Abstract
Small heat shock proteins (sHSPs) are ubiquitous ATP-independent chaperones that contribute to the maintenance of proteome integrity and functionality. Recent evidence suggests that sHSPs are ubiquitously expressed in numerous types of tumors and have been proposed to be implicated in oncogenesis and malignant progression. Heat shock protein family B member 2 (HSPB2) is a member of the sHSPs, which is found to be expressed, among others, in human breast cancer cell lines and constitutes an inhibitor of apical caspase activation in the extrinsic apoptotic pathway. In this study, we investigated the potential prognostic significance of HSPB2 mRNA expression levels in breast cancer, which represents the most frequent malignancy in females and one of the three most common cancer types worldwide. To this end, malignant breast tumors along with paired non-cancerous breast tissue specimens were used. HSPB2 expression levels were quantified in these two cohorts using a sensitive and accurate SYBR green-based quantitative real-time polymerase chain reaction (q-RT-PCR). Extensive biostatistical analyses were performed including Kaplan–Meier and Cox regression survival analyses for the assessment of the results. The significant downregulation of HSPB2 gene expression was revealed in breast tumors compared to their adjacent non-cancerous breast tissues. Notably, high HSPB2 mRNA expression predicts poor disease-free survival and overall survival of breast cancer patients. Multivariate Cox regression analysis revealed that HSPB2 mRNA overexpression is a significant predictor of poor prognosis in breast cancer, independent of other clinicopathological factors. In conclusion, high HSPB2 mRNA expression levels are associated with breast cancer patients’ relapse and poor survival.
1. Introduction
Breast cancer (BrCa) is the most frequent malignancy in females and one of the three most common cancers worldwide, followed by lung and colorectal cancer. In 2020, 2.3 million women were diagnosed with BrCa, and 685,000 deaths occurred globally [1,2]. Early-stage, non-metastatic BrCa, localized to the breast and local lymph nodes, is considered curable in ~70–80% of patients due to the improvements in multimodal therapy [3]. On the other hand, advanced BrCa with distant organ metastases (bones, lungs, liver, and brain), in addition to lymph nodes, is considered incurable by the currently available therapies, which aim to prolong the survival and maintain the patient’s quality of life [4,5].
BrCa is a complex heterogeneous disease classified into three major subtypes according to estrogen receptor (ER) or progesterone receptor (PR) expression and erb-b2 receptor tyrosine kinase 2 (ERBB2; formerly human epidermal growth factor 2, HER2) gene amplification. Notably, tumors expressing ER and/or PR are termed “hormone receptor-positive”, tumors expressing HER2 are called “HER2-positive”, and tumors lacking these three markers are considered “triple negative” [6,7]. Therapeutic approaches for BrCa differ according to the molecular subtype and generally include surgery, radiotherapy, chemotherapy, endocrine therapy, and/or targeted therapy [6,8].
Molecular chaperones, also known as heat shock proteins (HSPs) constitute a large family of molecular machines involved in the proper folding, unfolding, and assembly of polypeptides in order to maintain their structure and function [9,10]. In particular, HSPs curate the folding of nascent polypeptides into their native/functional configurations and prevent protein misfolding and aggregation [11,12,13]. They also target the misfolded or aggregated proteins for degradation, jointly with the protein quality control degradation machineries, i.e., the ubiquitin–proteasome system, and the autophagy–lysosome system, [14].
Several pieces of evidence suggest that HSPs are abnormally expressed in different types of cancer, including, among others, breast, colorectal, lung, prostate, pancreatic, bladder, and ovarian malignancies [15,16,17,18]. Recent advances in the field indicate that HSPs constitute potential biomarkers for cancer diagnosis and prognosis and are promising targets in cancer therapy [19,20].
Heat shock protein family B member 2 (HSPB2) is a member of the small HSPs (sHSPs), which consist of ubiquitous ATP-independent chaperones with low molecular weights [21,22,23]. HSPB2 is also named myotonic dystrophy protein kinase binding protein (MKBP), because it was found to bind to the myotonic dystrophy protein kinase (DMPK), thus increasing its activity and conferring thermal protection [24]. Although it is mainly expressed in cardiac and skeletal muscle [12,25], the HSPB2 gene was also reported to be expressed in human breast cancer cell lines and constitutes an inhibitor of apical caspase activation in the extrinsic apoptotic pathway [26]. However, the role of HSPB2 in breast tumorigenesis remains largely obscure. In the current study, we examined HSPB2 expression levels in BrCa tumors and matched adjacent normal tissue, and we also evaluated its potential association with patients’ relapse and overall survival.
2. Results
2.1. Downregulation of HSPB2 Gene Expression Levels during Oncogenic Transformation in Mammary Epithelial Cells
Firstly, we sought to examine whether the expression levels of the HSPB2 gene are affected during oncogenic transformation. To this end, we used a genetically defined model of stepwise carcinogenesis, in which human primary mammary epithelial cells (HMEC) were immortalized by the expression of the human telomerase catalytic subunit (hTERT) (HME cells) followed by p53/pRb inactivation due to concomitant simian virus 40 large T antigen (LT) expression (HMLE cells); HMLE cells were then transformed by the co-expression of the oncogene H-RasV12, resulting in the generation of breast metastatic malignant cells (HMLER cells) [27]. Our analysis showed that HSPB2 mRNA levels are significantly decreased in the HMLE and HMLER cell lines during oncogenesis as compared to normal HMEC cells (Figure 1); notably, this effect was maximized in HMLE cells suggesting a likely positive regulation of p53 and/or pRb in HSPB2 gene expression.
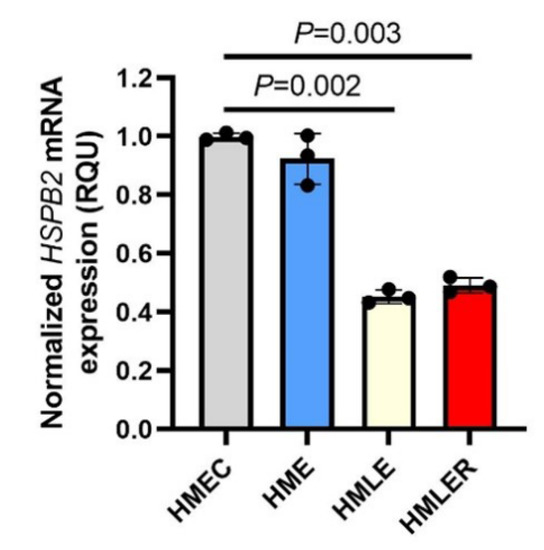
Figure 1.
