Overexpression of MsRCI2D and MsRCI2E Enhances Salt Tolerance in Alfalfa (Medicago sativa L.) by Stabilizing Antioxidant Activity and Regulating Ion Homeostasis
Abstract
:1. Introduction
2. Results
2.1. MsRCI2D and MsRCI2E Encode PMP3 Protein and Contain Two Putative Transmembrane Domains
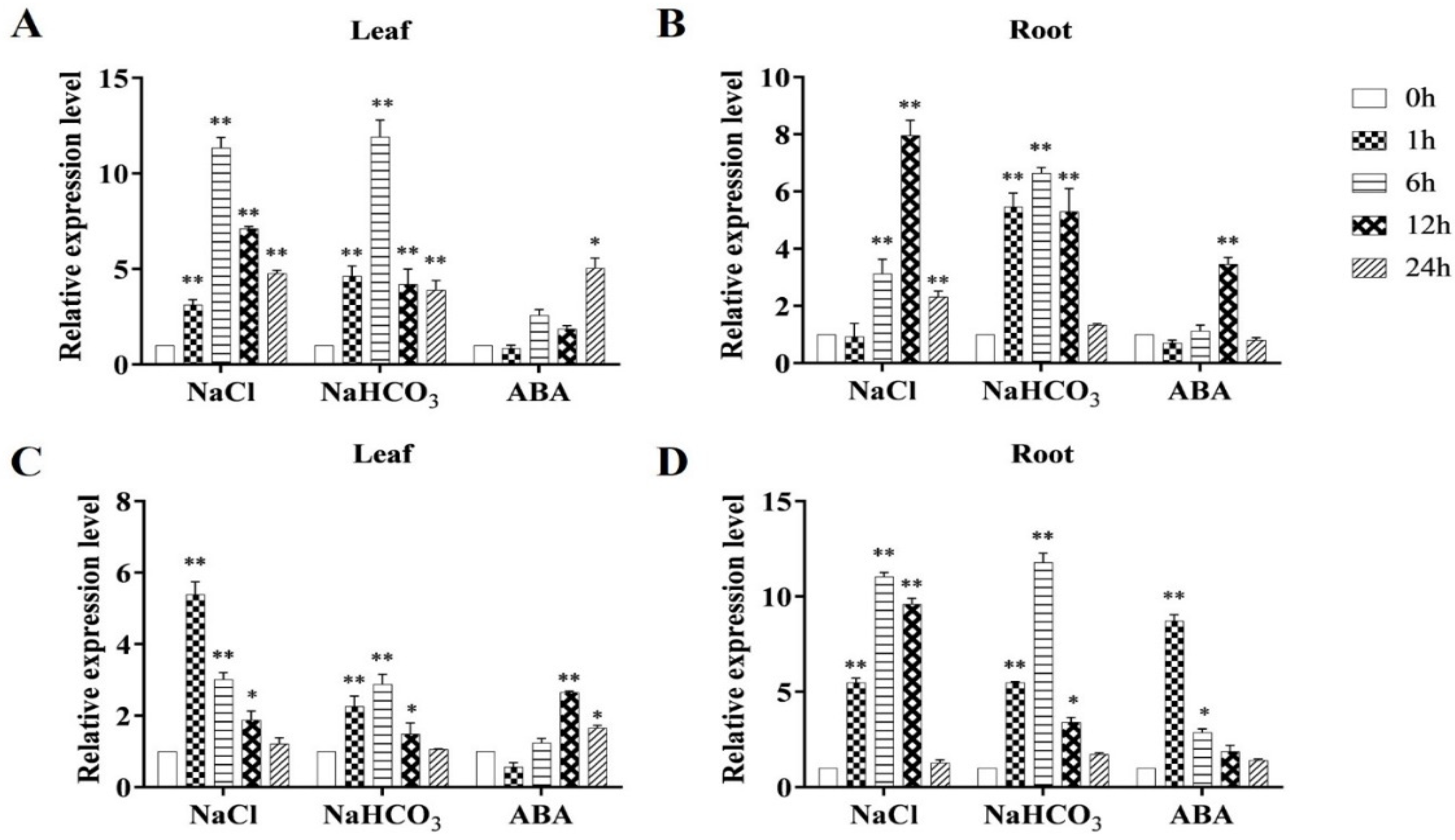
2.2. Differential Expression of the MsRCI2D,-E Genes under Abiotic Stress
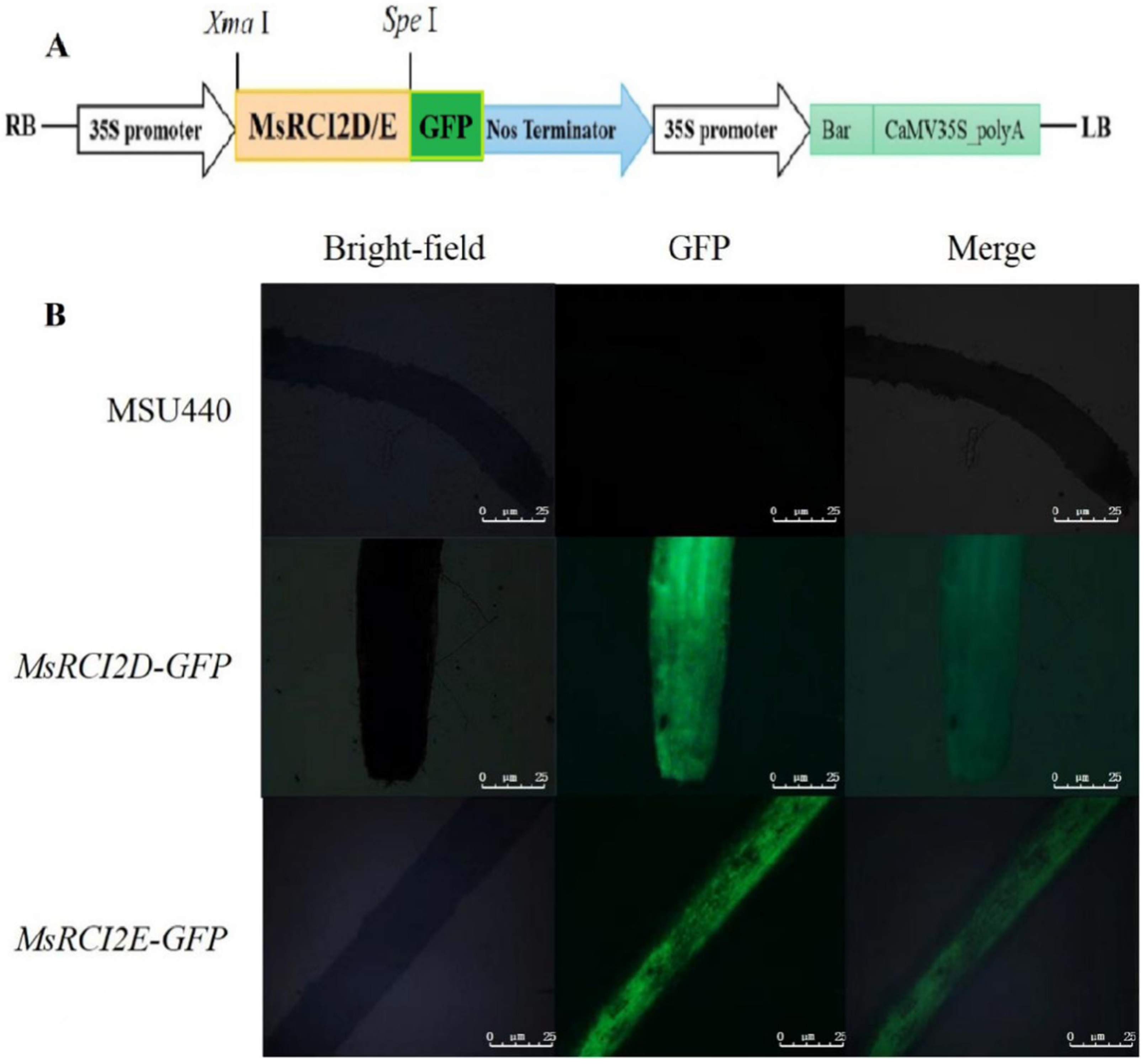
2.3. Subcellular Localization of MsRCI2D/-E
2.4. Generation of Transgenic Alfalfa Chimeras That Overexpress MsRCI2D/-E in Hairy Roots via Agrobacterium Rhizogenes
2.5. Overexpression of MsRCI2D/-E Gene Significantly Increases the Salt Tolerance of the Transgenic Alfalfa Chimera
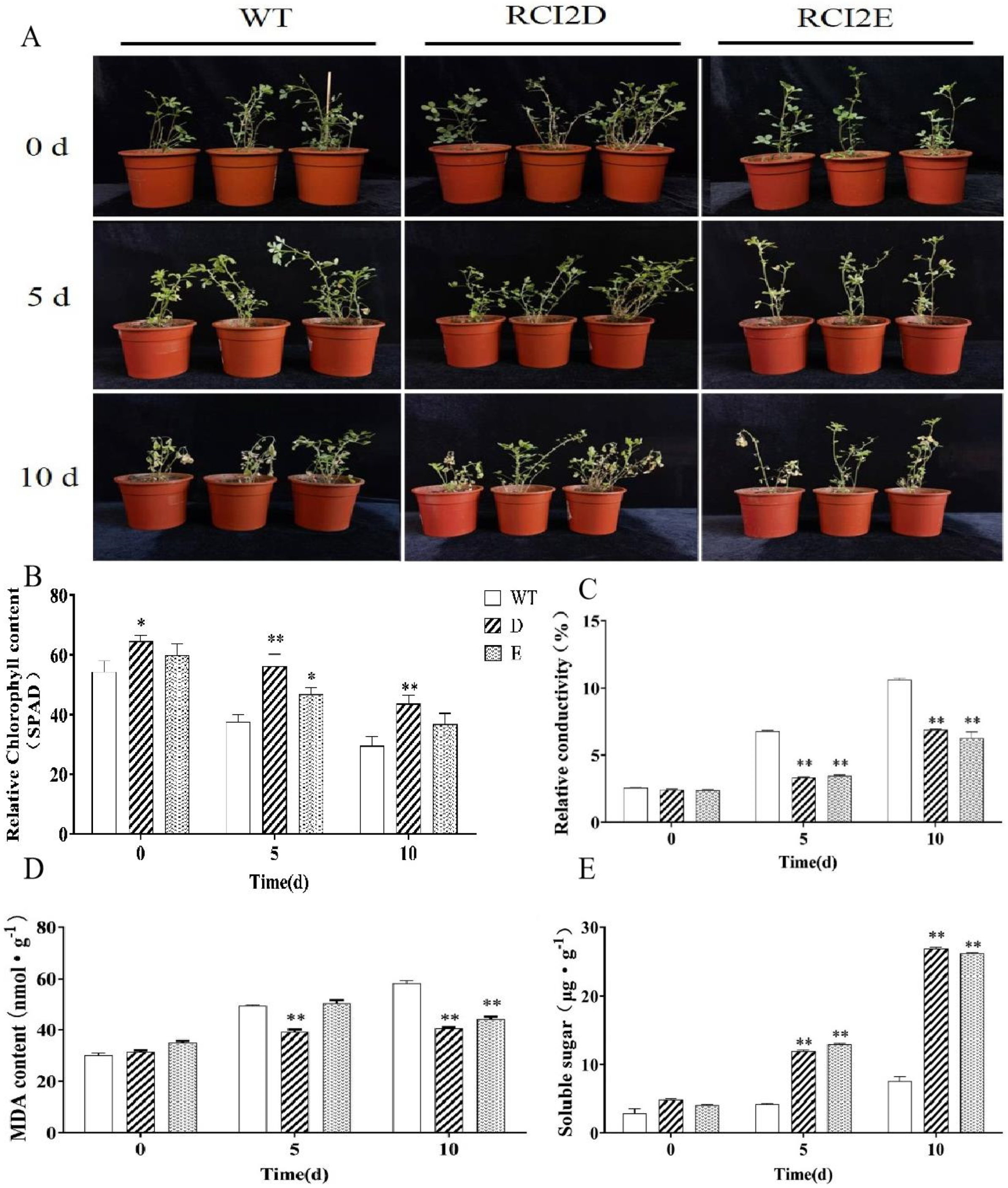
2.6. Changes in Physiological Indexes in the Transgenic MsRCI2D/-E Gene in Alfalfa under Salt Stress
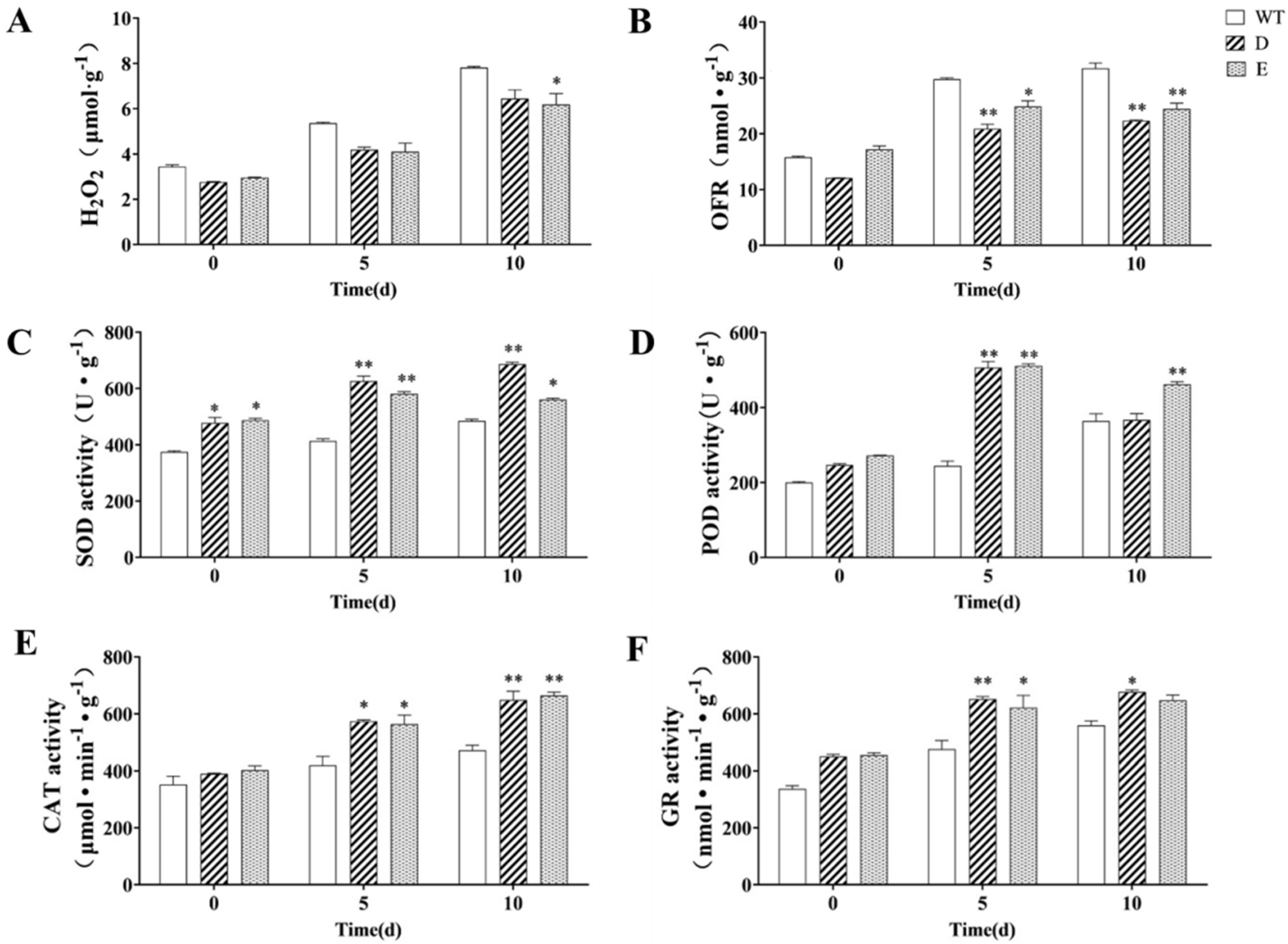
2.7. Change in the Antioxidant Capacity of MsRCI2D/-E Transgenic Alfalfa Hairy Roots under Salt Stress
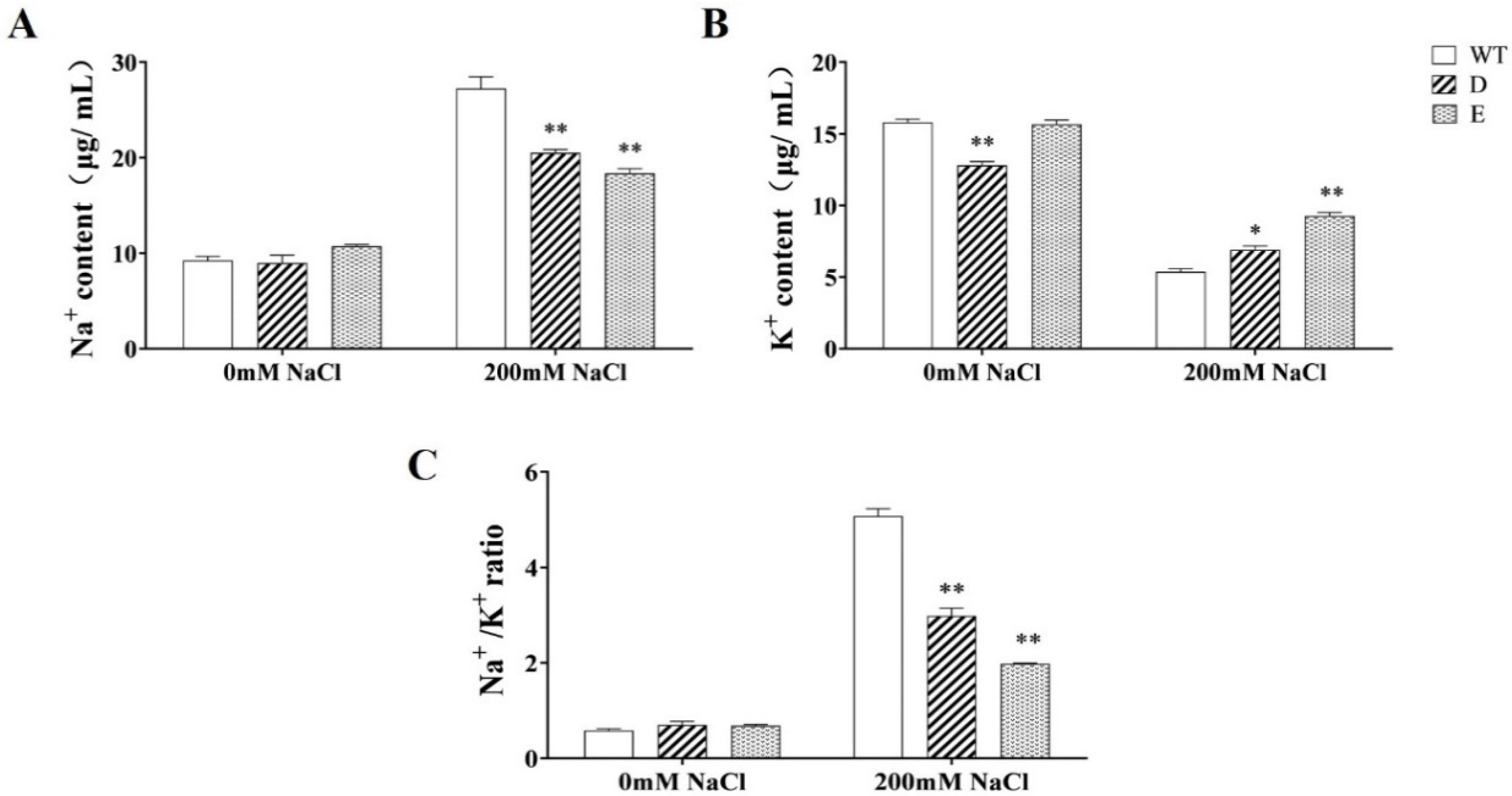
2.8. Changes in Na+ and K+ Ions in Transgenic Alfalfa Hairy Roots under Salt Stress
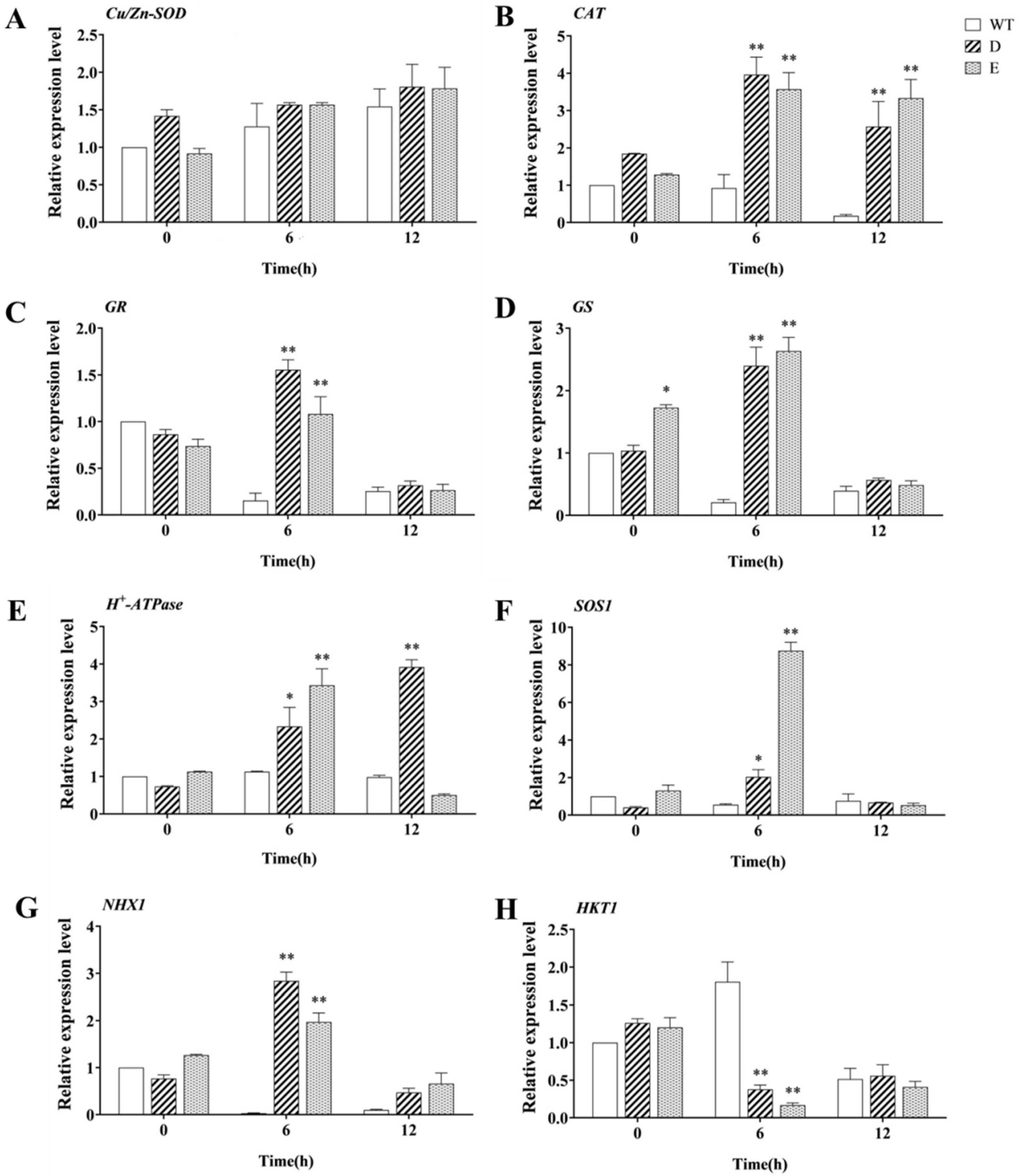
2.9. Differences in the Expression of Genes Related to Salt Stress in Transgenic Alfalfa Hairy Roots
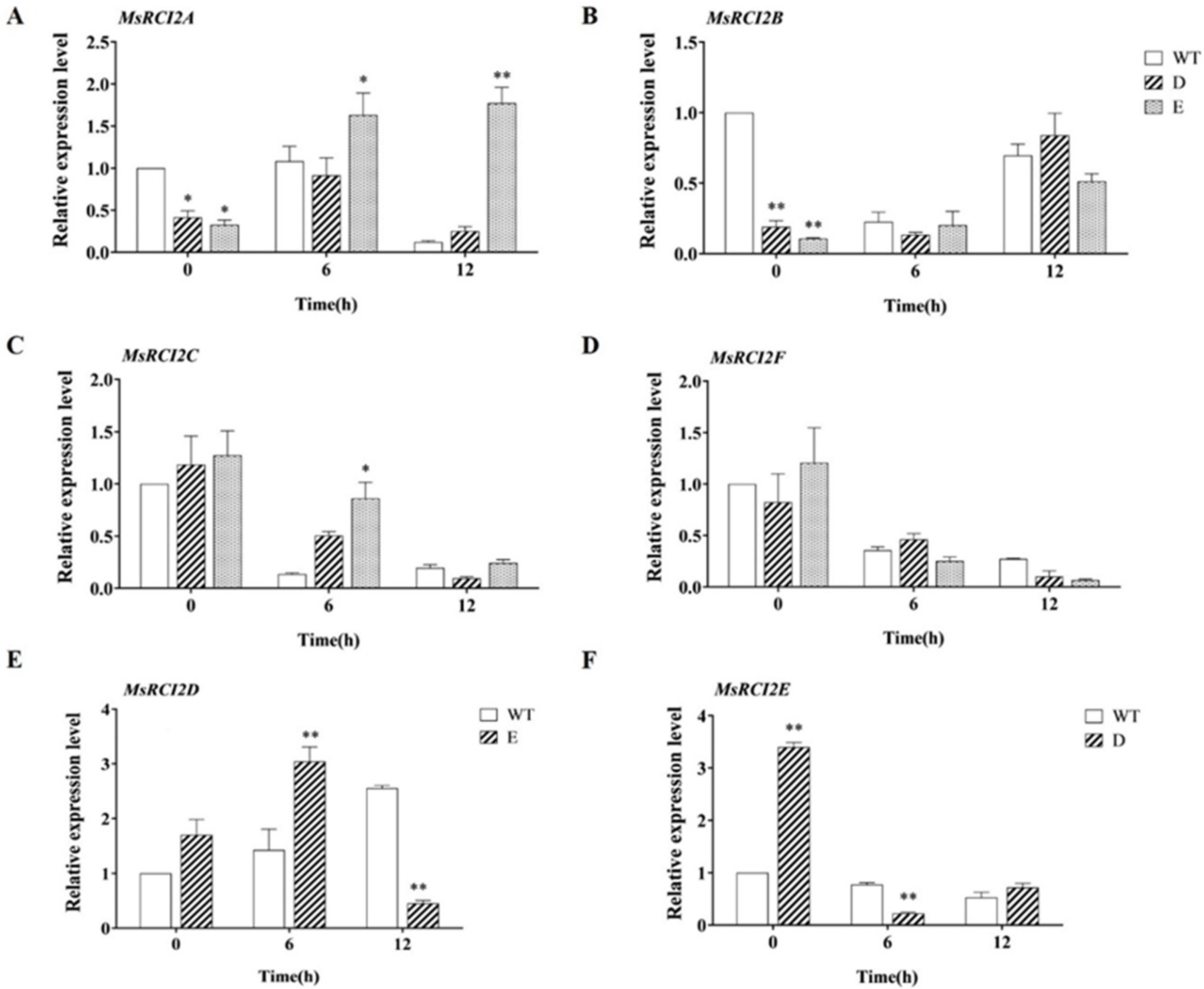
2.10. Differential Expression of MsRCI2 Genes in MsRCI2D,-E Transgenic Alfalfa
3. Discussion
3.1. Differences in Terms of Gene Function between MsRCI2D and MsRCI2E under Salt Stress
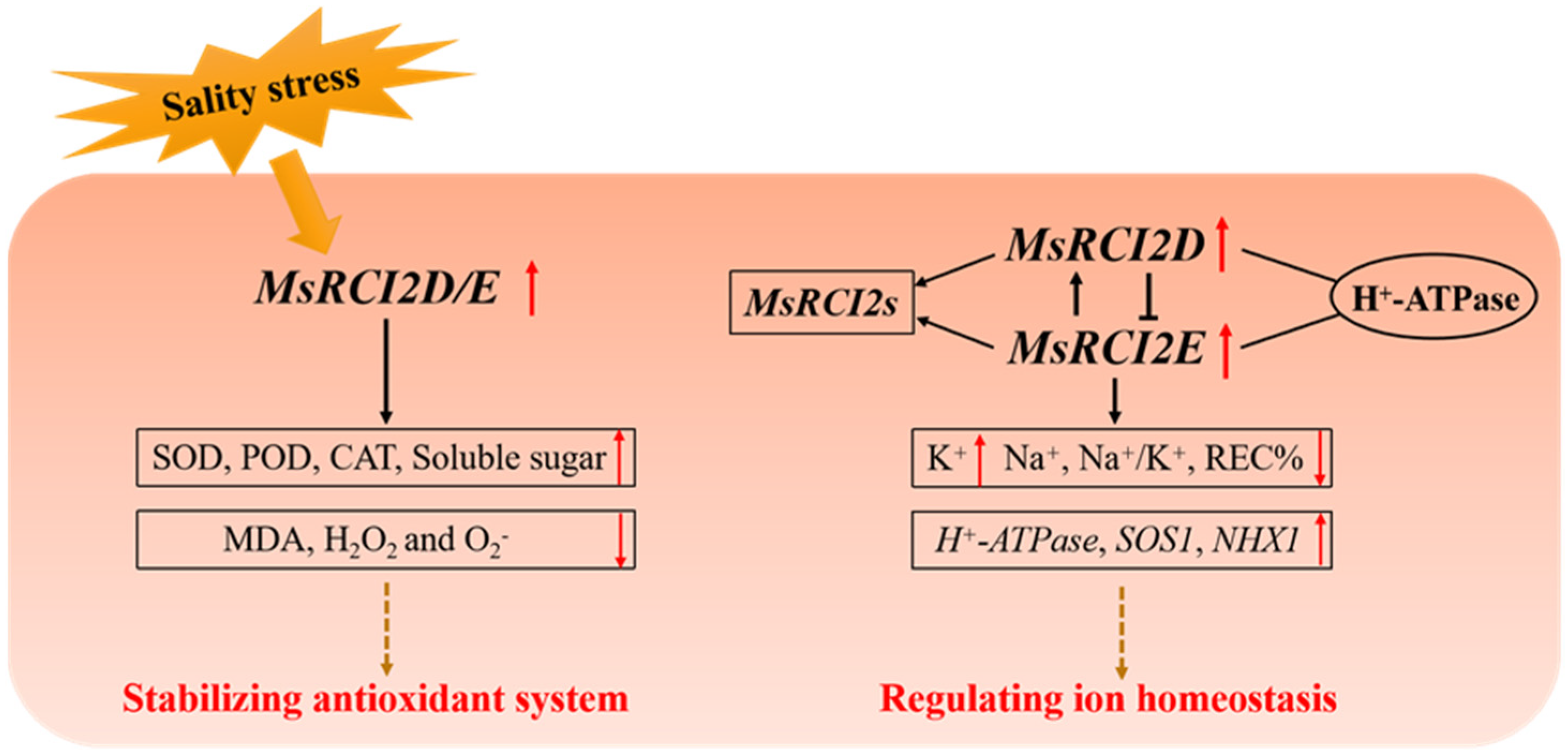
3.2. Molecular Mechanism of MsRCI2D and MsRCI2E Regulating Salt Tolerance
4. Materials and Methods
4.1. Cloning and Bioinformatic Analysis of MsRCI2 Genes
