Prime Editing: An All-Rounder for Genome Editing
Abstract
:1. Introduction
2. Origin of the PE System
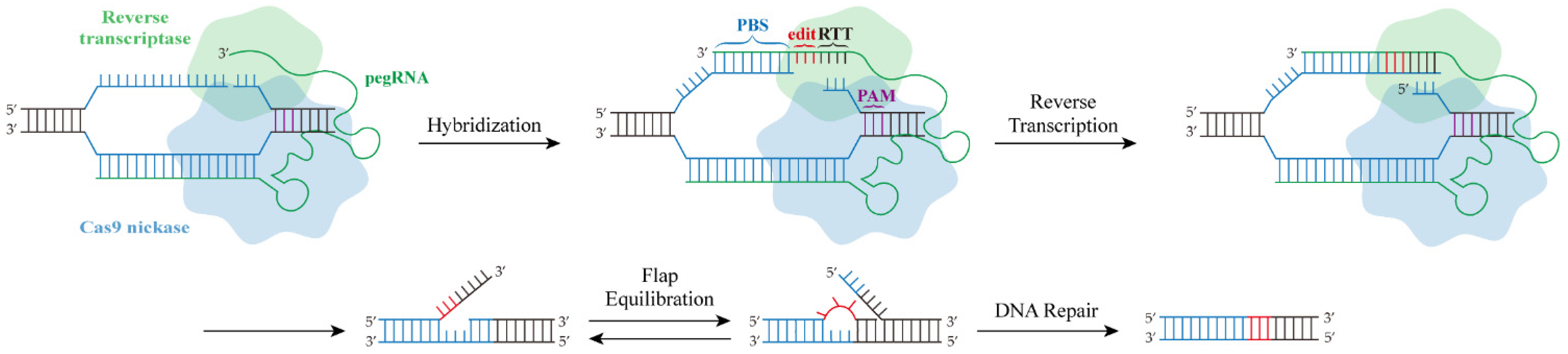
3. Principle of the PE System
4. Improvements in Prime Editors
4.1. Relaxed PAM Restriction for PE
4.2. Improvement of the Editing Efficiency
4.3. Delivery of PE Agents
4.4. Algorism Development for the Design of pegRNA
5. Applications of PE
5.1. Medicine
5.2. Agriculture
5.3. Biotechnological Applications in Different Species
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Gaj, T.; Gersbach, C.A.; Barbas, C.F., III. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013, 31, 397–405. [Google Scholar] [CrossRef] [PubMed]
- Porteus, M. Genome Editing: A New Approach to Human Therapeutics. Annu. Rev. Pharmacol. Toxicol. 2016, 56, 163–190. [Google Scholar] [CrossRef] [PubMed]
- Mussolino, C.; Cathomen, T. RNA guides genome engineering. Nat. Biotechnol. 2013, 31, 208–209. [Google Scholar] [CrossRef] [PubMed]
- Urnov, F.D. Prime Time for Genome Editing? N. Engl. J. Med. 2020, 382, 481–484. [Google Scholar] [CrossRef]
- Matsoukas, I.G. Prime Editing: Genome Editing for Rare Genetic Diseases Without Double-Strand Breaks or Donor DNA. Front. Genet. 2020, 11, 528. [Google Scholar] [CrossRef]
- Komor, A.C.; Kim, Y.B.; Packer, M.S.; Zuris, J.A.; Liu, D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 2016, 533, 420–424. [Google Scholar] [CrossRef]
- Rees, H.A.; Liu, D.R. Base editing: Precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 2018, 19, 770–788. [Google Scholar] [CrossRef]
- Huang, T.P.; Newby, G.A.; Liu, D.R. Precision genome editing using cytosine and adenine base editors in mammalian cells. Nat. Protoc. 2021, 16, 1089–1128. [Google Scholar] [CrossRef]
- Anzalone, A.V.; Randolph, P.B.; Davis, J.R.; Sousa, A.A.; Koblan, L.W.; Levy, J.M.; Chen, P.J.; Wilson, C.; Newby, G.A.; Raguram, A.; et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 2019, 576, 149–157. [Google Scholar] [CrossRef]
- Anzalone, A.V.; Koblan, L.; Liu, D.R. Genome editing with CRISPR–Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 2020, 38, 824–844. [Google Scholar] [CrossRef]
- Jiang, Y.-Y.; Chai, Y.-P.; Lu, M.-H.; Han, X.-L.; Lin, Q.; Zhang, Y.; Zhang, Q.; Zhou, Y.; Wang, X.-C.; Gao, C.; et al. Prime editing efficiently generates W542L and S621I double mutations in two ALS genes in maize. Genome Biol. 2020, 21, 257. [Google Scholar] [CrossRef]
- Doudna, J.A.; Charpentier, E. The new frontier of genome engineering with CRISPR-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef]
- Gilbert, L.; Larson, M.H.; Morsut, L.; Liu, Z.; Brar, G.A.; Torres, S.E.; Stern-Ginossar, N.; Brandman, O.; Whitehead, E.H.; Doudna, J.A.; et al. CRISPR-Mediated Modular RNA-Guided Regulation of Transcription in Eukaryotes. Cell 2013, 154, 442–451. [Google Scholar] [CrossRef]
- Wang, H.; La Russa, M.; Qi, L.S. CRISPR/Cas9 in Genome Editing and Beyond. Annu. Rev. Biochem. 2016, 85, 227–264. [Google Scholar] [CrossRef]
- Gaudelli, N.M.; Komor, A.C.; Rees, H.A.; Packer, M.S.; Badran, A.H.; Bryson, D.I.; Liu, D.R. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 2017, 551, 464–471. [Google Scholar] [CrossRef]
