Ionomic Combined with Transcriptomic and Metabolomic Analyses to Explore the Mechanism Underlying the Effect of Melatonin in Relieving Nutrient Stress in Apple
Abstract
:1. Introduction
2. Results
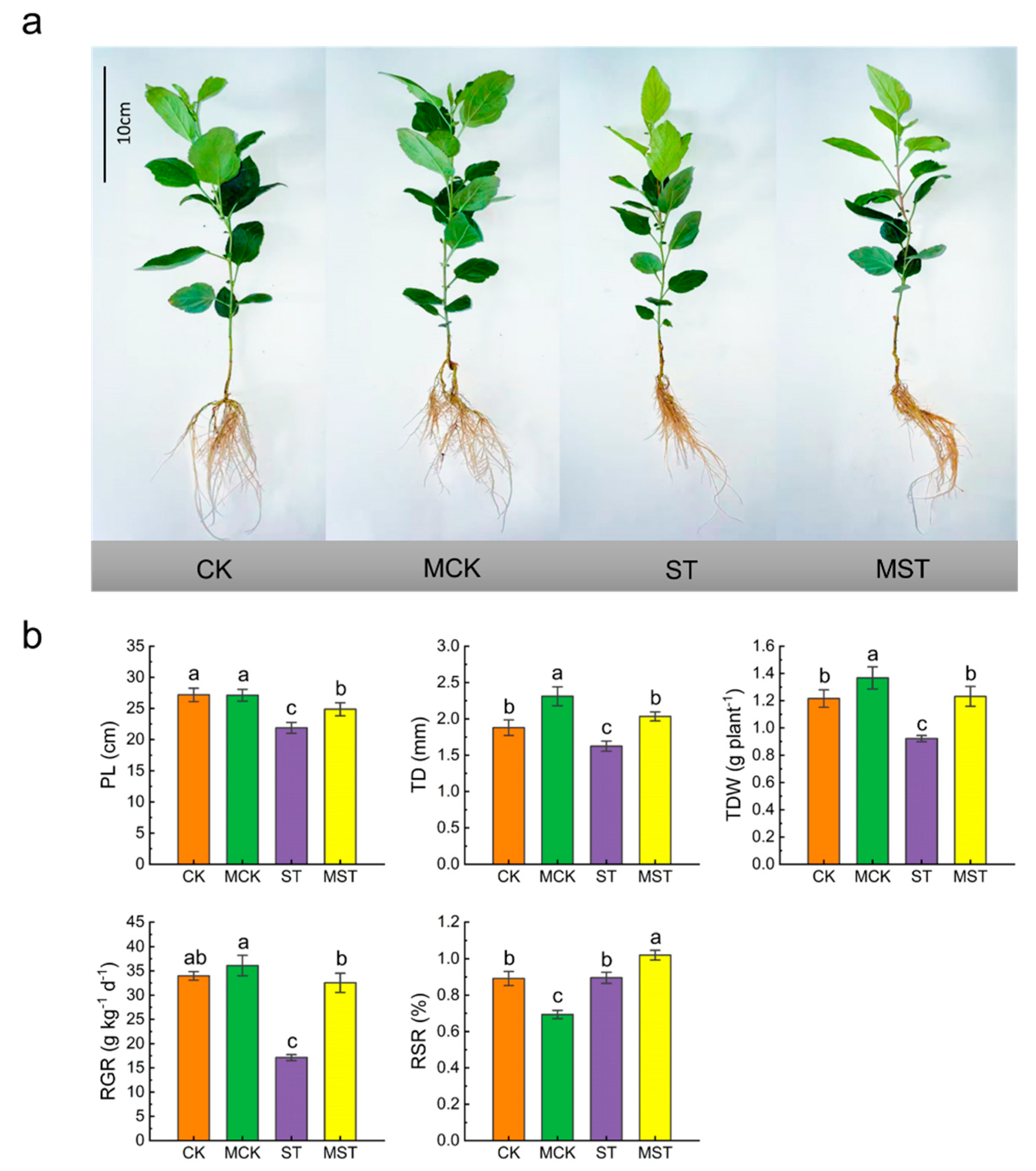
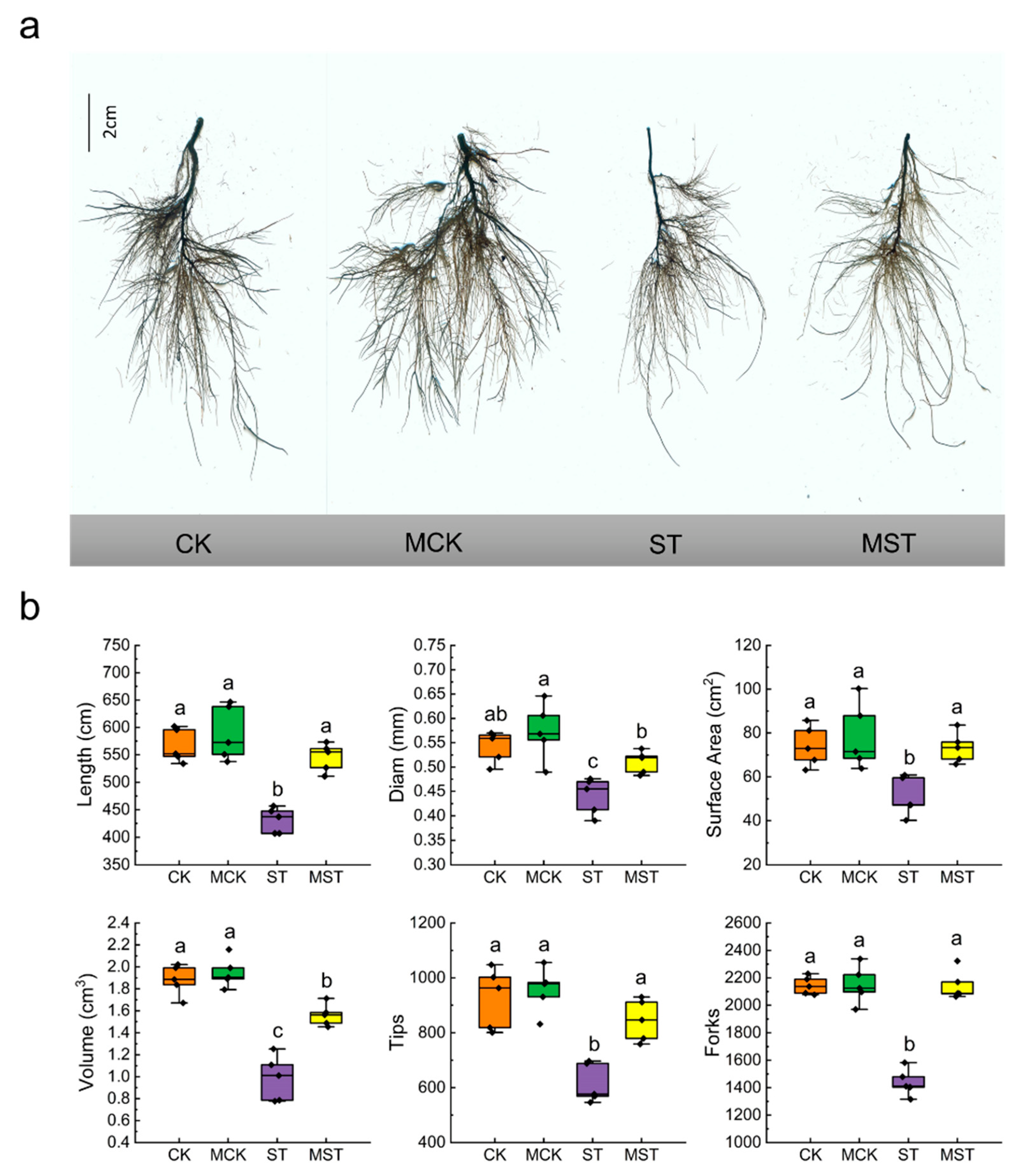
2.1. Exogenous Melatonin Regulates the Growth and Development of Apple Plants and Activates the Antioxidant System under Nutrient Stress
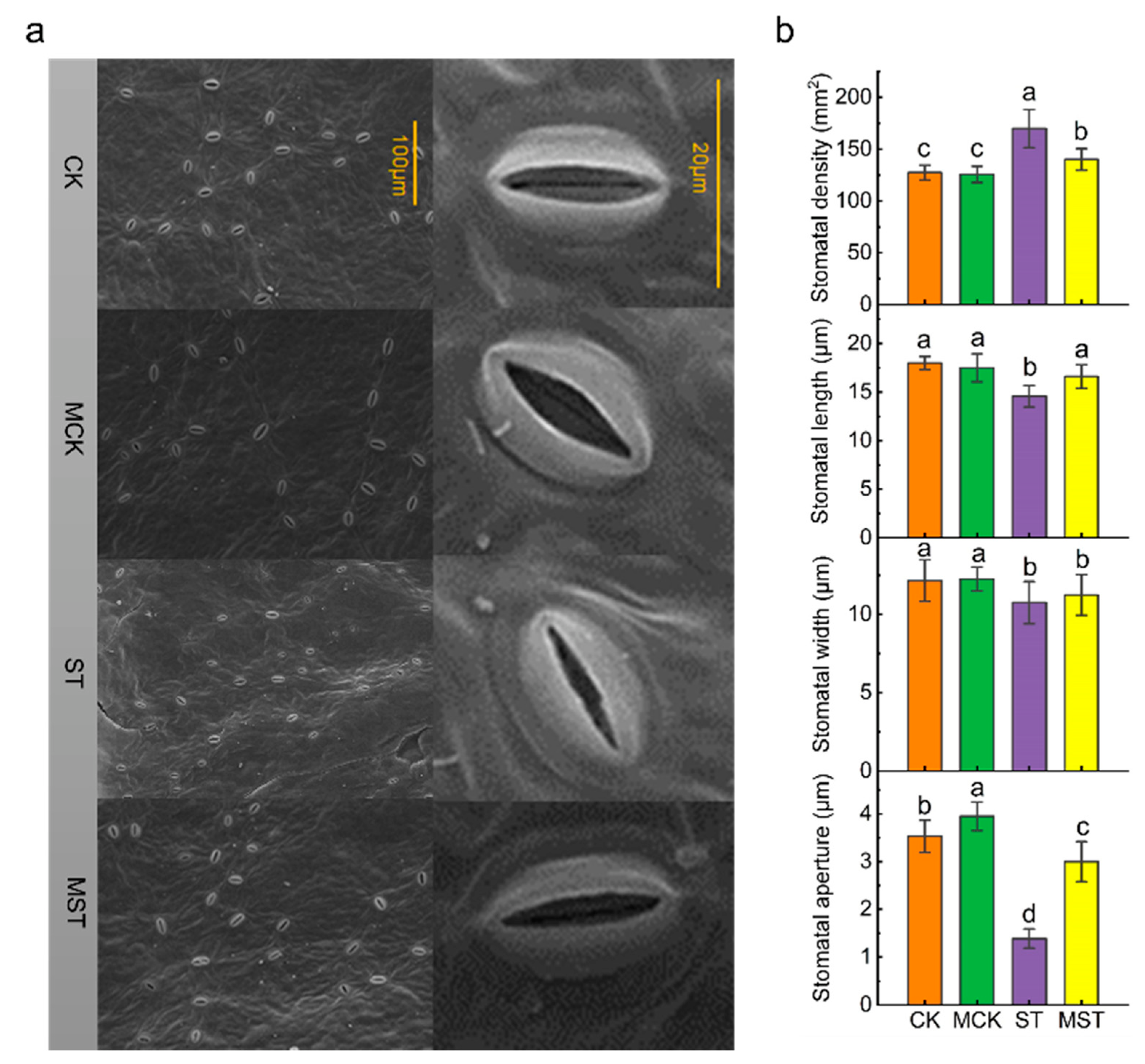
2.2. Exogenous Melatonin Mediates the Regulation of Stomatal Configuration under Nutrient Stress
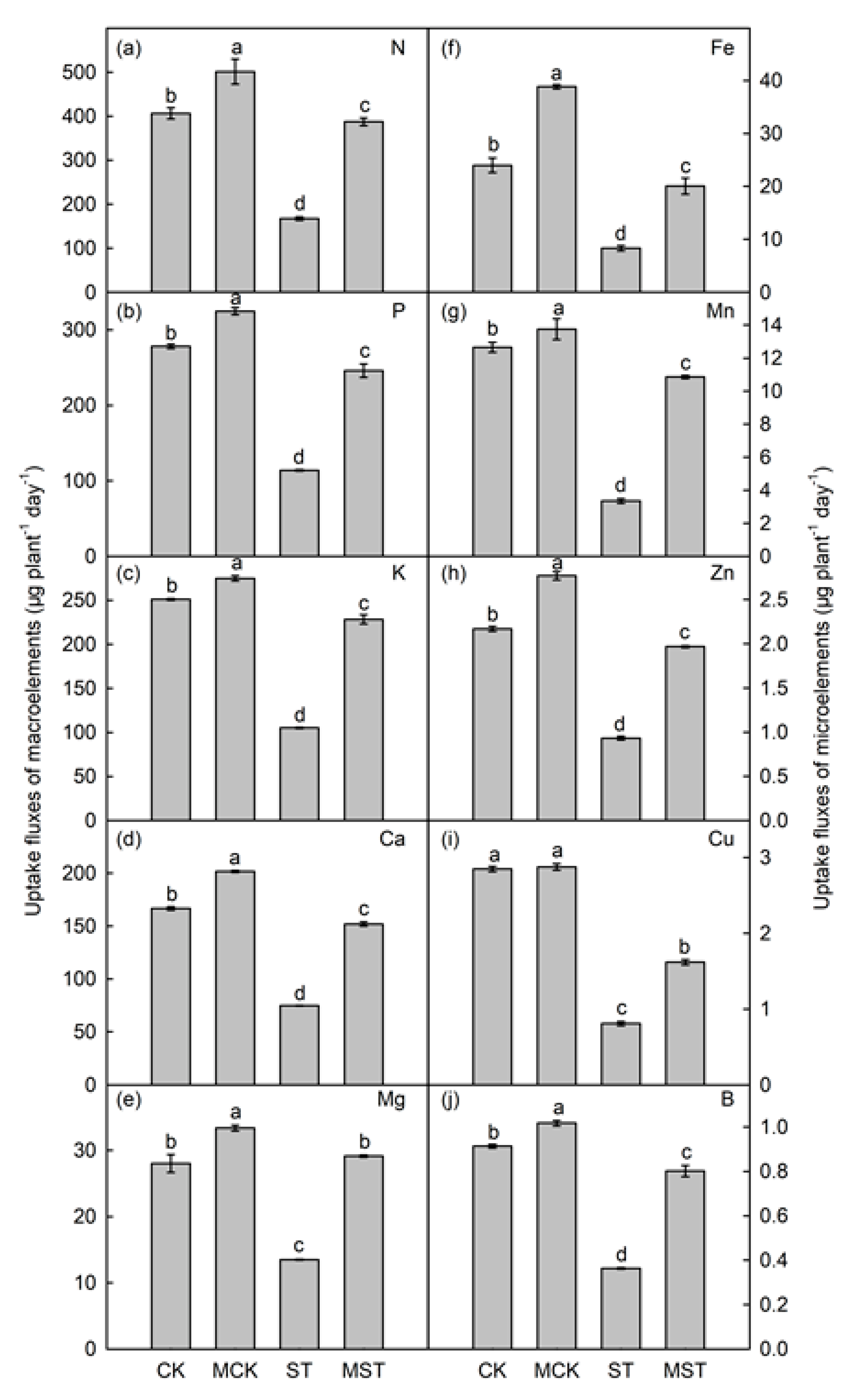
2.3. Exogenous Melatonin Mediates the Regulation of the Concentration, Uptake, Transfer, and Distribution of Minerals in Plants under Nutrient Stress
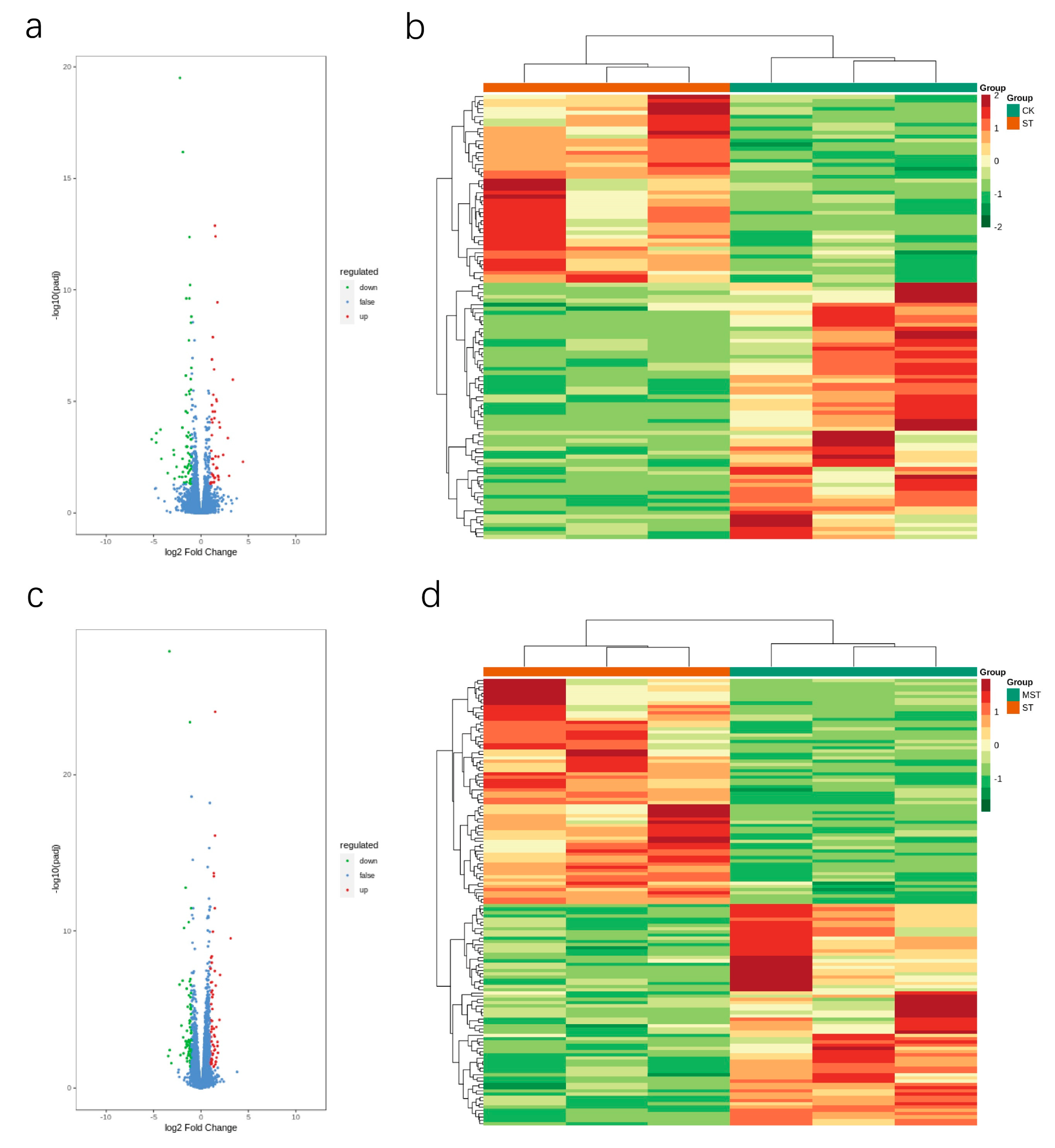
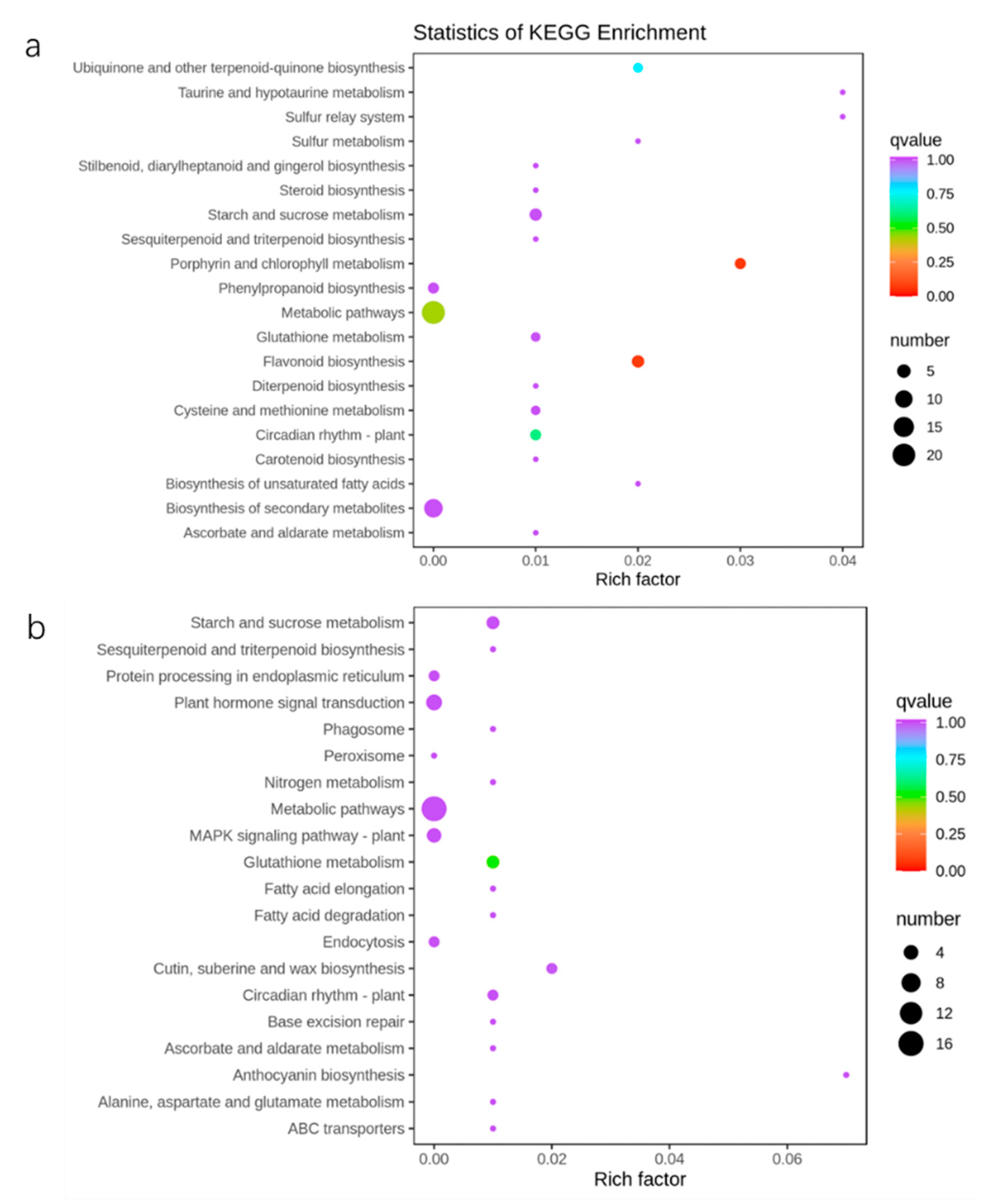
2.4. Differentially Expressed Gene Analysis
2.5. Validation of DEGs by qRT-PCR
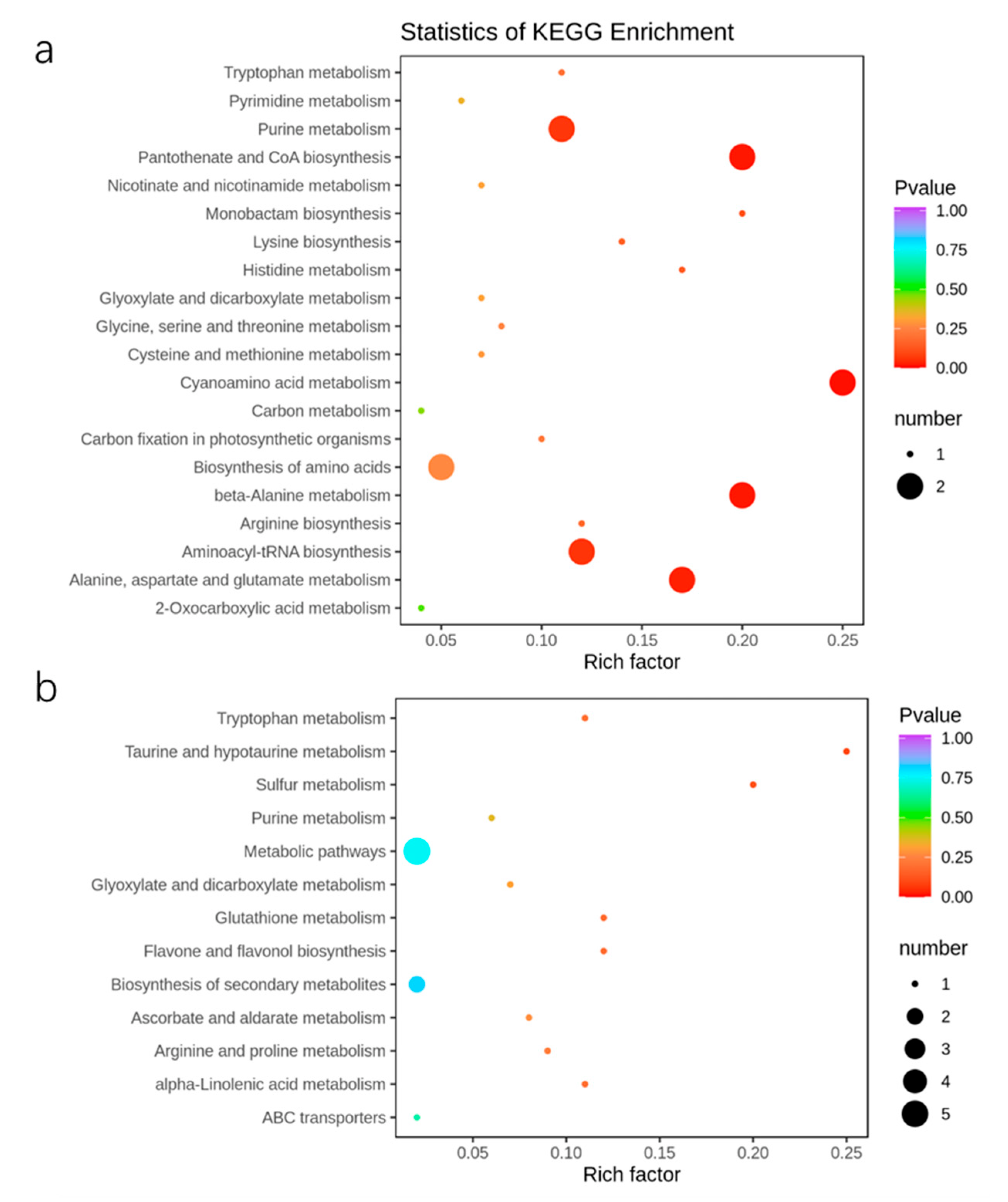
2.6. Metabolic Response of Apple Trees to Nutrient Stress
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Experimental Design
4.3. Measurement of Growth Characteristics and Root Architecture
4.4. Determination of TCC, TTC, REL, MDA, and Antioxidant Enzyme Activity
4.5. Determination of the Stomatal Configuration
4.6. Determination of Minerals
4.7. Extracting RNA and Transcriptome Sequencing
4.8. qRT-PCR Validation
4.9. Metabolite Analysis
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Williams, L.; Salt, D.E. The plant ionome coming into focus. Curr. Opin. Plant Biol. 2009, 12, 247–249. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.M.; Gao, T.T.; Zhang, Z.J.; Tan, K.X.; Jin, Y.B.; Zhao, Y.J.; Ma, F.W.; Li, C. The mitigation effects of exogenous dopamine on low nitrogen stress in Malus hupehensis. J. Integr. Agric. 2020, 19, 2709–2724. [Google Scholar] [CrossRef]