4.2. Analysis of the Expression of MsRCI2 Genes under Different Treatments
4.3. Subcellular Localization
4.4. Generation of MsRCI2-Overexpressing Transgenic Alfalfa Chimera
4.5. Determination of Salt Tolerance of Transgenic Alfalfa Chimera
4.6. Analysis of Na+ and K+ Ion Contents in Transgenic Plants
4.7. Analysis of the Expression Patterns of Related Genes in Transgenic Alfalfa Hairy Roots after Salt Treatment
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Li, C.; Song, T.; Zhan, L.; Cong, C.; Xu, H.; Dong, L.; Cai, H. Overexpression of MsRCI2A, MsRCI2B, and MsRCI2C in Alfalfa (Medicago sativa L.) Provides Different Extents of Enhanced Alkali and Salt Tolerance Due to Functional Specialization of MsRCI2s. Front. Plant Sci. 2021, 12, 702195. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yang, P.; Zhou, Y.; Hu, T.; Zhang, P.; Wu, Y. A proteomic approach to understand the impact of nodulation on salinity stress response in alfalfa (Medicago sativa L.). Plant Biol. 2022, 24, 323–332. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.; Raza, M.A.; Awan, S.A.; Shah, G.A.; Rizwan, M.; Ali, B.; Tariq, R.; Hassan, M.J.; Alyemeni, M.N.; Brestic, M. Amelioration of salt induced toxicity in pearl millet by seed priming with silver nanoparticles (AgNPs): The oxidative damage, antioxidant enzymes and ions uptake are major determinants of salt tolerant capacity. Plant Physiol. Biochem. 2020, 56, 221–232. [Google Scholar] [CrossRef]
- Iqbal, P.; Ghani, M.A.; Ali, B.; Shahid, M.; Iqbal, Q.; Ziaf, K.; Azam, M.; Noor, A.; Cheema, K.L.; Ahmad, J. Exogenous application of glutamic acid promotes cucumber (Cucumis sativus L.) growth under salt stress conditions. Emir. J. Food Agric. 2021, 33, 407–416. [Google Scholar] [CrossRef]