- Vakulskas, C.A.; Dever, D.P.; Rettig, G.R.; Turk, R.; Jacobi, A.M.; Collingwood, M.A.; Bode, N.M.; McNeill, M.S.; Yan, S.; Camarena, J.; et al. A high-fidelity Cas9 mutant delivered as a ribonucleoprotein complex enables efficient gene editing in human hematopoietic stem and progenitor cells. Nat. Med. 2018, 24, 1216–1224. [Google Scholar] [CrossRef]
- Kantor, A.; McClements, M.E.; MacLaren, R.E. CRISPR-Cas9 DNA Base-Editing and Prime-Editing. Int. J. Mol. Sci. 2020, 21, 6240. [Google Scholar] [CrossRef]
- Caso, F.; Davies, B. Base editing and prime editing in laboratory animals. Lab. Anim. 2021, 56, 35–49. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A Programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Hu, J.H.; Miller, S.M.; Geurts, M.H.; Tang, W.; Chen, L.; Sun, N.; Zeina, C.M.; Gao, X.; Rees, H.A.; Lin, Z.; et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 2018, 556, 57–63. [Google Scholar] [CrossRef]
- Kleinstiver, B.; Prew, M.S.; Tsai, S.Q.; Topkar, V.V.; Nguyen, N.T.; Zheng, Z.; Gonzales, A.P.W.; Li, Z.; Peterson, R.T.; Yeh, J.-R.J.; et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 2015, 523, 481–485. [Google Scholar] [CrossRef] [PubMed]
- Nishimasu, H.; Shi, X.; Ishiguro, S.; Gao, L.; Hirano, S.; Okazaki, S.; Noda, T.; Abudayyeh, O.O.; Gootenberg, J.S.; Mori, H.; et al. Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science 2018, 361, 1259–1262. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.M.; Wang, T.; Randolph, P.B.; Arbab, M.; Shen, M.W.; Huang, T.P.; Matuszek, Z.; Newby, G.A.; Rees, H.A.; Liu, D.R. Continuous evolution of SpCas9 variants compatible with non-G PAMs. Nat. Biotechnol. 2020, 38, 471–481. [Google Scholar] [CrossRef] [PubMed]
- Walton, R.T.; Christie, K.A.; Whittaker, M.N.; Kleinstiver, B.P. Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science 2020, 368, 290–296. [Google Scholar] [CrossRef]
- Kweon, J.; Yoon, J.-K.; Jang, A.-H.; Shin, H.R.; See, J.-E.; Jang, G.; Kim, J.-I.; Kim, Y. Engineered prime editors with PAM flexibility. Mol. Ther. 2021, 29, 2001–2007. [Google Scholar] [CrossRef]
- Kim, Y.; Hong, S.-A.; Yu, J.; Eom, J.; Jang, K.; Yoon, S.; Hong, D.H.; Seo, D.; Lee, S.-N.; Woo, J.-S.; et al. Adenine base editing and prime editing of chemically derived hepatic progenitors rescue genetic liver disease. Cell Stem Cell 2021, 28, 1614–1624. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, G.; Huang, S.; Li, X.; Wang, X.; Li, G.; Chi, T.; Chen, Y.; Huang, X.; Wang, X. Enhancing prime editing by Csy4-mediated processing of pegRNA. Cell Res. 2021, 31, 1134–1136. [Google Scholar] [CrossRef]
- Nelson, J.W.; Randolph, P.B.; Shen, S.P.; Everette, K.A.; Chen, P.J.; Anzalone, A.V.; An, M.; Newby, G.A.; Chen, J.C.; Hsu, A.; et al. Engineered pegRNAs improve prime editing efficiency. Nat. Biotechnol. 2022, 40, 402–410. [Google Scholar] [CrossRef]
- Roth, A.; Winkler, W.C.; E Regulski, E.; Lee, B.W.K.; Lim, J.; Jona, I.; E Barrick, J.; Ritwik, A.; Kim, J.N.; Welz, R.; et al. A riboswitch selective for the queuosine precursor preQ1 contains an unusually small aptamer domain. Nat. Struct. Mol. Biol. 2007, 14, 308–317. [Google Scholar] [CrossRef]