- Du, P.H.; Yin, B.Y.; Cao, Y.; Han, R.X.; Ji, J.H.; He, X.L.; Liang, B.W.; Xu, J.Z. Beneficial effects of exogenous melatonin and dopamine on low nitrate stress in Malus hupehensis. Front. Plant Sci. 2022, 12, 807472. [Google Scholar] [CrossRef] [PubMed]
- Du, P.H.; Yin, B.H.; Zhou, S.S.; Li, Z.Y.; Zhang, X.Y.; Cao, Y.; Han, R.X.; Shi, C.J.; Liang, B.W.; Xu, J.Z. Melatonin and dopamine mediate the regulation of nitrogen uptake and metabolism at low ammonium levels in Malus hupehensis. Plant Physiol. Biochem. 2022, 171, 182–190. [Google Scholar] [CrossRef]
- Sun, T.T.; Zhang, J.K.; Zhang, Q.; Li, X.Y.; Li, M.J.; Yang, Y.Z.; Zhou, J.; Wei, Q.P.; Zhou, B.B. Transcriptome and metabolome analyses revealed the response mechanism of apple to different phosphorus stresses. Plant Physiol. Biochem. 2021, 167, 639–650. [Google Scholar] [CrossRef]
- Li, C.; Liang, B.W.; Chang, C.; Wei, Z.W.; Zhou, S.S.; Ma, F.W. Exogenous melatonin improved potassium content in Malus under different stress conditions. J. Pineal Res. 2016, 61, 218–229. [Google Scholar] [CrossRef]
- Sun, T.T.; Zhang, J.K.; Zhang, Q.; Li, X.L.; Li, M.J.; Yang, Y.Z.; Zhou, J.; Wei, Q.P.; Zhou, B.B. Integrative physiological, transcriptome, and metabolome analysis reveals the effects of nitrogen sufficiency and deficiency conditions in apple leaves and roots. Environ. Exp. Bot. 2021, 192, 104633. [Google Scholar] [CrossRef]
- Liang, B.W.; Li, C.Y.; Ma, C.Q.; Wei, Z.W.; Wang, Q.; Huang, D.; Chen, Q.; Li, C.; Ma, F.W. Dopamine alleviates nutrient deficiency-induced stress in Malus hupehensis. Plant Physiol. Biochem. 2017, 119, 346–359. [Google Scholar] [CrossRef]
- Wang, L.; Feng, C.; Zheng, X.D.; Guo, Y.; Zhou, F.F.; Shan, D.Q.; Liu, X.; Kong, J. Plant mitochondria synthesize melatonin and enhance the tolerance of plants to drought stress. J. Pineal Res. 2017, 63, e12429. [Google Scholar] [CrossRef]