Downregulation of the HSPB2 mRNA levels during the oncogenic transformation of mammary epithelial cells. Relative HSPB2 mRNA expression levels in the genetically defined model of stepwise carcinogenesis consisting of HMEC (normal), HME, HMLE, and HMLER cells. Control HMEC cells were set to 1. GAPDH gene expression was used as a reference for RNA input. p-values were calculated using an unpaired t-test.
2.2. Clinicopathological and Biological Characteristics of BrCa Patients and Used Samples
In the current study, two groups of samples were used, namely one group of 150 cancerous tissue specimens and another group of 16 paired non-cancerous tissue specimens from patients with primary BrCa. The cohort of patients consisted of 150 women, with a total median age of 60 years, ranging from 31 to 90 years, at the time of diagnosis. Moreover, concerning the histological grade and according to the World Health Organization (WHO) classification system, 8 patients were diagnosed with grade I (well-differentiated), 97 with grade II (moderately differentiated), and 45 with grade III malignant tumors (poorly differentiated). Furthermore, according to the TNM staging system, 43 malignant lesions were characterized as stage I (28.7%), 89 as stage II (59.3%), and 18 as stage III (12.0%). The clinical and biological traits of patients with BrCa included in the current study are shown in Table 1.

Table 1.
Clinicopathological features of BrCa patients.
2.3. Reduced Expression Levels of HSPB2 mRNA in Breast Carcinoma Tissues as Compared with Paired Non-Cancerous Tissues
The initial comparison of HSPB2 mRNA levels among 16 pairs of malignant breast tumors and their adjacent non-cancerous breast tissues revealed the downregulation of the HSPB2 mRNA expression in the majority of malignant breast tumors (p < 0.001) (Figure 2). Specifically, the mean HSPB2 mRNA expression levels were equal to 2215.7 RQU in tumor samples with a standard error of 265.8, while in non-cancerous samples the mean HSPB2 mRNA expression was 11,234.5 RQU with a standard error of 3094.54 (Table 2). These findings corroborated our data in mammary epithelial cells suggesting that the HSPB2 gene is most likely downregulated in malignant tumors.
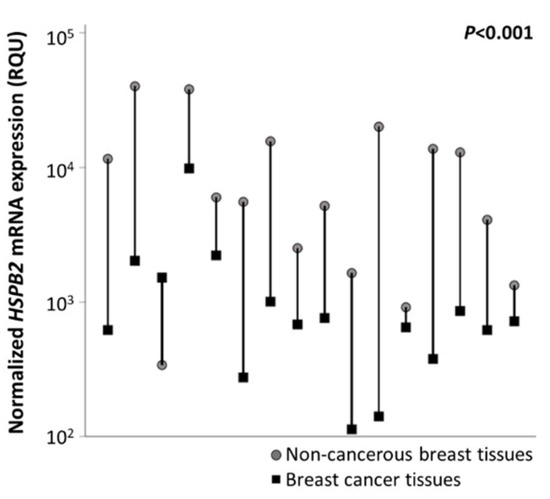
Figure 2.
Graphical illustration of HSPB2 mRNA expression levels in cancerous vs. non-cancerous tissues, after comparing 16 pairs of tissue specimens. The HSPB2 mRNA expression levels were downregulated, as compared to normal adjacent tissues, in almost all tumors. p-value was calculated by the Wilcoxon signed-rank test.

Table 2.
Distributions of HSPB2 mRNA expression levels in cancerous and non-cancerous tissue samples.
To classify the HSPB2 mRNA expression status in each tissue specimen as positive or negative, an optimal cut-off value was set, as described in the “Materials and Methods” section. We found that 75 (50%) samples were classified as HSPB2 mRNA-positive, when the relative HSPB2 mRNA expression was equal to or higher than 656.5 RQU and 75 (50%) samples as HSPB2 mRNA-negative (relative HSPB2 mRNA expression lower than 656.5 RQU). We then investigated the potential association of HSPB2 mRNA expression status with patients’ clinicopathological parameters including molecular subtype, anatomic stage, mitotic rate, and HER2, ER, and PR status. We found no significant association between HSPB2 gene expression levels and the mentioned clinicopathological parameters (not shown).
2.4. HSPB2 mRNA Overexpression Is a Reliable Predictor of Poor Prognosis in BrCa Patients, Independent of Other Clinicopathological Factors
The independent significance of HSPB2 mRNA expression regarding patients’ relapse was revealed by the univariate Cox regression analysis (Table 3). Specifically, we found that BrCa patients with an HSPB2-positive expression status entailed a 2.29-fold higher risk of tumor recurrence as compared to HSPB2-negative ones (HR = 2.29, 95% CI = 1.19–4.42, p = 0.014). Kaplan–Meier curves revealed that HSPB2-positive BrCa patients have a significantly lower DFS compared to those who are HSPB2-negative (p = 0.011) (Figure 3A). The significance of the HSPB2 mRNA expression status in the prognosis of patients’ DFS was also maintained in the multivariate Cox regression analysis (HR = 2.61, 95% CI = 1.34–5.08, p = 0.005). Consequently, high HSPB2 mRNA expression levels represent an independent prognostic indicator for tumor recurrence in BrCa patients.

Table 3.
HSPB2 mRNA expression status and disease-free survival (DFS) of BrCa patients.
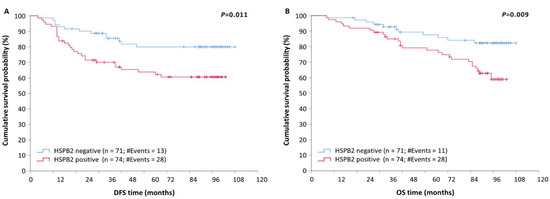
Figure 3.
Kaplan–Meier survival curves for disease-free survival (DFS) and overall survival (OS) of BrCa patients. (A) Patients with tumors being positive for HSPB2 mRNA expression had shorter DFS time intervals than patients with HSPB2-negative tumors (p = 0.011). (B) Patients with HSPB2-positive tumors had shorter OS than those with HSPB2-negative tumors (p < 0.009). p-value was calculated using the log-rank test.
Then, we sought to examine the potential prognostic significance of HSPB2 mRNA expression status for patients’ overall survival (OS). To this end, a univariable Cox regression analysis was performed and revealed that high HSPB2 mRNA expression predicts poor OS for BrCa patients. Specifically, BrCa patients bearing HSPB2-positive tumors had a significantly shorter OS time interval than those with lower HSPB2 mRNA levels (HR = 2.45, 95% CI = 1.23–4.96, p = 0.011). Furthermore, the molecular subtype and prognostic stage were found to be significant prognosticators of OS (Table 4).