- Ali, B.; Saleem, M.; Ali, S.; Shahid, M.; Sagir, M.; Tahir, M.B.; Ahmad, K.Q.; Jaremko, M.; Selim, S.; Hussain, A.; et al. Mitigation of Salinity Stress in Barley Genotypes with Variable Salt Tolerance by Application of Zinc Oxide Nanoparticles (ZnO NPs). Front. Plant Sci. 2022. [Google Scholar] [CrossRef]
- Chen, W.; Lin, F.; Lin, K.H.; Chen, C.; Xia, C.; Liao, Q.; Chen, S.; Kuo, Y. Growth promotion and salt-tolerance improvement of gerbera jamesonii by root colonization of Piriformospora indica. J. Plant Growth Regul. 2021, 41, 1219–1228. [Google Scholar] [CrossRef]
- Huang, X.Y.; Wang, C.K.; Zhao, Y.W.; Sun, C.H.; Hu, D.G. Mechanisms and regulation of organic acid accumulation in plant vacuoles. Hortic. Res. 2021, 8, 227. [Google Scholar] [CrossRef]
- Deinlein, U.; Stephan, A.B.; Horie, T.; Luo, W.; Xu, G.; Schroeder, J.I. Plant salt-tolerance mechanisms. Trends Plant Sci. 2014, 19, 371–379. [Google Scholar] [CrossRef]
- Nieves-Cordone, M.; Lara, A.; Ródenas, R.; Amo, J.; Rivero, M.R.; Martínez, V.; Rubio, F. Modulation of K+ translocation by AKT1 and AtHAK5 in Arabidopsis plants. Plant Cell Environ. 2019, 42, 2357–2371. [Google Scholar] [CrossRef]
- Kim, H.S.; Park, W.; Lee, H.S.; Shin, J.H.; Ahn, S.J. Subcellular Journey of Rare Cold Inducible 2 Protein in Plant Under Stressful Condition. Front. Plant Sci. 2021, 11, 610251. [Google Scholar] [CrossRef]
- Kim, Y.O.; Lim, H.G.; Kim, H.S.; Ahn, S.J. Overexpression of CsRCI2H enhances salt tolerance in Camelina sativa (L.). Plant Biotechnol. Rep. 2020, 14, 439–449. [Google Scholar] [CrossRef]