- Anzalone, A.V.; Lin, A.J.; Zairis, S.; Rabadan, R.; Cornish, V.W. Reprogramming eukaryotic translation with ligand-responsive synthetic RNA switches. Nat. Chem. Biol. 2016, 13, 453–458. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Zhou, L.; Gao, B.-Q.; Li, G.; Wang, X.; Wang, Y.; Wei, J.; Han, W.; Wang, Z.; Li, J.; et al. Highly efficient prime editing by introducing same-sense mutations in pegRNA or stabilizing its structure. Nat. Commun. 2022, 13, 1669. [Google Scholar] [CrossRef]
- Zhang, G.; Liu, Y.; Huang, S.; Qu, S.; Cheng, D.; Yao, Y.; Ji, Q.; Wang, X.; Huang, X.; Liu, J. Enhancement of prime editing via xrRNA motif-joined pegRNA. Nat. Commun. 2022, 13, 1856. [Google Scholar] [CrossRef]
- Li, X.; Wang, X.; Sun, W.; Huang, S.; Zhong, M.; Yao, Y.; Ji, Q.; Huang, X. Enhancing prime editing efficiency by modified pegRNA with RNA G-quadruplexes. J. Mol. Cell Biol. 2022, 14, mjac022. [Google Scholar] [CrossRef]
- Hussmann, J.A.; Ling, J.; Ravisankar, P.; Yan, J.; Cirincione, A.; Xu, A.; Simpson, D.; Yang, D.; Bothmer, A.; Cotta-Ramusino, C.; et al. Mapping the genetic landscape of DNA double-strand break repair. Cell 2021, 184, 5653–5669. [Google Scholar] [CrossRef]
- Chen, P.J.; Hussmann, J.A.; Yan, J.; Knipping, F.; Ravisankar, P.; Chen, P.-F.; Chen, C.; Nelson, J.W.; Newby, G.A.; Sahin, M.; et al. Enhanced prime editing systems by manipulating cellular determinants of editing outcomes. Cell 2021, 184, 5635–5652. [Google Scholar] [CrossRef]
- Wu, J.; Corbett, A.H.; Berland, K.M. The Intracellular Mobility of Nuclear Import Receptors and NLS Cargoes. Biophys. J. 2009, 96, 3840–3849. [Google Scholar] [CrossRef]
- Dang, C.V.; Lee, W.M. Identification of the human c-myc protein nuclear translocation signal. Mol. Cell. Biol. 1988, 8, 4048–4054. [Google Scholar] [CrossRef]
- Spencer, J.M.; Zhang, X. Deep mutational scanning of S. pyogenes Cas9 reveals important functional domains. Sci. Rep. 2017, 7, 16836. [Google Scholar] [CrossRef]
- Lin, Q.; Jin, S.; Zong, Y.; Yu, H.; Zhu, Z.; Liu, G.; Kou, L.; Wang, Y.; Qiu, J.-L.; Li, J.; et al. High-efficiency prime editing with optimized, paired pegRNAs in plants. Nat. Biotechnol. 2021, 39, 923–927. [Google Scholar] [CrossRef]
- Zong, Y.; Liu, Y.; Xue, C.; Li, B.; Li, X.; Wang, Y.; Li, J.; Liu, G.; Huang, X.; Cao, X.; et al. An engineered prime editor with enhanced editing efficiency in plants. Nat. Biotechnol. 2022, 1–9. [Google Scholar] [CrossRef] [PubMed]
- van Haasteren, J.; Li, J.; Scheideler, O.J.; Murthy, N.; Schaffer, D.V. The delivery challenge: Fulfilling the promise of therapeutic genome editing. Nat. Biotechnol. 2020, 38, 845–855. [Google Scholar] [CrossRef] [PubMed]
- Mendell, J.R.; Al-Zaidy, S.; Shell, R.; Arnold, W.D.; Rodino-Klapac, L.R.; Prior, T.W.; Lowes, L.; Alfano, L.; Berry, K.; Church, K.; et al. Single-Dose Gene-Replacement Therapy for Spinal Muscular Atrophy. N. Engl. J. Med. 2017, 377, 1713–1722. [Google Scholar] [CrossRef] [PubMed]
- Goldschmidt, D.S. FDA Approves Gene Therapy for a Type of Blindness. Available online: https://www.cnn.com/2017/12/20/health/fda-gene-therapy-blindness-bn/index.html (accessed on 21 December 2017).