Table 4.
HSPB2 mRNA expression status and overall survival (OS) of BrCa patients.
The prognostic value of HSPB2-positive mRNA expression status in BrCa patients was also depicted by the Kaplan–Meier curves (p = 0.009) (Figure 3B). Additionally, multivariable Cox regression analysis revealed that HSPB2 mRNA expression status remained a statistically significant factor of poor OS in BrCa, independent of molecular subtype and prognostic stage (HR = 2.69, 95% CI = 1.33–5.42, p = 0.006). Therefore, the HSPB2-positive mRNA expression status in BrCa could be considered a novel independent indicator of poor OS.
2.5. Prognostic Value of HSPB2 mRNA Expression in Patients with Breast Adenocarcinoma, Stratified According to Molecular Subtype, Anatomic Stage, or Prognostic Stage
Determination of tumor characteristics such as molecular subtype and tumor grade is crucial in the prognosis of patients diagnosed with BrCa [28]. Thus, patients were stratified according to these variables in order to evaluate the potential additional impact of HSPB2 mRNA expression status. The stratification according to molecular subtype revealed that patients with triple-negative or HER2-positive tumors had significantly lower DFS (p < 0.001) and OS (p < 0.001) rates (Figure S1) compared to those with luminal A or luminal B tumors, regardless of the HSPB2 expression status. Moreover, as depicted in Figure 4A, triple-negative patients positive for HSPB2 mRNA expression showed an increased probability of poorer OS, as compared to those bearing HSPB2-negative tumors (p = 0.026).
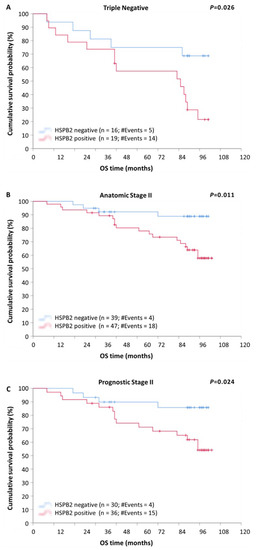
Figure 4.
Stratified Kaplan–Meier survival curves for overall survival (OS) of BrCa patients, according to molecular subtype, anatomic, and prognostic stage. (A) Patients with triple-negative tumors being positive for HSPB2 mRNA expression had shorter OS time intervals than patients with HSPB2-negative tumors. (B) Patients with HSPB2-positive tumors of anatomic stage II had poorer OS than those with HSPB2-negative tumors. (C) Patients with HSPB2-positive tumors of prognostic stage II showed an increased probability of poorer OS compared to those with HSPB2-negative tumors. p-value was calculated using the log-rank test.
Additionally, after stratification, according to the anatomic stage, patients with tumors of anatomic stage III were found to have a remarkably shorter DFS (p < 0.001) and OS (p < 0.001), compared to those with tumors of anatomic stage I or II, regardless of the HSPB2 expression status (Figure S2). Furthermore, as illustrated in Kaplan–Meier curves, patients of anatomic stage II with HSPB2-positive tumors had remarkably lower DFS (Figure 5A; p = 0.006) and OS (Figure 4B; p = 0.011) rates, in comparison with those with HSPB2-negative tumors.
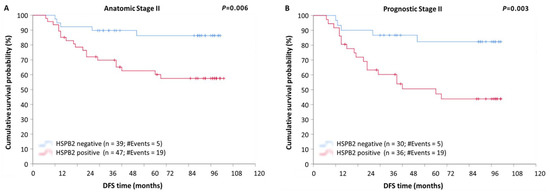
Figure 5.
Stratified Kaplan–Meier survival curves for disease-free survival (DFS) of BrCa patients, according to anatomic and prognostic stage. (A) Patients with HSPB2-positive tumors of anatomic stage II had significantly poorer DFS than those with HSPB2-negative tumors. (B) Patients with HSPB2-positive tumors of prognostic stage II had shorter DFS time intervals than those with HSPB2-negative tumors. p-value was calculated using the log-rank test.
The stratification of patients according to prognostic stage revealed that patients of prognostic stage III had significantly shorter DFS (p < 0.001) and OS (p < 0.001) intervals, as compared to patients of stage I or II tumors, regardless of the HSPB2 expression status (Figure S3). Furthermore, patients of prognostic stage II with HSPB2-positive tumors showed an increased probability of a poorer DFS (Figure 5B; p = 0.003) and OS (Figure 4C; p = 0.024), as compared to those with HSPB2-negative tumors.
3. Discussion
The network of molecular chaperones, also referred to as HSPs, plays a central role in the maintenance of proteome integrity and functionality (collectively referred to as proteostasis), and is of the utmost importance for cell homeodynamics and survival [29,30]. HSPs are involved in numerous cellular functions including protein degradation, stress tolerance, cell signaling, cell differentiation, and apoptosis [31,32,33]. Under stress conditions, cells induce the expression of HSPs, thus activating the heat shock response (HSR) [9,16,34]. Notably, sustained activation of HSR is often observed in cancer cells, as they experience increased levels of proteotoxic stress, which has been proposed as a stress hallmark of cancer [16,35,36].
BrCa is a highly heterogeneous disease characterized by not only various phenotypes and molecular subgroups, but also by different responses to treatment [37]. Depending on the tumor subtype, different kinds of therapies are applied, such as endocrine therapy for hormone receptor-positive disease [38] or anti-HER2 therapy in HER2-positive cases [39]. Despite significant medical achievements in its diagnosis, the biomarkers that are used in clinical practice today lack sensitivity and specificity [40,41]. Therefore, it is crucial to find novel non-invasive biomarkers that could ameliorate the estimation of patients’ recurrence and survival.
Over the last decades, growing evidence has shown that sHSPs expression is frequently deregulated in diverse cancer types and is proposed to profoundly impact malignant progression. In particular, sHSPs have been associated with several hallmarks of cancer, including tumorigenesis, cell growth, the evasion of apoptosis, immune surveillance, angiogenesis, metastasis, and chemoresistance [19,20,25,36,42,43,44].
HSPB2 expression is particularly elevated (despite ubiquitous expression) in cardiac and skeletal muscle [12,25] and was also found to be expressed in human BrCa cell lines [26]. Considering that BrCa is one of the most frequent malignancies, accounting for 11.7% of the total number of new cases diagnosed in 2020 [1,2] and that the role of HSPB2 in breast tumorigenesis or cancer progression has not yet been investigated, our study was focused on the potential prognostic significance of HSPB2 mRNA expression levels in BrCa patients.