- Arnao, M.B.; Hernandez-Ruiz, J. Melatonin: Plant growth regulator and/or biostimulator during stress? Trends Plant Sci. 2014, 19, 789–797. [Google Scholar] [CrossRef]
- Arnao, M.B.; Hernandez-Ruiz, J. Melatonin: A new plant hormone and/or a plant master regulator? Trends Plant Sci. 2019, 24, 38–48. [Google Scholar] [CrossRef]
- Mao, J.P.; Niu, C.D.; Li, K.; Chen, S.Y.; Tahir, M.M.; Han, M.Y.; Zhang, D. Melatonin promotes adventitious root formation in apple by promoting the function of MdWOX11. BMC Plant Biol. 2020, 20, 536. [Google Scholar] [CrossRef]
- Verde, A.; Míguez, J.M.; Gallardo, M. Role of Melatonin in Apple Fruit during Growth and Ripening: Possible Interaction with Ethylene. Plants 2022, 11, 688. [Google Scholar] [CrossRef]
- Sharma, A.; Wang, J.; Xu, D.; Tao, S.; Chong, S.; Yan, D.; Li, Z.; Yuan, H.; Zheng, B. Melatonin regulates the functional components of photosynthesis, antioxidant system, gene expression, and metabolic pathways to induce drought resistance in grafted Carya cathayensis plants. Sci. Total Environ. 2020, 713, 136675. [Google Scholar] [CrossRef]
- Wei, W.; Li, Q.T.; Chu, Y.N.; Reiter, R.J.; Yu, X.M.; Zhu, D.H.; Zhang, W.K.; Ma, B.; Lin, Q.; Zhang, J.S.; et al. Melatonin enhances plant growth and abiotic stress tolerance in soybean plants. J. Exp. Bot. 2015, 66, 695–707. [Google Scholar] [CrossRef]
- Liu, J.Y.; Yang, J.; Zhang, H.Q.; Cong, L.; Zhai, R.; Yang, C.Q.; Wang, Z.G.; Ma, F.W.; Xu, L.F. Melatonin inhibits ethylene synthesis via nitric oxide regulation to delay postharvest senescence in pears. J. Agric. Food Chem. 2019, 67, 2279–2288. [Google Scholar] [CrossRef]
- Li, J.P.; Liu, J.; Zhu, T.T.; Zhao, C.; Li, L.Y.; Chen, M. The role of melatonin in salt stress responses. Int. J. Mol. Sci. 2019, 20, 1375. [Google Scholar] [CrossRef]