- Grieger, J.C.; Samulski, R.J. Packaging Capacity of Adeno-Associated Virus Serotypes: Impact of Larger Genomes on Infectivity and Postentry Steps. J. Virol. 2005, 79, 9933–9944. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Wu, Z. Ocular delivery of CRISPR/Cas genome editing components for treatment of eye diseases. Adv. Drug Deliv. Rev. 2020, 168, 181–195. [Google Scholar] [CrossRef]
- McClements, M.E.; MacLaren, R.E. Adeno-associated Virus (AAV) Dual Vector Strategies for Gene Therapy Encoding Large Transgenes. Yale J. Biol. Med. 2017, 90, 611–623. [Google Scholar]
- Zhi, S.; Chen, Y.; Wu, G.; Wen, J.; Wu, J.; Liu, Q.; Li, Y.; Kang, R.; Hu, S.; Wang, J.; et al. Dual-AAV delivering split prime editor system for in vivo genome editing. Mol. Ther. 2022, 30, 283–294. [Google Scholar] [CrossRef]
- Tornabene, P.; Trapani, I. Can Adeno-Associated Viral Vectors Deliver Effectively Large Genes? Hum. Gene Ther. 2020, 31, 47–56. [Google Scholar] [CrossRef]
- Trapani, I.; Colella, P.; Sommella, A.; Iodice, C.; Cesi, G.; de Simone, S.; Marrocco, E.; Rossi, S.; Giunti, M.; Palfi, A.; et al. Effective delivery of large genes to the retina by dual AAV vectors. EMBO Mol. Med. 2013, 6, 194–211. [Google Scholar] [CrossRef]
- Zheng, C.; Liang, S.-Q.; Liu, B.; Liu, P.; Kwan, S.-Y.; Wolfe, S.A.; Xue, W. A flexible split prime editor using truncated reverse transcriptase improves dual-AAV delivery in mouse liver. Mol. Ther. 2022, 30, 1343–1351. [Google Scholar] [CrossRef]
- Ran, F.A.; Cong, L.; Yan, W.X.; Scott, D.A.; Gootenberg, J.; Kriz, A.J.; Zetsche, B.; Shalem, O.; Wu, X.; Makarova, K.S.; et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature 2015, 520, 186–191. [Google Scholar] [CrossRef]
- Esvelt, K.M.; Mali, P.; Braff, J.L.; Moosburner, M.; Yaung, S.; Church, G.M. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat. Methods 2013, 10, 1116–1121. [Google Scholar] [CrossRef] [Green Version]
- Kim, E.; Koo, T.; Park, S.W.; Kim, D.; Kim, K.; Cho, H.-Y.; Song, D.W.; Lee, K.J.; Jung, M.H.; Kim, S.; et al. In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat. Commun. 2017, 8, 14500. [Google Scholar] [CrossRef]
- Hu, Z.; Wang, S.; Zhang, C.; Gao, N.; Li, M.; Wang, D.; Wang, D.; Liu, D.; Liu, H.; Ong, S.-G.; et al. A compact Cas9 ortholog from Staphylococcus Auricularis (SauriCas9) expands the DNA targeting scope. PLoS Biol. 2020, 18, e3000686. [Google Scholar] [CrossRef]
- Hirano, S.; Abudayyeh, O.O.; Gootenberg, J.S.; Horii, T.; Ishitani, R.; Hatada, I.; Zhang, F.; Nishimasu, H.; Nureki, O. Structural basis for the promiscuous PAM recognition by Corynebacterium diphtheriae Cas9. Nat. Commun. 2019, 10, 1968. [Google Scholar] [CrossRef]
- Pausch, P.; Al-Shayeb, B.; Bisom-Rapp, E.; Tsuchida, C.A.; Li, Z.; Cress, B.F.; Knott, G.J.; Jacobsen, S.E.; Banfield, J.F.; Doudna, J.A. CRISPR-CasΦ from huge phages is a hypercompact genome editor. Science 2020, 369, 333–337. [Google Scholar] [CrossRef]
- Kim, D.Y.; Lee, J.M.; Bin Moon, S.; Chin, H.J.; Park, S.; Lim, Y.; Kim, D.; Koo, T.; Ko, J.-H.; Kim, Y.-S. Efficient CRISPR editing with a hypercompact Cas12f1 and engineered guide RNAs delivered by adeno-associated virus. Nat. Biotechnol. 2021, 40, 94–102. [Google Scholar] [CrossRef]
- Yang, H.; Patel, D.J. CasX: A new and small CRISPR gene-editing protein. Cell Res. 2019, 29, 345–346. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, H.; Xu, X.; Wang, Y.; Chen, W.; Wang, Y.; Wu, Z.; Tang, N.; Wang, Y.; Zhao, S.; et al. Catalytic-state structure and engineering of Streptococcus thermophilus Cas9. Nat. Catal. 2020, 3, 813–823. [Google Scholar] [CrossRef]
