activation, leading to release of cytotoxic fragments that are clients for extracellular chaperones. J. Biol. Chem. 2017, 292, 14425–14437. [Google Scholar] [CrossRef] [PubMed]
- Crill, W.D.; Roehrig, J.T. Monoclonal antibodies that bind to domain III of dengue virus E glycoprotein are the most efficient blockers of virus adsorption to Vero cells. J. Virol. 2001, 75, 7769–7773. [Google Scholar] [CrossRef] [PubMed]
- Saotome, T.; Nakamura, S.; Islam, M.M.; Nakazawa, A.; Dellarole, M.; Arisaka, F.; Kidokoro, S.; Kuroda, Y. Unusual reversible oligomerization of unfolded dengue envelope protein domain 3 at high temperatures and its abolition by a point mutation. Biochemistry 2016, 55, 4469–4475. [Google Scholar] [CrossRef]
- Saotome, T.; Doret, M.; Kulkarni, M.; Yang, Y.-S.; Barthe, P.; Kuroda, Y.; Roumestand, C. Folding of the Ig-like domain of the envelope protein from dengue virus analyzed by high hydrostatic pressure NMR at a residue-level resolution. Biomolecules 2019, 9, 309. [Google Scholar] [CrossRef]
- Saotome, T.; Yamazaki, T.; Kuroda, Y. Misfolding of a Single Disulfide Bonded Globular Protein into a Low-Solubility Species Conformationally and Biophysically Distinct from the Native One. Biomolecules 2019, 9, 250. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, M.R.; Numoto, N.; Ito, N.; Kuroda, Y. Modeling, and experimental assessment of a buried Leu–Ile mutation in dengue envelope domain III. Biochem. Biophys. Res. Commun. 2016, 471, 163–168. [Google Scholar] [CrossRef]
- Islam, M.M.; Miura, S.; Hasan, M.N.; Rahman, N.; Kuroda, Y. Anti-Dengue ED3 Long-Term Immune Response with T-Cell Memory Generated Using Solubility Controlling Peptide Tags. Front. Immunol. 2020, 11, 333. [Google Scholar] [CrossRef]
- Nieminen, A.L.; Saylor, A.K.; Tesfai, S.A.; Herman, B.; Lemasters, J.J. Contribution of the mitochondrial permeability transition to lethal injury after exposure of hepatocytes to t-butylhydroperoxide. Biochem. J. 1995, 307, 99–106. [Google Scholar] [CrossRef] [Green Version]
- Shibata, T.; Yoneda, M.; Morikawa, D.; Ohta, Y. Time-lapse imaging of Ca2+-induced swelling and permeability transition: Single mitochondrion study. Arch. Biochem. Biophys. 2019, 663, 288–296. [Google Scholar] [CrossRef] [PubMed]
- Zou, H.; Li, Y.; Liu, X.; Wang, X. An APAF-1 cytochrome c multimeric complex is a functional apoptosome that activates procaspase-9. J. Biol. Chem. 1999, 274, 11549–11556. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, S.; Kibria, M.G.; Unzai, S.; Kuroda, Y.; Kidokoro, S.-I. Reversible Oligomerization and Reverse Hydrophobic Effect Induced by Isoleucine Tags Attached at the C-Terminus of a Simplified BPTI Variant. Biochemistry 2020, 59, 3660–3668. [Google Scholar] [CrossRef] [PubMed]
- Kabir, G.M.; Islam, M.M.; Kuroda, Y. Reversible association of proteins into sub-visible amorphous aggregates using short solubility controlling peptide tags. Biochim. Biophys. Acta 2018, 1866, 366–372. [Google Scholar] [CrossRef] [PubMed]
- Sandra, M.C.; Russell, H.S.; Catarina, R.O. Induction of cytochrome c-mediated apoptosis by amyloid-beta 25–35 requires functional mitochondria. Brain Res. 2002, 931, 117–125. [Google Scholar]
- Ly, J.D.; Grubb, R.; Lawen, A. The mitochondrial membrane potential (Δψm) in apoptosis; An update. Apoptosis 2003, 8, 115–128. [Google Scholar] [CrossRef]
- Aklima, J.; Onojima, T.; Kimura, S.; Umiuchi, K.; Shibata, T.; Kuraoka, Y.; Oie, Y.; Suganuma, Y.; Ohta, Y. Effects of Matrix pH on Spontaneous Transient Depolarization and Reactive Oxygen Species Production in Mitochondria. Front. Cell Dev. Biol. 2021, 9, 692776. [Google Scholar] [CrossRef]
- Elahi, M.; Islam, M.M.; Noguchi, K.; Yohda, M.; Kuroda, Y. High resolution crystal structure of dengue-3 envelope protein domain III suggests possible molecular mechanisms for serospecific antibody recognition. Proteins 2013, 81, 1090–1095. [Google Scholar] [CrossRef]