- Brunetti, S.C.; Arseneault, M.K.M.; Gulick, P.J. Characterization of the Esi3/RCI2/PMP3 gene family in the Triticeae. BMC Genom. 2018, 19, 898. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.-S.; Park, W.; Lim, H.-G.; Eom, S.; Lee, J.-H.; Carlson, J.E. NaCl-induced CsRCI2E and CsRCI2F interact with aquaporin CsPIP2; 1 to reduce water transport in Camelina sativa L. Biochem. Biophys. Res. Commol. 2019, 513, 213–218. [Google Scholar] [CrossRef]
- Medina, J.; Ballesteros, M.L.; Salinas, J. Phylogenetic and functional analysis of Arabidopsis RCI2 genes. J. Exp. Bot. 2007, 58, 4333–4346. [Google Scholar] [CrossRef] [PubMed]
- Kwok, A.C.; Zhang, F.; Ma, Z.; Chan, W.S.; Yu, V.C.; Tsang, J.S. Functional responses between PMP3 small membrane proteins and membrane potential. Environ. Microbiol. 2020, 22, 3066–3080. [Google Scholar] [CrossRef]
- Rocha Pedro, S.C.F. Plant abiotic stress-related RCI2/PMP3s: Multigenes for multiple roles. Planta 2016, 243, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Walid, B.R.; Rania, B.S.; Donaldo, M.; Jean-Luc, V.; Jalel, A.; Nabil, Z.; Fki, L.; Guiderdoni, E.; Al-Doss, A.; Hassairi, A. Ectopic expression of Aeluropus littoralis plasma membrane protein gene AlTMP1 confers abiotic stress tolerance in transgenic tobacco by improving water status and cation homeostasis. Int. J. Mol. Sci. 2017, 18, 692. [Google Scholar]
- Ben, R.W.; Ben, S.R.; Meynard, D.; Zouari, N.; Mahjoub, A.; Fki, L.; Guiderdoni, E.; Al-Doss, A.; Hassairi, A. Overexpression of AlTMP2 gene from the halophyte grass Aeluropus littoralis in transgenic tobacco enhances tolerance to different abiotic stresses by improving membrane stability and deregulating some stress-related genes. Protoplasma 2018, 255, 1161–1177. [Google Scholar]