- Tran, M.T.N.; Khalid, M.K.N.M.; Wang, Q.; Walker, J.K.R.; Lidgerwood, G.E.; Dilworth, K.L.; Lisowski, L.; Pébay, A.; Hewitt, A.W. Engineering domain-inlaid SaCas9 adenine base editors with reduced RNA off-targets and increased on-target DNA editing. Nat. Commun. 2020, 11, 1–10. [Google Scholar] [CrossRef]
- Tolmachova, T.; Tolmachov, O.E.; Wavre-Shapton, S.T.; Tracey-White, D.; Futter, C.E.; Seabra, M.C. CHM/REP1 cDNA delivery by lentiviral vectors provides functional expression of the transgene in the retinal pigment epithelium of choroideremia mice. J. Gene Med. 2012, 14, 158–168. [Google Scholar] [CrossRef]
- Maus, M.V.; Fraietta, J.A.; Levine, B.L.; Kalos, M.; Zhao, Y.; June, C.H. Adoptive Immunotherapy for Cancer or Viruses. Annu. Rev. Immunol. 2014, 32, 189–225. [Google Scholar] [CrossRef] [Green Version]
- Naldini, L.; Trono, D.; Verma, I.M. Lentiviral vectors, two decades later. Science 2016, 353, 1101–1102. [Google Scholar] [CrossRef] [PubMed]
- Naso, M.F.; Tomkowicz, B.; Perry, W.L., III; Strohl, W.R. Adeno-Associated Virus (AAV) as a Vector for Gene Therapy. BioDrugs 2017, 31, 317–334. [Google Scholar] [CrossRef] [PubMed]
- Vandendriessche, T.; Thorrez, L.; Acosta-Sanchez, A.; Petrus, I.; Wang, L.; Ma, L.; De Waele, L.; Iwasaki, Y.; Gillijns, V.; Wilson, J.; et al. Efficacy and safety of adeno-associated viral vectors based on serotype 8 and 9 vs. lentiviral vectors for hemophilia B gene therapy. J. Thromb. Haemost. 2006, 5, 16–24. [Google Scholar] [CrossRef]
- Harvey, A.R.; Kamphuis, W.; Eggers, R.; Symons, N.A.; Blits, B.; Niclou, S.; Boer, G.J.; Verhaagen, J. Intravitreal Injection of Adeno-associated Viral Vectors Results in the Transduction of Different Types of Retinal Neurons in Neonatal and Adult Rats: A Comparison with Lentiviral Vectors. Mol. Cell. Neurosci. 2002, 21, 141–157. [Google Scholar] [CrossRef] [PubMed]
- Segel, M.; Lash, B.; Song, J.; Ladha, A.; Liu, C.C.; Jin, X.; Mekhedov, S.L.; Macrae, R.K.; Koonin, E.V.; Zhang, F. Mammalian retrovirus-like protein PEG10 packages its own mRNA and can be pseudotyped for mRNA delivery. Science 2021, 373, 882–889. [Google Scholar] [CrossRef] [PubMed]
- Banskota, S.; Raguram, A.; Suh, S.; Du, S.W.; Davis, J.R.; Choi, E.H.; Wang, X.; Nielsen, S.C.; Newby, G.A.; Randolph, P.B.; et al. Engineered virus-like particles for efficient in vivo delivery of therapeutic proteins. Cell 2022, 185, 250–265. [Google Scholar] [CrossRef]
- Roth, T.L.; Puig-Saus, C.; Yu, R.; Shifrut, E.; Carnevale, J.; Li, P.J.; Hiatt, J.; Saco, J.; Krystofinski, P.; Li, H.; et al. Reprogramming human T cell function and specificity with non-viral genome targeting. Nature 2018, 559, 405–409. [Google Scholar] [CrossRef]
- Yin, H.; Kanasty, R.L.; Eltoukhy, A.A.; Vegas, A.J.; Dorkin, J.R.; Anderson, D.G. Non-viral vectors for gene-based therapy. Nat. Rev. Genet. 2014, 15, 541–555. [Google Scholar] [CrossRef]
- Petri, K.; Zhang, W.; Ma, J.; Schmidts, A.; Lee, H.; Horng, J.E.; Kim, D.Y.; Kurt, I.C.; Clement, K.; Hsu, J.Y.; et al. CRISPR prime editing with ribonucleoprotein complexes in zebrafish and primary human cells. Nat. Biotechnol. 2022, 40, 189–193. [Google Scholar] [CrossRef]
- Nayerossadat, N.; Ali, P.A.; Maedeh, T. Viral and nonviral delivery systems for gene delivery. Adv. Biomed. Res. 2012, 1, 27. [Google Scholar] [CrossRef]