- Moustafa-Farag, M.; Mahmoud, A.; Arnao, M.B.; Sheteiwy, M.S.; Dafea, M.; Soltan, M.; Elkelish, A.; Hasanuzzaman, M.; Ai, S.Y. Melatonin-induced water stress tolerance in plants: Recent advances. Antioxidants 2020, 9, 809. [Google Scholar] [CrossRef]
- Liang, B.W.; Ma, C.Q.; Zhang, Z.J.; Wei, Z.W.; Gao, T.T.; Zhao, Q.; Ma, F.W.; Li, C. Long-term exogenous application of melatonin improves nutrient uptake fluxes in apple plants under moderate drought stress. Environ. Exp. Bot. 2018, 155, 650–661. [Google Scholar] [CrossRef]
- Wang, Y.L.; Lysoe, E.; Armarego-Marriott, T.; Erban, A.; Paruch, L.; Van Eerde, A.; Bock, R.; Liu-Clarke, J. Transcriptome and metabolome analyses provide insights into root and root-released organic anion responses to phosphorus deficiency in oat. J. Exp. Bot. 2018, 69, 3759–3771. [Google Scholar] [CrossRef] [Green Version]
- Hafsi, C.; Debez, A.; Abdelly, C. Potassium deficiency in plants: Effects and signaling cascades. Acta Physiol. Plant. 2014, 36, 1055–1070. [Google Scholar] [CrossRef]
- Aldon, D.; Mbengue, M.; Mazars, C.; Galaud, J.P. Calcium signalling in plant biotic interactions. Int. J. Mol. Sci. 2018, 19, 665. [Google Scholar] [CrossRef] [PubMed]
- Thor, K. Calcium-nutrient and messenger. Front. Plant Sci. 2019, 10, 440. [Google Scholar] [CrossRef] [PubMed]
- de Bang, T.C.; Husted, S.; Laursen, K.H.; Persson, D.P.; Schjoerring, J.K. The molecular-physiological functions of mineral macronutrients and their consequences for deficiency symptoms in plants. New Phytol. 2021, 229, 2446–2469. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.D.; Tan, D.X.; Allan, A.C.; Zuo, B.X.; Zhao, Y.; Reiter, R.J.; Wang, L.; Wang, Z.; Guo, Y.; Zhou, J.Z.; et al. Chloroplastic biosynthesis of melatonin and its involvement in protection of plants from salt stress. Sci. Rep 2017, 7, 41236. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Li, D.X.; Zhang, J.R.; Shan, C.; Rengel, Z.; Song, Z.B.; Chen, Q. Phytomelatonin receptor PMTR1-mediated signaling regulates stomatal closure in Arabidopsis thaliana. J. Pineal Res. 2018, 65, e12500. [Google Scholar] [CrossRef]