- Kim, H.S.; Shin, J.H.; Lee, H.S.; Kim, S.; Jang, H.Y.; Kim, E. CsRCI2D enhances high-temperature stress tolerance in Camelina sativa L. through endo-membrane trafficking from the plasma membrane. Plant Sci. 2022, 320, 111294. [Google Scholar] [CrossRef]
- Mitsuya, S.; Taniguchi, M.; Miyake, H.; Takabe, T. Disruption of RCI2A leads to over-accumulation of Na+ and increased salt sensitivity in Arabidopsis thaliana plants. Planta 2005, 222, 1001–1009. [Google Scholar] [CrossRef]
- Mitsuya, S.; Taniguchi, M.; Miyake, H.; Takabe, T. Overexpression of RCI2A decreases Na+ uptake and mitigates salinity-induced damages in Arabidopsis thaliana plants. Physiol. Plant 2006, 128, 95–102. [Google Scholar] [CrossRef]
- Liu, B.; Feng, D.; Zhang, B.; Mu, P.; Zhang, Y.; He, Y.; Qi, K.; Walng, J.; Wang, H. Musa paradisica RCI complements AtRCI and confers Na+ tolerance and K+ sensitivity in Arabidopsis. Plant Sci. 2012, 184, 102–111. [Google Scholar] [CrossRef]
- Yu, Y.; Cui, Y.; Ren, C.; Rocha, P.; Peng, M.; Xu, G.; Wang, M.; Xia, X. Transgenic rice expressing a cassava (Manihot esculenta Crantz) plasma membrane gene MePMP3-2 exhibits enhanced tolerance to salt and drought stresses. Genet. Mol. Res. 2016, 15, 15017336. [Google Scholar] [CrossRef]
- Kim, H.S.; Lee, J.E.; Jang, H.Y.; Jin, K.K.; Ahn, S.J. CsRCI2A and CsRCI2E genes show opposite salt sensitivity reaction due to membrane potential control. Acta Physiol. Plant. 2016, 38, 50. [Google Scholar] [CrossRef]
- Sivankalyani, V.; Geetha, M.; Subramanyam, K.; Girija, S. Ectopic expression of Arabidopsis RCI2A gene contributes to cold tolerance in tomato. Transgenic Res. 2015, 24, 237–251. [Google Scholar] [CrossRef]