Despite the fact that many sHSPs are overexpressed in a wide range of solid tumors [45], we observed that HSPB2 gene transcription is predominantly downregulated in malignant breast tumors compared to their adjacent non-cancerous breast tissues. In line with our findings, a recent comprehensive transcriptomic study of the HSP gene family in BrCa patients from both The Cancer Genome Atlas (TCGA) and the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) cohorts revealed that HSPB2 gene expression levels are profoundly downregulated in all BrCa molecular subtypes as compared to normal breast tissues [46]. Salhia et al. have also demonstrated the underexpression of HSPB2 in BrCa samples, due to HSPB2 gene deletion [47]. Other studies support the downregulation of HSPB2 in cancer, as well; specifically, HSPB2 mRNA was lower in esophageal squamous cell carcinoma cell lines, due to hypermethylation of the promoter of the HSPB2 gene [48], while HSPB2 mRNA was also found to be barely expressed in pancreatic cancer [49]. Since all the aforementioned studies note the lower mRNA expression of HSPB2, whereas most studies stating the overexpression of HSPs have examined their protein levels, a possible explanation is that HSPB2 protein expression levels do not perfectly correlate with HSPB2 mRNA levels. Indeed, as shown by transcriptomics and proteomics data of HSPB2 expression in various BrCa cell lines, deposited to the Expression Atlas database [50], the lower mRNA levels are not reflected by lower protein expression levels of this molecule.
Multivariate Cox regression analysis revealed that HSPB2 mRNA overexpression is a significant predictor of poor prognosis in BrCa, independent of other clinicopathological factors, including molecular subtype and prognostic stage. Additionally, Kaplan–Meier survival curves revealed that patients with HSPB2-positive tumors were more likely to have poor outcomes, such as relapse and death. Consistently, the elevated levels of diverse HSPs expression in specific malignancies have been associated with a poor prognosis and an increased resistance to therapies [44,45,51,52,53].
Heat shock factor 1 (HSF1) constitutes the most robust regulator of HSPs expression to maintain (among others) proteome stability [54]. A growing number of studies support that HSF1 is implicated in the initiation, promotion, and progression of cancer and is widely exploited as a potential therapeutic target in a broad spectrum of malignancies [16,55]. In line with our findings, increased expression levels of HSF1 were found to be associated with a poor prognosis in BrCa patients [16,56,57].
Furthermore, although the role of HSPB2 has not been investigated so widely as the role of other HSPs, it was found to confer resistance to apoptosis in human BrCa cell lines, as it inhibited the apical caspase activation in the extrinsic apoptotic pathway [26], as well as to inhibit pancreatic cancer cell proliferation via activating targets of the TP53 signaling pathway, such as the RPRM, ADGRB1, and STEAP3 genes [49]. In addition, it has recently been hypothesized that the inhibition of HSPB2 by miR-17-5p promotes cell proliferation, migration, and invasion of colon cancer cell lines [58]. The constitutive activation of HSPs in various malignancies is proposed to confer a survival advantage to cancer cells [59,60]. Therefore, HSPs could exert oncogenic functions and mediate the “non-oncogene addiction” of cancer cells being crucial for tumor development and survival [36,59,60,61,62].
The major implication of HSPs in cell transformation and tumor progression largely supports the notion that HSPs-targeting drugs could constitute a promising approach in cancer therapy [52,60,63,64]. Notably, HER2 is a protein client of the heat shock protein 90 (HSP90) and thus, HSP90 inhibitors have already been used in clinical trials in HER2 positive BrCa patients [65,66]. Additionally, trastuzumab (anti-HER2 monoclonal antibody) resistance, which is associated with a poorer prognosis, was attenuated in HER2-positive BrCa through HSP90 inhibition [58,67] and could serve as an adverse, independent prognostic biomarker for this malignancy. Nevertheless, our study is characterized by some limitations that need to be addressed. Firstly, our cohort size is of a medium size, and the number of non-cancerous breast tissue specimens is rather small. In addition, the patients’ cohort was not equivalently stratified in the defined subgroups, which could diminish the obtained findings. Future studies should be conducted to further evaluate the role of HSPB2 in BrCa prognosis.
Molecular biomarkers represent biological molecules used to infer disease risk, diagnosis, prognosis, and therapeutic response [68]. It is evident that the identification of accurate and precise non-invasive molecular biomarkers constitutes the main tool for paving the way toward “individualized biomarker-driven cancer therapy” or otherwise “precision medicine”, so that optimal treatment decisions can be made [69,70]. Consequently, HSPB2 mRNA expression status could be combined with other well-validated clinical biomarkers, which could tailor the therapeutic options aiming to improve the outcome of cancer therapy and a patient’s overall survival, while minimizing the associated risk.
4. Materials and Methods
4.1. Cell Lines and Cell Culture Conditions
Human mammary epithelial cells HMEC were obtained from Lonza Group AG (Basel, Switzerland), while the human mammary adenocarcinoma cell line MCF-7 was obtained from the American Tissue Culture Collection (Manassas, VA, USA). The HMEC-derived cell lines HME, HMLE, and HMLER were a kind gift by Prof. Robert A. Weinberg (Massachusetts Institute of Technology, Cambridge, MA, USA). HMEC, HME, HMLE, and HMLER cells were grown in MEGM™ Mammary Epithelial Cell Growth Medium BulletKit™ (Lonza Group AG, #CC-3150). MCF-7 cells were cultured in DMEM (Thermo Fisher Scientific Inc., Waltham, MA, USA) supplemented with 2 g/L glucose, 10% FBS, and 1% penicillin/streptomycin. All cell lines were maintained in a humidified incubator at 5% CO2, 95% humidity, and 37 °C.
4.2. Patients and Tissue Collection
One hundred and fifty (150) BrCa samples and 16 paired, non-cancerous tissue samples were collected from patients with primary BrCa, subjected to surgery at the “Saint Savvas” Cancer Hospital of Athens, Athens, Greece. The biological and clinicopathological data collected included the age of patients, the dimensions of the resected tumor, the infiltration of regional lymph nodes, the presence of distant metastasis, the histological and intrinsic (molecular) subtypes, as well as the histological grade of the tumor, the expression status of PR, ER, HER2, and the mitotic rate based on the Ki-67 index. All tumors were independently characterized by two pathologists. The anatomic (TNM) and prognostic stages were determined, based on these data. All breast tissue specimens were stored in liquid nitrogen immediately after surgery. Survival data were available for 145 out of 150 patients, and the median follow-up time was 97 months.