- Fan, J.B.; Xie, Y.; Zhang, Z.C.; Chen, L. Melatonin: A multifunctional factor in plants. Int. J. Mol. Sci. 2018, 19, 1528. [Google Scholar] [CrossRef] [PubMed]
- Amtmann, A.; Armengaud, P. Effects of N, P, K and S on metabolism: New knowledge gained from multi-level analysis. Curr. Opin. Plant Biol. 2009, 12, 275–283. [Google Scholar] [CrossRef]
- O’Rourke, J.A.; McCabe, C.E.; Graham, M.A. Dynamic gene expression changes in response to micronutrient, macronutrient, and multiple stress exposures in soybean. Funct. Integr. Genom. 2020, 20, 321–341. [Google Scholar] [CrossRef]
- Williamson, L.C.; Ribrioux, S.P.C.P.; Fitter, A.H.; Leyser, H.M.O. Phosphate availability regulates root system architecture in Arabidopsis. Plant Physiol. 2001, 126, 875–882. [Google Scholar] [CrossRef] [Green Version]
- Jia, X.M.; Zhu, Y.F.; Zhang, R.; Zhu, Z.L.; Zhao, T.; Cheng, L.; Gao, L.Y.; Liu, B.; Zhang, X.Y.; Wang, Y.X. Ionomic and metabolomic analyses reveal the resistance response mechanism to saline-alkali stress in Malus halliana seedlings. Plant Physiol. Biochem. 2020, 147, 77–90. [Google Scholar] [CrossRef]
- Wang, J.C.; Chen, X.F.; Chu, S.H.; You, Y.M.; Chi, Y.W.; Wang, R.Y.; Yang, X.J.; Hayat, K.; Zhang, D.; Zhou, P. Comparative cytology combined with transcriptomic and metabolomic analyses of Solanum nigrum L. in response to Cd toxicity. J. Hazard. Mater. 2022, 423, 127168. [Google Scholar] [CrossRef] [PubMed]
- Ma, T.L.; Wu, W.H.; Wang, Y. Transcriptome analysis of rice root responses to potassium deficiency. BMC Plant Biol. 2012, 12, 161. [Google Scholar] [CrossRef]