- Long, R.; Zhang, F.; Li, Z.; Li, M.; Cong, L.; Kang, J.; Zhang, T.; Zhao, Z.; Sun, Y.; Yang, Q. Isolation and functional characterization of salt-stress induced RCI2-like genes from Medicago sativa and Medicago truncatula. J. Plant Res. 2015, 128, 697–707. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Zhang, H.; Zhang, L.; Xu, F.; Shi, L.; Wang, S.; Hong, J.; Ding, G. Identification and Comprehensive Analysis of the Nuclear Factor-Y Family Genes Reveal Their Multiple Roles in Response to Nutrient Deficiencies in Brassica napus. Int. J. Mol. Sci. 2021, 22, 10354. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Liu, X.; Wang, X.; Sun, M.; Song, R.; Mao, P.; Jia, S. Genome-Wide Identification of GRAS Gene Family and Their Responses to Abiotic Stress in Medicago sativa. Int. J. Mol. Sci. 2021, 22, 7729. [Google Scholar] [CrossRef]
- Smith, H. Signal perception, differential expression within multigene families and the molecular basis of phenotypic plasticity. Plant Cell Environ. 2010, 13, 585–594. [Google Scholar] [CrossRef]
- Zhao, Y.; Tong, H.; Cai, R.; Peng, X.; Li, X.; Gan, D.; Zhu, S. Identification and characterization of the RCI2 gene family in maize (Zea mays). J. Genet. 2014, 93, 655–666. [Google Scholar]
- Mohamad, H.; Sheikh, M.; Nematollah, E.; Mostafa, A.; Mohammad, P. Salt stress triggers augmented levels of Na+, K+ and ROS alters salt-related gene expression in leaves and roots of tall wheatgrass (Agropyron elongatum). Plant Physiol. Biochem. 2022, 183, 9–22. [Google Scholar] [CrossRef]
- Wang, B.; Zou, Q. Research progress of plasma membrane transporters and their relationship with plant salt tolerance. J. Bot. 2000, 17, 17–26. [Google Scholar]