- Chow, R.D.; Chen, J.S.; Shen, J.; Chen, S. A web tool for the design of prime-editing guide RNAs. Nat. Biomed. Eng. 2020, 5, 190–194. [Google Scholar] [CrossRef] [PubMed]
- Hwang, G.-H.; Jeong, Y.K.; Habib, O.; Hong, S.-A.; Lim, K.; Kim, J.-S.; Bae, S. PE-Designer and PE-Analyzer: Web-based design and analysis tools for CRISPR prime editing. Nucleic Acids Res. 2021, 49, W499–W504. [Google Scholar] [CrossRef]
- Standage-Beier, K.; Tekel, S.J.; Brafman, D.A.; Wang, X. Prime Editing Guide RNA Design Automation Using PINE-CONE. ACS Synth. Biol. 2021, 10, 422–427. [Google Scholar] [CrossRef]
- Hsu, J.Y.; Grünewald, J.; Szalay, R.; Shih, J.; Anzalone, A.V.; Lam, K.C.; Shen, M.W.; Petri, K.; Liu, D.R.; Joung, J.K.; et al. PrimeDesign software for rapid and simplified design of prime editing guide RNAs. Nat. Commun. 2021, 12, 1034. [Google Scholar] [CrossRef]
- Siegner, S.M.; Karasu, M.E.; Schröder, M.S.; Kontarakis, Z.; Corn, J.E. PnB Designer: A web application to design prime and base editor guide RNAs for animals and plants. BMC Bioinform. 2021, 22, 101. [Google Scholar] [CrossRef]
- Liu, Y.; Li, X.; He, S.; Huang, S.; Li, C.; Chen, Y.; Liu, Z.; Huang, X.; Wang, X. Efficient generation of mouse models with the prime editing system. Cell Discov. 2020, 6, 1. [Google Scholar] [CrossRef]
- Schene, I.F.; Joore, I.P.; Oka, R.; Mokry, M.; van Vugt, A.H.M.; van Boxtel, R.; van der Doef, H.P.J.; van der Laan, L.J.W.; Verstegen, M.M.A.; van Hasselt, P.M.; et al. Prime editing for functional repair in patient-derived disease models. Nat. Commun. 2020, 11, 1–8. [Google Scholar] [CrossRef]
- Jang, H.; Jo, D.H.; Cho, C.S.; Shin, J.H.; Seo, J.H.; Yu, G.; Gopalappa, R.; Kim, D.; Cho, S.-R.; Kim, J.H.; et al. Application of prime editing to the correction of mutations and phenotypes in adult mice with liver and eye diseases. Nat. Biomed. Eng. 2021, 6, 181–194. [Google Scholar] [CrossRef]
- Lin, J.; Liu, X.; Lu, Z.; Huang, S.; Wu, S.; Yu, W.; Liu, Y.; Zheng, X.; Huang, X.; Sun, Q.; et al. Modeling a cataract disorder in mice with prime editing. Mol. Ther.-Nucleic Acids 2021, 25, 494–501. [Google Scholar] [CrossRef]
- Sürün, D.; Schneider, A.; Mircetic, J.; Neumann, K.; Lansing, F.; Paszkowski-Rogacz, M.; Hänchen, V.; Lee-Kirsch, M.A.; Buchholz, F. Efficient Generation and Correction of Mutations in Human iPS Cells Utilizing mRNAs of CRISPR Base Editors and Prime Editors. Genes 2020, 11, 511. [Google Scholar] [CrossRef] [PubMed]
- Fiaz, S.; Wang, X.; Khan, S.A.; Ahmar, S.; Noor, M.A.; Riaz, A.; Ali, K.; Abbas, F.; Mora-Poblete, F.; Figueroa, C.R.; et al. Novel plant breeding techniques to advance nitrogen use efficiency in rice: A review. GM Crop. Food 2021, 12, 627–646. [Google Scholar] [CrossRef]
- Xu, R.; Li, J.; Liu, X.; Shan, T.; Qin, R.; Wei, P. Development of Plant Prime-Editing Systems for Precise Genome Editing. Plant Commun. 2020, 1, 100043. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Sretenovic, S.; Ren, Q.; Jia, X.; Li, M.; Fan, T.; Yin, D.; Xiang, S.; Guo, Y.; Liu, L.; et al. Plant Prime Editors Enable Precise Gene Editing in Rice Cells. Mol. Plant 2020, 13, 667–670. [Google Scholar] [CrossRef] [PubMed]
- Butt, H.; Rao, G.S.; Sedeek, K.; Aman, R.; Kamel, R.; Mahfouz, M. Engineering herbicide resistance via prime editing in rice. Plant Biotechnol. J. 2020, 18, 2370–2372. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Li, J.; Chen, J.; Yan, L.; Xia, L. Precise Modifications of Both Exogenous and Endogenous Genes in Rice by Prime Editing. Mol. Plant 2020, 13, 671–674. [Google Scholar] [CrossRef] [PubMed]