- Sung, J.; Lee, S.; Lee, Y.; Ha, S.; Song, B.; Kim, T.; Waters, B.M.; Krishnan, H.B. Metabolomic profiling from leaves and roots of tomato (Solanum lycopersicum L.) plants grown under nitrogen, phosphorus or potassium-deficient condition. Plant Sci. 2015, 241, 55–64. [Google Scholar] [CrossRef]
- Onodera, J.; Ohsumi, Y. Autophagy is required for maintenance of amino acid levels and protein synthesis under nitrogen starvation. J. Biol. Chem. 2005, 280, 31582–31586. [Google Scholar] [CrossRef]
- Yang, J.; Fu, M.; Ji, C.; Huang, Y.; Wu, Y. Maize oxalyl-CoA decarboxylase1 degrades oxalate and affects the seed metabolome and nutritional quality. Plant Cell 2018, 30, 2447–2462. [Google Scholar] [CrossRef]
- Nussbaumer, T.; Warth, B.; Sharma, S.; Ametz, C.; Bueschl, C.; Parich, A.; Pfeifer, M.; Siegwart, G.; Steiner, B.; Lemmens, M.; et al. Joint Transcriptomic and metabolomic analyses reveal changes in the primary metabolism and imbalances in the subgenome orchestration in the bread wheat molecular response to fusarium graminearum. G3-Genes Genomes Genet. 2015, 5, 2579–2592. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.Q.; Yuan, H.J.; Dong, X.K.; Peng, M.; Jing, X.Y.; Xu, Q.J.; Tang, T.; Wang, Y.L.; Zha, S.; Gao, M.; et al. Genome-wide dissection of co-selected uv-b responsive pathways in the UV-B adaptation of qingke. Mol. Plant 2020, 13, 112–127. [Google Scholar] [CrossRef]
- Schulz, E.; Tohge, T.; Zuther, E.; Fernie, A.R.; Hincha, D.K. Natural variation in flavonol and anthocyanin metabolism during cold acclimation in Arabidopsis thaliana accessions. Plant Cell Environ. 2015, 38, 1658–1672. [Google Scholar] [CrossRef] [PubMed]
- Nakabayashi, R.; Yonekura-Sakakibara, K.; Urano, K.; Suzuki, M.; Yamada, Y.; Nishizawa, T.; Matsuda, F.; Kojima, M.; Sakakibara, H.; Shinozaki, K.; et al. Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids. Plant J. 2014, 77, 367–379. [Google Scholar] [CrossRef] [Green Version]