- Morikawa, D.; Kanematsu, K.; Shibata, T.; Haseda, K.; Umeda, N.; Ohta, Y. Detection of swelling of single isolated mitochondrion with optical microscopy. Biomed. Opt. Express 2014, 5, 848–857. [Google Scholar] [CrossRef]
- Shibata, T.; Yamashita, S.; Hirusaki, K.; Katoh, K.; Ohta, Y. Isolation of mitochondria by gentle cell membrane disruption, and their subsequent characterization. Biochem. Biophys. Res. Commun. 2015, 463, 563–568. [Google Scholar] [CrossRef]
- Nakayama, S.; Sakuyama, T.; Mitaku, S.; Ohta, Y. Fluorescence imaging of metabolic responses in single mitochondria. Biochem. Biophys. Res. Commun. 2002, 290, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Ott, M.; Robertson, J.D.; Gogvadze, V.; Zhivotovsky, B.; Orrenius, S. Cytochrome c release from mitochondria proceeds by a two-step process. Proc. Natl. Acad. Sci. USA 2002, 99, 1259–1263. [Google Scholar] [CrossRef] [PubMed]
- Morozova-Roche, L.A.; Zurdo, J.; Spencer, A.; Noppe, W.; Receveur, V.; Archer, D.B.; Joniau, M.; Dobson, C.M. Amyloid Fibril Formation and Seeding by Wild-Type Human Lysozyme and Its Disease-Related Mutational Variants. J. Struct. Biol. 2000, 130, 339–351. [Google Scholar] [CrossRef] [PubMed]
- Greenfield, N.J. Using circular dichroism collected as a function of temperature to determine the thermodynamics of protein unfolding and binding interactions. Nat. Protoc. 2006, 1, 2527–2535. [Google Scholar] [CrossRef]


| Type | Variants | Short Name | Description of the Variants |
|---|---|---|---|
| Wild type | 3ED3 | WT | Domain 3 of the envelope protein of the DENV from serotype 3 (DENV3) |
| Tag | 3ED3-C4I | C4I | 4-Isoleucine tag added after two |
| Glycine-spacer at the C-terminus of 3ED3 | |||
| Mutation | 3ED3-I380A | IA | Isoleucine 380 was replaced by Alanine in 3ED3 |
| 3ED3-V310M I380A | VMIA | Valine 310 was replaced by Methionine | |
| and Isoleucine 380 was replaced by Alanine in 3ED3 | |||
| 3ED3-V310M I318T I380A | VMITIA | Valine 310 was replaced by Methionine, | |
| Isoleucine 318 was replaced by Threonine | |||
| and Isoleucine 380 was replaced by Alanine in 3ED3 |
| Protein | MMP | IMM | OMM | Oligomer Sizes | Fiber (F) or, Amorphous (A) | Molten Globule-like Properties | Thermal Stability | Protein Digestion |
|---|---|---|---|---|---|---|---|---|
| WT | N | N | N | S | A | Lo | St | St |
| C4I | D | D | D | L | A | H | U | St |
| IA | N | N | N | S | A | Lo | St | St |
| VMIA | D | D | D | S | A | H | U | U |
| VMITIA | D | D | D | S | A | Lo | U | U |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aklima, J.; Onchaiya, S.; Saotome, T.; Velmurugan, P.; Motoichi, T.; Naima, J.; Kuroda, Y.; Ohta, Y. Direct Analysis of Mitochondrial Damage Caused by Misfolded/Destabilized Proteins. Int. J. Mol. Sci. 2022, 23, 9881. https://doi.org/10.3390/ijms23179881
Aklima J, Onchaiya S, Saotome T, Velmurugan P, Motoichi T, Naima J, Kuroda Y, Ohta Y. Direct Analysis of Mitochondrial Damage Caused by Misfolded/Destabilized Proteins. International Journal of Molecular Sciences. 2022; 23(17):9881. https://doi.org/10.3390/ijms23179881
Chicago/Turabian StyleAklima, Jannatul, Sawaros Onchaiya, Tomonori Saotome, Punitha Velmurugan, Taihei Motoichi, Jannatul Naima, Yutaka Kuroda, and Yoshihiro Ohta. 2022. "Direct Analysis of Mitochondrial Damage Caused by Misfolded/Destabilized Proteins" International Journal of Molecular Sciences 23, no. 17: 9881. https://doi.org/10.3390/ijms23179881
APA StyleAklima, J., Onchaiya, S., Saotome, T., Velmurugan, P., Motoichi, T., Naima, J., Kuroda, Y., & Ohta, Y. (2022). Direct Analysis of Mitochondrial Damage Caused by Misfolded/Destabilized Proteins. International Journal of Molecular Sciences, 23(17), 9881. https://doi.org/10.3390/ijms23179881