- Nylander, M.; Heino, P.; Helenius, E.; Palva, T.E.; Ronne, H.; Welin, B.V. The low-temperature- and salt-induced RCI2A gene of Arabidopsis complements the sodium sensitivity caused by a deletion of the homologous yeast gene SNA1. Plant. Mol. Biol 2001, 45, 341–352. [Google Scholar] [CrossRef]
- Heidi, P.O.; Ana, G.; Verena, K.; Evrim, S.; Huang, S.; Fang, H.; Lang, V.; Sydow, K.; Pöckl, M.; Schulze, W.X.; et al. PH modulates interaction of 14-3-3 proteins with pollen plasma membrane H+-ATPases independently from phosphorylation. J. Exp. Bot. 2022, 73, 168–181. [Google Scholar]
- Zhou, S.; Wang, P.; Ding, Y.; Xie, L.; Li, A. Modification of plasma membrane H+-ATPase in Masson pine (Pinus massoniana Lamb.) seedling roots adapting to acid deposition. Tree Physiol. 2022, 7, 432–1449. [Google Scholar] [CrossRef]
- Gupta, A.; Shaw, B.P.; Roychoudhury, A. NHX1, HKT, and monovalent cation transporters regulate K+ and Na+ transport during abiotic stress. In Transporters and Plant Osmotic Stress; Academic Press: Cambridge, MA, USA, 2021; pp. 1–27. [Google Scholar]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant. 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Beard, K.M.; Boling, A.; Bargmann, B. Protoplast isolation, transient transformation, and flow-cytometric analysis of reporter-gene activation in Cannabis sativa L. Ind. Crops Prod. 2021, 164, 113360. [Google Scholar] [CrossRef]
- Sun, N.; Song, T.; Ma, Z.; Dong, L.; Zhan, L.; Xing, Y.; Liu, J.; Song, J.; Wang, S.; Cai, H. Overexpression of MsSiR enhances alkali tolerance in alfalfa (Medicago sativa L.) by increasing the glutathione content. Plant Physiol. Biochem. 2020, 154, 538–546. [Google Scholar] [CrossRef]