- Hill, C.B.; Taylor, J.D.; Edwards, J.; Mather, D.; Bacic, A.; Langridge, P.; Roessner, U. Whole-genome mapping of agronomic and metabolic traits to identify novel quantitative trait loci in bread wheat grown in a water-limited environment. Plant Physiol. 2013, 162, 1266–1281. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.Q.; Li, T.X.; Jiao, L.C.; Jiang, C.; Chen, S.Y.L.; Huang, L.Q.; Liu, J. Metabolome and transcriptome analyses identify the plant immunity systems that facilitate sesquiterpene and lignan biosynthesis in Syringa pinnatifolia Hemsl. BMC Plant Biol. 2022, 22, 132. [Google Scholar] [CrossRef] [PubMed]
- Liang, B.W.; Gao, T.T.; Zhao, Q.; Ma, C.Q.; Chen, Q.; Wei, Z.W.; Li, C.Y.; Li, C.; Ma, F.W. Effects of exogenous dopamine on the uptake, transport, and resorption of apple ionome under moderate drought. Front. Plant Sci. 2018, 9, 755. [Google Scholar] [CrossRef] [PubMed]
- Dionisio-Sese, M.L.; Tobita, S. Antioxidant responses of rice seedlings to salinity stress. Plant Sci. 1998, 135, 1–9. [Google Scholar] [CrossRef]
- Mahmood, U.; Hussain, S.; Hussain, S.; Ali, B.; Ashraf, U.; Zamir, S.; Al-Robai, S.A.; Alzahrani, F.O.; Hano, C.; El-Esawi, M.A. Morpho-physio-biochemical and molecular responses of maize hybrids to salinity and waterlogging during stress and recovery phase. Plants 2021, 10, 1345. [Google Scholar] [CrossRef]
- Hu, L.Y.; Zhou, K.; Li, Y.T.S.; Chen, X.F.; Liu, B.B.; Li, C.Y.; Gong, X.Q.; Ma, F.W. Exogenous myo-inositol alleviates salinity-induced stress in Malus hupehensis Rehd. Plant Physiol. Biochem. 2018, 133, 116–126. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Ying, H.; Shi, J.; Zhang, S.S.; Pingcuo, G.S.; Wang, S.; Zhao, F.; Cui, Y.N.; Zeng, X.L. Transcriptomic and metabolomic profiling provide novel insights into fruit development and flesh coloration in Prunus mira Koehne, a special wild peach species. BMC Plant Biol. 2019, 19, 463. [Google Scholar] [CrossRef]