The study was conducted in compliance with the 1964 Declaration of Helsinki and its later amendments and was approved by the institutional Ethics Committee of the “Saint Savvas” Cancer Hospital of Athens, Athens, Greece. Written informed consent was obtained from all patients.
4.3. Total RNA Extraction and First-Strand cDNA Synthesis
Total RNA was extracted from each sample using the TRI reagent® (Molecular Research Center, Inc., Cincinnati, OH, USA), following the manufacturer’s protocol instructions. Spectrophotometric evaluation of the concentration and purity of the isolated RNA samples was conducted using a BioSpec-nano Micro-volume UV–Vis Spectrophotometer (Shimadzu, Kyoto, Japan). Two micrograms μg of total RNA were subjected to reverse transcription, using M-MLV reverse transcriptase (Life Technologies Ltd., Carlsbad, CA, USA) and oligo-dT 5 μM, according to the manufacturer’s instructions, to obtain first-strand cDNA.
4.4. Quantitative Real-Time PCR (qPCR)
A real-time qPCR assay was developed and carried out, using the SYBR Green Chemistry, in an ABI 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). Specific primers were designed for HSPB2; moreover, specific primer pairs were designed for housekeeping genes encoding glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and hydroxymethylbilane synthase (HMBS), which were chosen among others as reference genes, according to supporting literature data [71] and our preliminary results in a random sample of our cohort of BrCa specimens. The sequences of HSPB2 primers were 5′-CCGAGTACGAATTTGCCAACC-3′ and 5′-AGGCCGGACATAGTAGCCAT-3′. Those of GAPDH were 5′-GTCAAGGCTGAGAACGGGAA-3′ and 5′-TCGCCCCACTTGATTTTGGA-3′, and those of HMBS were 5′-AAGAGACCATGCAGGCTACCA-3′ and 5′-ACAAGTTGGCCAGGCTGATG-3′.
The qPCR mixture contained KAPA SYBR FAST qPCR Master Mix Universal, supplemented with ROX as a passive reference dye, forward and reverse primers at a final concentration of 200 nM, 2.5 μL DEPC-treated H2O, and 0.5 μL of 10-fold diluted cDNA. We optimized the qPCR assays by standardizing the primer concentration and the thermal protocol, so as to observe a unique melting curve for each assay. The uniqueness and specificity of each amplicon were also assessed by agarose gel electrophoresis. The cDNA derived from the MCF-7 cells was used as a calibrator to render “−∆Ct” values calculated in distinct qPCR runs comparable. The comparative Ct method (2−∆∆Ct) was applied for relative quantification [72,73]. The normalized HSPB2 expression of each sample was defined as the ratio of HSPB2 molecules to GAPDH and HMBS molecules, divided by the same ratio that was calculated for the calibrator (MCF-7 cells) and was determined in relative quantification units (RQU).
4.5. Biostatistical Analysis
The distribution in the patients’ cohort was not normal; thus, non-parametric tests were used. Firstly, descriptive statistics were performed. The Wilcoxon signed-rank test was carried out to assess the difference in HSPB2 mRNA expression between paired samples. Next, the BrCa patients were categorized into two distinct groups, namely HSPB2-positive and HSPB2-negative patients; this categorization was done based on the median value of HSPB2 mRNA expression (656.5 RQU). More specifically, patients with an HSPB2 mRNA expression value higher than 656.5 RQU were characterized as HSPB2-positive, whereas those with an HSPB2 mRNA expression value lower than 656.5 RQU were characterized as HSPB2-negative. Potential associations between the expression of HSPB2 mRNA and other categorical clinicopathological variables were investigated, using a two-tailed χ2-test.
Kaplan–Meier survival analysis was then applied, with regard to disease-free survival (DFS) and overall survival (OS). Differences between Kaplan–Meier curves were assessed using the log-rank (Mantel–Cox) test. Bootstrap (1000 random samples) univariate and multivariable Cox regression analyses were carried out, concerning also DFS and OS; further, the bias-corrected and accelerated (BCa) 95% confidence interval (CI) of each hazard ratio (HR) was estimated. Next, Kaplan–Meier survival analysis was performed in subgroups of the cohort, in which patients were stratified according to specific clinicopathological characteristics. Each outcome was considered statistically significant if the p-value < 0.050.
5. Conclusions
Our findings suggest that the HSPB2 molecular chaperone gene transcription is predominantly downregulated in BrCa, whereas increased HSPB2 gene expression levels are associated with patients’ relapse and poor survival. Consequently, HSPB2 mRNA expression status could be potentially used to assess the clinical prognosis of patients with this frequent malignancy, independent of other clinicopathological factors.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/ijms23179758/s1.
Author Contributions
Conceptualization, design, and ervision of the study, C.K.K., A.S. and I.P.T.; Methodology, C.K.K.; Investigation, A.D.S., D.D.G., C.C., G.P. and P.K.; Data curation, Formal analysis, P.K.; Validation, C.K.K.; Visualization, A.D.S. and P.K.; Resources, G.P., C.K.K., A.S. and I.P.T.; Writing—Original Draft Preparation, A.D.S.; Writing—Review and Editing, D.D.G., P.K., C.K.K., A.S. and I.P.T.; Funding Acquisition, C.K.K., A.S. and I.P.T.; Project administration, A.S. and I.P.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the institutional Ethics Committee of the “Saint Savvas” Cancer Hospital of Athens, Athens, Greece (approval number: 29; date of approval: 15 March 2006).
Informed Consent Statement
Written informed consent was obtained from all patients.
Data Availability Statement
The data presented in this study are available on reasonable request from the corresponding authors.