- Song, T.; Sun, N.; Dong, L.; Cai, H. Enhanced alkali tolerance of rhizobia-inoculated alfalfa correlates with altered proteins and metabolic processes as well as decreased oxidative damage. Plant Physiol. Biochem. 2021, 159, 301–311. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, D.; Zhang, Z.; Li, C.; Xing, Y.; Luo, Y.; Wang, X.; Li, D.; Ma, Z.; Cai, H. Overexpression of MsRCI2D and MsRCI2E Enhances Salt Tolerance in Alfalfa (Medicago sativa L.) by Stabilizing Antioxidant Activity and Regulating Ion Homeostasis. Int. J. Mol. Sci. 2022, 23, 9810. https://doi.org/10.3390/ijms23179810
Zhang D, Zhang Z, Li C, Xing Y, Luo Y, Wang X, Li D, Ma Z, Cai H. Overexpression of MsRCI2D and MsRCI2E Enhances Salt Tolerance in Alfalfa (Medicago sativa L.) by Stabilizing Antioxidant Activity and Regulating Ion Homeostasis. International Journal of Molecular Sciences. 2022; 23(17):9810. https://doi.org/10.3390/ijms23179810
Chicago/Turabian StyleZhang, Depeng, Zhenyue Zhang, Chunxin Li, Yimei Xing, Yaqin Luo, Xinsheng Wang, Donghuan Li, Zhiyun Ma, and Hua Cai. 2022. "Overexpression of MsRCI2D and MsRCI2E Enhances Salt Tolerance in Alfalfa (Medicago sativa L.) by Stabilizing Antioxidant Activity and Regulating Ion Homeostasis" International Journal of Molecular Sciences 23, no. 17: 9810. https://doi.org/10.3390/ijms23179810
APA StyleZhang, D., Zhang, Z., Li, C., Xing, Y., Luo, Y., Wang, X., Li, D., Ma, Z., & Cai, H. (2022). Overexpression of MsRCI2D and MsRCI2E Enhances Salt Tolerance in Alfalfa (Medicago sativa L.) by Stabilizing Antioxidant Activity and Regulating Ion Homeostasis. International Journal of Molecular Sciences, 23(17), 9810. https://doi.org/10.3390/ijms23179810