- Zhong, Y.D.; Chen, C.H.; Gong, X.; Luan, X.Y.; Wu, Z.X.; Li, H.H.; Liu, Q.L.; Xu, M.; Yu, F.X. Transcriptome and metabolome analyses reveal a key role of the anthocyanin biosynthetic pathway cascade in the pigmentation of a Cinnamomum camphora red bark mutant (‘Gantong 1′). Ind. Crops Prod. 2022, 175, 114236. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, Y.; Du, P.; Ji, J.; He, X.; Zhang, J.; Shang, Y.; Liu, H.; Xu, J.; Liang, B. Ionomic Combined with Transcriptomic and Metabolomic Analyses to Explore the Mechanism Underlying the Effect of Melatonin in Relieving Nutrient Stress in Apple. Int. J. Mol. Sci. 2022, 23, 9855. https://doi.org/10.3390/ijms23179855
Cao Y, Du P, Ji J, He X, Zhang J, Shang Y, Liu H, Xu J, Liang B. Ionomic Combined with Transcriptomic and Metabolomic Analyses to Explore the Mechanism Underlying the Effect of Melatonin in Relieving Nutrient Stress in Apple. International Journal of Molecular Sciences. 2022; 23(17):9855. https://doi.org/10.3390/ijms23179855
Chicago/Turabian StyleCao, Yang, Peihua Du, Jiahao Ji, Xiaolong He, Jiran Zhang, Yuwei Shang, Huaite Liu, Jizhong Xu, and Bowen Liang. 2022. "Ionomic Combined with Transcriptomic and Metabolomic Analyses to Explore the Mechanism Underlying the Effect of Melatonin in Relieving Nutrient Stress in Apple" International Journal of Molecular Sciences 23, no. 17: 9855. https://doi.org/10.3390/ijms23179855
APA StyleCao, Y., Du, P., Ji, J., He, X., Zhang, J., Shang, Y., Liu, H., Xu, J., & Liang, B. (2022). Ionomic Combined with Transcriptomic and Metabolomic Analyses to Explore the Mechanism Underlying the Effect of Melatonin in Relieving Nutrient Stress in Apple. International Journal of Molecular Sciences, 23(17), 9855. https://doi.org/10.3390/ijms23179855