Acknowledgments
The authors would like to sincerely thank Robert A. Weinberg (Massachusetts Institute of Technology, Cambridge, MA, USA) for donating the HMEC-derived cell lines HME, HMLE and HMLER.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Parkin, D.M.; Pineros, M.; Znaor, A.; Bray, F. Cancer statistics for the year 2020: An overview. Int. J. Cancer 2021, 149, 778–789. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Lukasiewicz, S.; Czeczelewski, M.; Forma, A.; Baj, J.; Sitarz, R.; Stanislawek, A. Breast Cancer-Epidemiology, Risk Factors, Classification, Prognostic Markers, and Current Treatment Strategies-An Updated Review. Cancers 2021, 13, 4287. [Google Scholar] [CrossRef] [PubMed]
- Harbeck, N.; Penault-Llorca, F.; Cortes, J.; Gnant, M.; Houssami, N.; Poortmans, P.; Ruddy, K.; Tsang, J.; Cardoso, F. Breast cancer. Nat. Rev. Dis. Prim. 2019, 5, 66. [Google Scholar] [CrossRef] [PubMed]
- Riggio, A.I.; Varley, K.E.; Welm, A.L. The lingering mysteries of metastatic recurrence in breast cancer. Br. J. Cancer 2021, 124, 13–26. [Google Scholar] [CrossRef]
- Harbeck, N.; Gnant, M. Breast cancer. Lancet 2017, 389, 1134–1150. [Google Scholar] [CrossRef]
- Waks, A.G.; Winer, E.P. Breast Cancer Treatment: A Review. JAMA 2019, 321, 288–300. [Google Scholar] [CrossRef]
- Loibl, S.; Poortmans, P.; Morrow, M.; Denkert, C.; Curigliano, G. Breast cancer. Lancet 2021, 397, 1750–1769. [Google Scholar] [CrossRef]
- Jolly, C.; Morimoto, R.I. Role of the heat shock response and molecular chaperones in oncogenesis and cell death. J. Natl. Cancer Inst. 2000, 92, 1564–1572. [Google Scholar] [CrossRef]
- Niforou, K.; Cheimonidou, C.; Trougakos, I.P. Molecular chaperones and proteostasis regulation during redox imbalance. Redox Biol. 2014, 2, 323–332. [Google Scholar] [CrossRef] [Green Version]
- Freilich, R.; Arhar, T.; Abrams, J.L.; Gestwicki, J.E. Protein-Protein Interactions in the Molecular Chaperone Network. Acc Chem. Res. 2018, 51, 940–949. [Google Scholar] [CrossRef] [PubMed]
- Tedesco, B.; Cristofani, R.; Ferrari, V.; Cozzi, M.; Rusmini, P.; Casarotto, E.; Chierichetti, M.; Mina, F.; Galbiati, M.; Piccolella, M.; et al. Insights on Human Small Heat Shock Proteins and Their Alterations in Diseases. Front. Mol. Biosci. 2022, 9, 842149. [Google Scholar] [CrossRef] [PubMed]
- Treweek, T.M.; Meehan, S.; Ecroyd, H.; Carver, J.A. Small heat-shock proteins: Important players in regulating cellular proteostasis. Cell. Mol. Life Sci. 2015, 72, 429–451. [Google Scholar] [CrossRef] [PubMed]
- Sala, A.J.; Bott, L.C.; Morimoto, R.I. Shaping proteostasis at the cellular, tissue, and organismal level. J. Cell Biol. 2017, 216, 1231–1241. [Google Scholar] [CrossRef]
- Artemaki, P.I.; Sklirou, A.D.; Kontos, C.K.; Liosi, A.A.; Gianniou, D.D.; Papadopoulos, I.N.; Trougakos, I.P.; Scorilas, A. High clusterin (CLU) mRNA expression levels in tumors of colorectal cancer patients predict a poor prognostic outcome. Clin. Biochem. 2020, 75, 62–69. [Google Scholar] [CrossRef]
- Cyran, A.M.; Zhitkovich, A. Heat Shock Proteins and HSF1 in Cancer. Front. Oncol. 2022, 12, 860320. [Google Scholar] [CrossRef]
- Kalioraki, M.A.; Artemaki, P.I.; Sklirou, A.D.; Kontos, C.K.; Adamopoulos, P.G.; Papadopoulos, I.N.; Trougakos, I.P.; Scorilas, A. Heat shock protein beta 3 (HSPB3) is an unfavorable molecular biomarker in colorectal adenocarcinoma. Mol. Carcinog. 2020, 59, 116–125. [Google Scholar] [CrossRef]
- Sheng, B.; Qi, C.; Liu, B.; Lin, Y.; Fu, T.; Zeng, Q. Increased HSP27 correlates with malignant biological behavior of non-small cell lung cancer and predicts patient’s survival. Sci. Rep. 2017, 7, 13807. [Google Scholar] [CrossRef]
- Yun, C.W.; Kim, H.J.; Lim, J.H.; Lee, S.H. Heat Shock Proteins: Agents of Cancer Development and Therapeutic Targets in Anti-Cancer Therapy. Cells 2019, 9, 60. [Google Scholar] [CrossRef]
- Zhang, Z.; Jing, J.; Ye, Y.; Chen, Z.; Jing, Y.; Li, S.; Hong, W.; Ruan, H.; Liu, Y.; Hu, Q.; et al. Characterization of the dual functional effects of heat shock proteins (HSPs) in cancer hallmarks to aid development of HSP inhibitors. Genome Med. 2020, 12, 101. [Google Scholar] [CrossRef]
- Bakthisaran, R.; Tangirala, R.; Rao Ch, M. Small heat shock proteins: Role in cellular functions and pathology. Biochim. Biophys. Acta 2015, 1854, 291–319. [Google Scholar] [CrossRef]
- Janowska, M.K.; Baughman, H.E.R.; Woods, C.N.; Klevit, R.E. Mechanisms of Small Heat Shock Proteins. Cold Spring Harb. Perspect Biol. 2019, 11, a034025. [Google Scholar] [CrossRef] [PubMed]
- Webster, J.M.; Darling, A.L.; Uversky, V.N.; Blair, L.J. Small Heat Shock Proteins, Big Impact on Protein Aggregation in Neurodegenerative Disease. Front. Pharmacol. 2019, 10, 1047. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, A.; Sugiyama, Y.; Hayashi, Y.; Nyu-i, N.; Yoshida, M.; Nonaka, I.; Ishiura, S.; Arahata, K.; Ohno, S. MKBP, a novel member of the small heat shock protein family, binds and activates the myotonic dystrophy protein kinase. J. Cell Biol. 1998, 140, 1113–1124. [Google Scholar] [CrossRef] [PubMed]
- Xiong, J.; Li, Y.; Tan, X.; Fu, L. Small Heat Shock Proteins in Cancers: Functions and Therapeutic Potential for Cancer Therapy. Int. J. Mol. Sci. 2020, 21, 6611. [Google Scholar] [CrossRef]