- Hua, K.; Jiang, Y.; Tao, X.; Zhu, J. Precision genome engineering in rice using prime editing system. Plant Biotechnol. J. 2020, 18, 2167–2169. [Google Scholar] [CrossRef]
- Lin, Q.; Zong, Y.; Xue, C.; Wang, S.; Jin, S.; Zhu, Z.; Wang, Y.; Anzalone, A.V.; Raguram, A.; Doman, J.L.; et al. Prime genome editing in rice and wheat. Nat. Biotechnol. 2020, 38, 582–585. [Google Scholar] [CrossRef]
- Lu, Y.; Tian, Y.; Shen, R.; Yao, Q.; Zhong, D.; Zhang, X.; Zhu, J. Precise genome modification in tomato using an improved prime editing system. Plant Biotechnol. J. 2020, 19, 415–417. [Google Scholar] [CrossRef]
- Hassan, M.; Yuan, G.; Chen, J.-G.; Tuskan, G.A.; Yang, X. Prime Editing Technology and Its Prospects for Future Applications in Plant Biology Research. BioDesign Res. 2020, 2020, 1–14. [Google Scholar] [CrossRef]
- Bosch, J.A.; Birchak, G.; Perrimon, N. Precise genome engineering in Drosophila using prime editing. Proc. Natl. Acad. Sci. USA 2020, 118, e2021996118. [Google Scholar] [CrossRef]
- Kolodny, E.H. Molecular genetics of the βxosaminidase isoenzymes: An introduction. In Advances in Genetics; Academic Press: Cambridge, MA, USA, 2001; Volume 44, pp. 101–126. [Google Scholar] [CrossRef]
- Qian, Y.; Zhao, D.; Sui, T.; Chen, M.; Liu, Z.; Liu, H.; Zhang, T.; Chen, S.; Lai, L.; Li, Z. Efficient and precise generation of Tay–Sachs disease model in rabbit by prime editing system. Cell Discov. 2021, 7, 501. [Google Scholar] [CrossRef]
- Tong, Y.; Jørgensen, T.S.; Whitford, C.M.; Weber, T.; Lee, S.Y. A versatile genetic engineering toolkit for E. coli based on CRISPR-prime editing. Nat. Commun. 2021, 12, 5206. [Google Scholar] [CrossRef]
- Doudna, J.A. The promise and challenge of therapeutic genome editing. Nature 2020, 578, 229–236. [Google Scholar] [CrossRef]
- Jin, S.; Lin, Q.; Luo, Y.; Zhu, Z.; Liu, G.; Li, Y.; Chen, K.; Qiu, J.-L.; Gao, C. Genome-wide specificity of prime editors in plants. Nat. Biotechnol. 2021, 39, 1292–1299. [Google Scholar] [CrossRef]
- Kim, D.Y.; Bin Moon, S.; Ko, J.-H.; Kim, Y.-S.; Kim, D. Unbiased investigation of specificities of prime editing systems in human cells. Nucleic Acids Res. 2020, 48, 10576–10589. [Google Scholar] [CrossRef]
- Newby, G.A.; Liu, D.R. In vivo somatic cell base editing and prime editing. Mol. Ther. 2021, 29, 3107–3124. [Google Scholar] [CrossRef]


| Designation of PE Systems | Components of PE | Efficiency | ||
|---|---|---|---|---|
| Cas9 Nickcase | Reverse Transcriptase | pegRNA | ||
| PE1 | Cas9(H840A) nickase | M-MLV RT | the original pegRNA | 0.2–17% |
| PE2 | M-MLV RT (D200N/L603W/ T330P/T306K/W31S) | 1.6–5.1-fold | ||
| PE3 | + a sgRNA for nicking the non-edited strand | 3-fold compared to PE2 | ||
| ePE | fusing Csy-T2A | + a sgRNA & fusing a Csy4 recognition site into the 3′ end | 1.9-fold * | |
| unnamed | Cas9(H840A) nickase | epegRNA: incorporating evopreQ1 | 3–4-fold * | |
| aPE | apegRNA: inserting a C/G pair or changing each non-C/G pair to a C/G pair | 2.77-fold in indel-editing * | ||
| sPE | spegRNA: introducing same-sense mutations | 353-fold in base-editing * | ||
| x rPE | xr-pegRNA: appending a viral exoribonuclease-resistant RNA motif | 3.1-fold in base conversion * | ||
| G-PE | incorporating a hTR G-quadruplex | similar to using epegRNA | ||
| PE4/PE5 | same as PE2 but transient expressing MLH1d | epegRNA | 7.7-fold compared to PE2/ 2.0-fold * | |
| PE4max/PE5max | based on PE4/PE5, using a human codon-optimized RT/adding a linker | higher than PE2/PE3 | ||