- Oshita, S.E.; Chen, F.; Kwan, T.; Yehiely, F.; Cryns, V.L. The small heat shock protein HspB2 is a novel anti-apoptotic protein that inhibits apical caspase activation in the extrinsic apoptotic pathway. Breast Cancer Res. Treat. 2010, 124, 307–315. [Google Scholar] [CrossRef]
- Elenbaas, B.; Spirio, L.; Koerner, F.; Fleming, M.D.; Zimonjic, D.B.; Donaher, J.L.; Popescu, N.C.; Hahn, W.C.; Weinberg, R.A. Human breast cancer cells generated by oncogenic transformation of primary mammary epithelial cells. Genes Dev. 2001, 15, 50–65. [Google Scholar] [CrossRef]
- Colzani, E.; Liljegren, A.; Johansson, A.L.; Adolfsson, J.; Hellborg, H.; Hall, P.F.; Czene, K. Prognosis of patients with breast cancer: Causes of death and effects of time since diagnosis, age, and tumor characteristics. J. Clin. Oncol. 2011, 29, 4014–4021. [Google Scholar] [CrossRef]
- Hartl, F.U.; Bracher, A.; Hayer-Hartl, M. Molecular chaperones in protein folding and proteostasis. Nature 2011, 475, 324–332. [Google Scholar] [CrossRef]
- Kim, Y.E.; Hipp, M.S.; Bracher, A.; Hayer-Hartl, M.; Hartl, F.U. Molecular chaperone functions in protein folding and proteostasis. Annu. Rev. Biochem. 2013, 82, 323–355. [Google Scholar] [CrossRef]
- Bakthisaran, R.; Akula, K.K.; Tangirala, R.; Rao Ch, M. Phosphorylation of alphaB-crystallin: Role in stress, aging and patho-physiological conditions. Biochim. Biophys. Acta 2016, 1860, 167–182. [Google Scholar] [CrossRef] [PubMed]
- Carra, S.; Alberti, S.; Arrigo, P.A.; Benesch, J.L.; Benjamin, I.J.; Boelens, W.; Bartelt-Kirbach, B.; Brundel, B.; Buchner, J.; Bukau, B.; et al. The growing world of small heat shock proteins: From structure to functions. Cell Stress Chaperones 2017, 22, 601–611. [Google Scholar] [CrossRef] [PubMed]
- Shevtsov, M.; Balogi, Z.; Khachatryan, W.; Gao, H.; Vigh, L.; Multhoff, G. Membrane-Associated Heat Shock Proteins in Oncology: From Basic Research to New Theranostic Targets. Cells 2020, 9, 1263. [Google Scholar] [CrossRef] [PubMed]
- Richter, K.; Haslbeck, M.; Buchner, J. The heat shock response: Life on the verge of death. Mol. Cell. 2010, 40, 253–266. [Google Scholar] [CrossRef]
- Luo, J.; Solimini, N.L.; Elledge, S.J. Principles of cancer therapy: Oncogene and non-oncogene addiction. Cell 2009, 136, 823–837. [Google Scholar] [CrossRef]
- Sklirou, A.; Papanagnou, E.D.; Fokialakis, N.; Trougakos, I.P. Cancer chemoprevention via activation of proteostatic modules. Cancer Lett. 2018, 413, 110–121. [Google Scholar] [CrossRef]
- Testa, U.; Castelli, G.; Pelosi, E. Breast Cancer: A Molecularly Heterogenous Disease Needing Subtype-Specific Treatments. Med. Sci. 2020, 8, 18. [Google Scholar] [CrossRef]
- Rugo, H.S.; Rumble, R.B.; Macrae, E.; Barton, D.L.; Connolly, H.K.; Dickler, M.N.; Fallowfield, L.; Fowble, B.; Ingle, J.N.; Jahanzeb, M.; et al. Endocrine Therapy for Hormone Receptor-Positive Metastatic Breast Cancer: American Society of Clinical Oncology Guideline. J. Clin. Oncol. 2016, 34, 3069–3103. [Google Scholar] [CrossRef]
- Wang, J.; Xu, B. Targeted therapeutic options and future perspectives for HER2-positive breast cancer. Signal Transduct. Target. Ther. 2019, 4, 34. [Google Scholar] [CrossRef]
- Afzal, S.; Hassan, M.; Ullah, S.; Abbas, H.; Tawakkal, F.; Khan, M.A. Breast Cancer; Discovery of Novel Diagnostic Biomarkers, Drug Resistance, and Therapeutic Implications. Front. Mol. Biosci. 2022, 9, 783450. [Google Scholar] [CrossRef]
- Wu, H.J.; Chu, P.Y. Recent Discoveries of Macromolecule- and Cell-Based Biomarkers and Therapeutic Implications in Breast Cancer. Int. J. Mol. Sci. 2021, 22, 636. [Google Scholar] [CrossRef] [PubMed]
- Caruso Bavisotto, C.; Marino Gammazza, A.; Campanella, C.; Bucchieri, F.; Cappello, F. Extracellular heat shock proteins in cancer: From early diagnosis to new therapeutic approach. Semin. Cancer Biol. 2021; accepted. [Google Scholar] [CrossRef] [PubMed]
- Lianos, G.D.; Alexiou, G.A.; Mangano, A.; Mangano, A.; Rausei, S.; Boni, L.; Dionigi, G.; Roukos, D.H. The role of heat shock proteins in cancer. Cancer Lett. 2015, 360, 114–118. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Xiao, H.; Cao, L. Recent advances in heat shock proteins in cancer diagnosis, prognosis, metabolism and treatment. Biomed. Pharmacother. 2021, 142, 112074. [Google Scholar] [CrossRef]
- Ciocca, D.R.; Calderwood, S.K. Heat shock proteins in cancer: Diagnostic, prognostic, predictive, and treatment implications. Cell Stress Chaperones 2005, 10, 86–103. [Google Scholar] [CrossRef]
- Zoppino, F.C.M.; Guerrero-Gimenez, M.E.; Castro, G.N.; Ciocca, D.R. Comprehensive transcriptomic analysis of heat shock proteins in the molecular subtypes of human breast cancer. BMC Cancer 2018, 18, 700. [Google Scholar] [CrossRef] [PubMed]
- Salhia, B.; Kiefer, J.; Ross, J.T.; Metapally, R.; Martinez, R.A.; Johnson, K.N.; DiPerna, D.M.; Paquette, K.M.; Jung, S.; Nasser, S.; et al. Integrated genomic and epigenomic analysis of breast cancer brain metastasis. PLoS ONE 2014, 9, e85448. [Google Scholar] [CrossRef]
- Chang, X.; Yamashita, K.; Sidransky, D.; Kim, M.S. Promoter methylation of heat shock protein B2 in human esophageal squamous cell carcinoma. Int. J. Oncol. 2011, 38, 1129–1135. [Google Scholar] [CrossRef]
- Yu, Z.; Wang, H.; Fang, Y.; Lu, L.; Li, M.; Yan, B.; Nie, Y.; Teng, C. Molecular chaperone HspB2 inhibited pancreatic cancer cell proliferation via activating p53 downstream gene RPRM, BAI1, and TSAP6. J. Cell Biochem. 2020, 121, 2318–2329. [Google Scholar] [CrossRef]
- Papatheodorou, I.; Fonseca, N.A.; Keays, M.; Tang, Y.A.; Barrera, E.; Bazant, W.; Burke, M.; Füllgrabe, A.; Fuentes, A.M.-P.; George, N.; et al. Expression Atlas: Gene and protein expression across multiple studies and organisms. Nucleic Acids Res. 2018, 46, D246–D251. [Google Scholar] [CrossRef]
- Calderwood, S.K.; Khaleque, M.A.; Sawyer, D.B.; Ciocca, D.R. Heat shock proteins in cancer: Chaperones of tumorigenesis. Trends Biochem. Sci. 2006, 31, 164–172. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S.; Burns, T.F. Targeting Heat Shock Proteins in Cancer: A Promising Therapeutic Approach. Int. J. Mol. Sci. 2017, 18, 1978. [Google Scholar] [CrossRef] [PubMed]
- Zoubeidi, A.; Gleave, M. Small heat shock proteins in cancer therapy and prognosis. Int. J. Biochem. Cell Biol. 2012, 44, 1646–1656. [Google Scholar] [CrossRef] [PubMed]
- Labbadia, J.; Morimoto, R.I. The biology of proteostasis in aging and disease. Annu. Rev. Biochem. 2015, 84, 435–464. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Cao, P.; Fan, Y.; Tan, K. Emerging roles of HSF1 in cancer: Cellular and molecular episodes. Biochim. Biophys. Acta Rev. Cancer 2020, 1874, 188390. [Google Scholar] [CrossRef]
- Mendillo, M.L.; Santagata, S.; Koeva, M.; Bell, G.W.; Hu, R.; Tamimi, R.M.; Fraenkel, E.; Ince, T.A.; Whitesell, L.; Lindquist, S. HSF1 drives a transcriptional program distinct from heat shock to support highly malignant human cancers. Cell 2012, 150, 549–562. [Google Scholar] [CrossRef]
- Santagata, S.; Hu, R.; Lin, N.U.; Mendillo, M.L.; Collins, L.C.; Hankinson, S.E.; Schnitt, S.J.; Whitesell, L.; Tamimi, R.M.; Lindquist, S.; et al. High levels of nuclear heat-shock factor 1 (HSF1) are associated with poor prognosis in breast cancer. Proc. Natl. Acad. Sci. USA 2011, 108, 18378–18383. [Google Scholar] [CrossRef]
- Yu, W.; Wang, J.; Li, C.; Xuan, M.; Han, S.; Zhang, Y.; Liu, P.; Zhao, Z. miR-17-5p promotes the invasion and migration of colorectal cancer by regulating HSPB2. J. Cancer 2022, 13, 918–931. [Google Scholar] [CrossRef]
- Calderwood, S.K.; Gong, J. Heat Shock Proteins Promote Cancer: It’s a Protection Racket. Trends Biochem. Sci. 2016, 41, 311–323. [Google Scholar] [CrossRef]
- Jego, G.; Hazoume, A.; Seigneuric, R.; Garrido, C. Targeting heat shock proteins in cancer. Cancer Lett. 2013, 332, 275–285. [Google Scholar] [CrossRef]
- Chang, H.R.; Jung, E.; Cho, S.; Jeon, Y.J.; Kim, Y. Targeting Non-Oncogene Addiction for Cancer Therapy. Biomolecules 2021, 11, 129. [Google Scholar] [CrossRef] [PubMed]
- Nagel, R.; Semenova, E.A.; Berns, A. Drugging the addict: Non-oncogene addiction as a target for cancer therapy. EMBO Rep. 2016, 17, 1516–1531. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Chen, M.; Zhou, J.; Zhang, X. HSP27, 70 and 90, anti-apoptotic proteins, in clinical cancer therapy (Review). Int. J. Oncol. 2014, 45, 18–30. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Liu, T.; Rios, Z.; Mei, Q.; Lin, X.; Cao, S. Heat Shock Proteins and Cancer. Trends Pharmacol. Sci. 2017, 38, 226–256. [Google Scholar] [CrossRef]
- De Mattos-Arruda, L.; Cortes, J. Breast cancer and HSP90 inhibitors: Is there a role beyond the HER2-positive subtype? Breast 2012, 21, 604–607. [Google Scholar] [CrossRef]
- Modi, S.; Stopeck, A.; Linden, H.; Solit, D.; Chandarlapaty, S.; Rosen, N.; D’Andrea, G.; Dickler, M.; Moynahan, M.E.; Sugarman, S.; et al. HSP90 inhibition is effective in breast cancer: A phase II trial of tanespimycin (17-AAG) plus trastuzumab in patients with HER2-positive metastatic breast cancer progressing on trastuzumab. Clin. Cancer Res. 2011, 17, 5132–5139. [Google Scholar] [CrossRef]
- Park, J.M.; Kim, Y.J.; Park, S.; Park, M.; Farrand, L.; Nguyen, C.T.; Ann, J.; Nam, G.; Park, H.J.; Lee, J.; et al. A novel HSP90 inhibitor targeting the C-terminal domain attenuates trastuzumab resistance in HER2-positive breast cancer. Mol. Cancer 2020, 19, 161. [Google Scholar] [CrossRef]
- Henry, N.L.; Hayes, D.F. Cancer biomarkers. Mol. Oncol. 2012, 6, 140–146. [Google Scholar] [CrossRef]
- Kamel, H.F.M.; Al-Amodi, H. Exploitation of Gene Expression and Cancer Biomarkers in Paving the Path to Era of Personalized Medicine. Genom. Proteom. Bioinform. 2017, 15, 220–235. [Google Scholar] [CrossRef]
- Vargas, A.J.; Harris, C.C. Biomarker development in the precision medicine era: Lung cancer as a case study. Nat. Rev. Cancer 2016, 16, 525–537. [Google Scholar] [CrossRef]
- de Kok, J.B.; Roelofs, R.W.; Giesendorf, B.A.; Pennings, J.L.; Waas, E.T.; Feuth, T.; Swinkels, D.W.; Span, P.N. Normalization of gene expression measurements in tumor tissues: Comparison of 13 endogenous control genes. Lab. Investig. 2005, 85, 154–159. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).