| unnamed | based on PPE (similar to PE2) | PBS with a melting temperature of 30℃ and using dual pegRNAs | at least 2.9-fold compared to PPE | |
| ePPE | based on PPE but removing RT’s RNase H domain and incorporating a viral nucleocapsid protein | the original pegRNA | 5.8-fold compared to PPE | |
| Ca9 Variants & Others | Cas Size (kb) | Predicted PE Size (kb) | AAV Delivery | Reference |
|---|---|---|---|---|
| SpCas9 | 4.2 | 6.2 | [10] | |
| St1Cas9 | 3.3 | 5.3 | [23] | |
| SaCas9 | 3.2 | 5.2 | √ | [6] |
| SauriCas9 | 3.1 | 5.3 | [18] | |
| NmeCas9 | 3.2 | 5.3 | [16] | |
| CjCas9 | 3.0 | 5.0 | √ | [17] |
| CdCas9 | 3.3 | 5.3 | [19] | |
| CasФ | 2.1~2.4 | 4.1~4.4 | √ | [20] |
| Cas12f | 1.2~1.8 | 3.2~3.8 | √ | [21] |
| CasX | <3.0 | <5.0 | √ | [22] |
| Fields | Targets | Details | References | |
|---|---|---|---|---|
| Applications of PE | Medicine | HEK293T cells | correcting the mutant HBB allele to wild-type HBB, T•A to A•T (Sickle cell disease) | [9] |
| deleting a 4-bp insertion in HEXA (Tay-Sachs disease) | ||||
| mouse neuro-2a (N2a) cells/mouse embryos | generating base conversion in Ar gene and Hoxd13 gene | [78] | ||
| liver- and intestine-derived organoid cells/HEK293T cells/Caco-2 cells | promoting a biallelic 3-bp deletion in DGAT1, creating in-frame deletions in CTNNB1 (liver cancer) and repairing a 1-bp duplication in ATP7B (Wilson disease) | [79] | ||
| Fahmut/mut mice | rescuing a homozygous G-to-A point mutation in Fah gene (hereditary tyrosinemia type 1) and correcting a C-to-T transition in RPE65 gene (Leber congenital amaurosis) | [80] | ||
| mouse N2a cells | installing a G-deletion mutation in Crygc gene (cataract disorder) | [81] | ||
| human induced pluripotent stem cells (iPSC) | repairing a missense mutation in SAMHD1 gene (Aicardi–Goutières syndrome) | [82] | ||
| Fah−/− mouse primary hepatocytes | correcting a Fah mutation (Hereditary tyrosinemia type 1) | [26] | ||
| Agriculture | rice (Oryza sativa) | editing the OsALS (herbicide resistance), OsIPA (rice yield) and OsTB1 gene (lateral braching) | [86] | |
| the Japonica rice (Oryza sativa) variety Zhonghua11 and the winter wheat variety Kenong199 protoplasts | producing a wide variety of edits at genomic sites (including C-to-T, G-to-T, A-to-G, G-to-A, T-to-A, and C-to-A substitutions in rice; including A-to-T, C-to-G, G-to-C, T-to-G, and C-to-A substitutions in wheat) | [89] | ||
| tomato | GAI, ALS2 and PDS1 | [90] | ||
| maize | introducing W542L and S621I double mutations in ZmALS1 and ZmALS2 (herbicide resistance) | [11] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, C.; Kuang, J.; Shao, T.; Xie, S.; Li, M.; Zhu, L.; Zhu, L. Prime Editing: An All-Rounder for Genome Editing. Int. J. Mol. Sci. 2022, 23, 9862. https://doi.org/10.3390/ijms23179862
Lu C, Kuang J, Shao T, Xie S, Li M, Zhu L, Zhu L. Prime Editing: An All-Rounder for Genome Editing. International Journal of Molecular Sciences. 2022; 23(17):9862. https://doi.org/10.3390/ijms23179862
Chicago/Turabian StyleLu, Chenyu, Jingyu Kuang, Tong Shao, Sisi Xie, Ming Li, Lingyun Zhu, and Lvyun Zhu. 2022. "Prime Editing: An All-Rounder for Genome Editing" International Journal of Molecular Sciences 23, no. 17: 9862. https://doi.org/10.3390/ijms23179862
APA StyleLu, C., Kuang, J., Shao, T., Xie, S., Li, M., Zhu, L., & Zhu, L. (2022). Prime Editing: An All-Rounder for Genome Editing. International Journal of Molecular Sciences, 23(17), 9862. https://doi.org/10.3390/ijms23179862






