A Transcription Factor SlNAC10 Gene of Suaeda liaotungensis Regulates Proline Synthesis and Enhances Salt and Drought Tolerance
Abstract
1. Introduction
2. Results
2.1. SlNAC10 Encodes a Protein Containing the NAC Domain
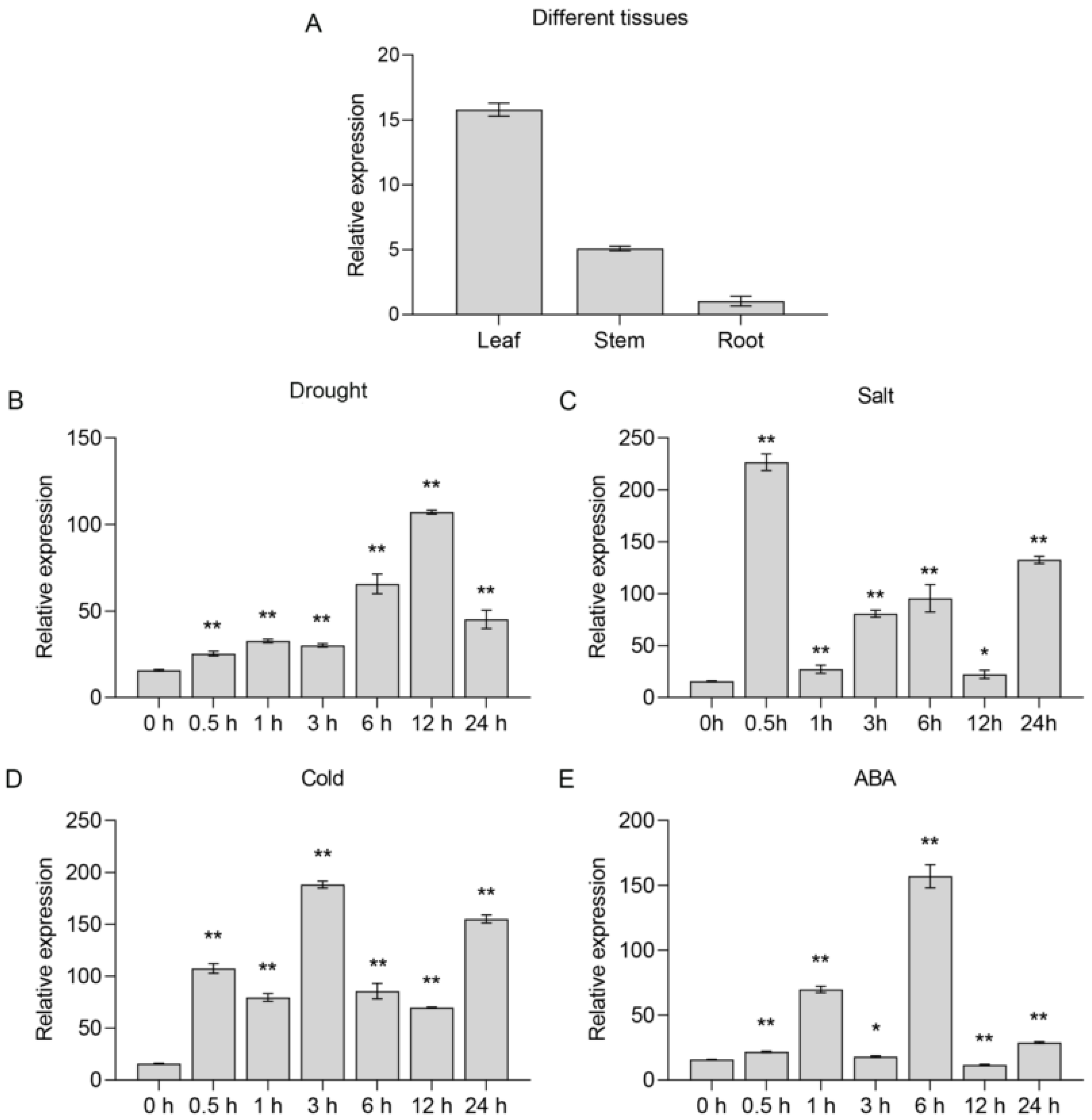
2.2. SlNAC10 Is a Stress-Responsive Gene
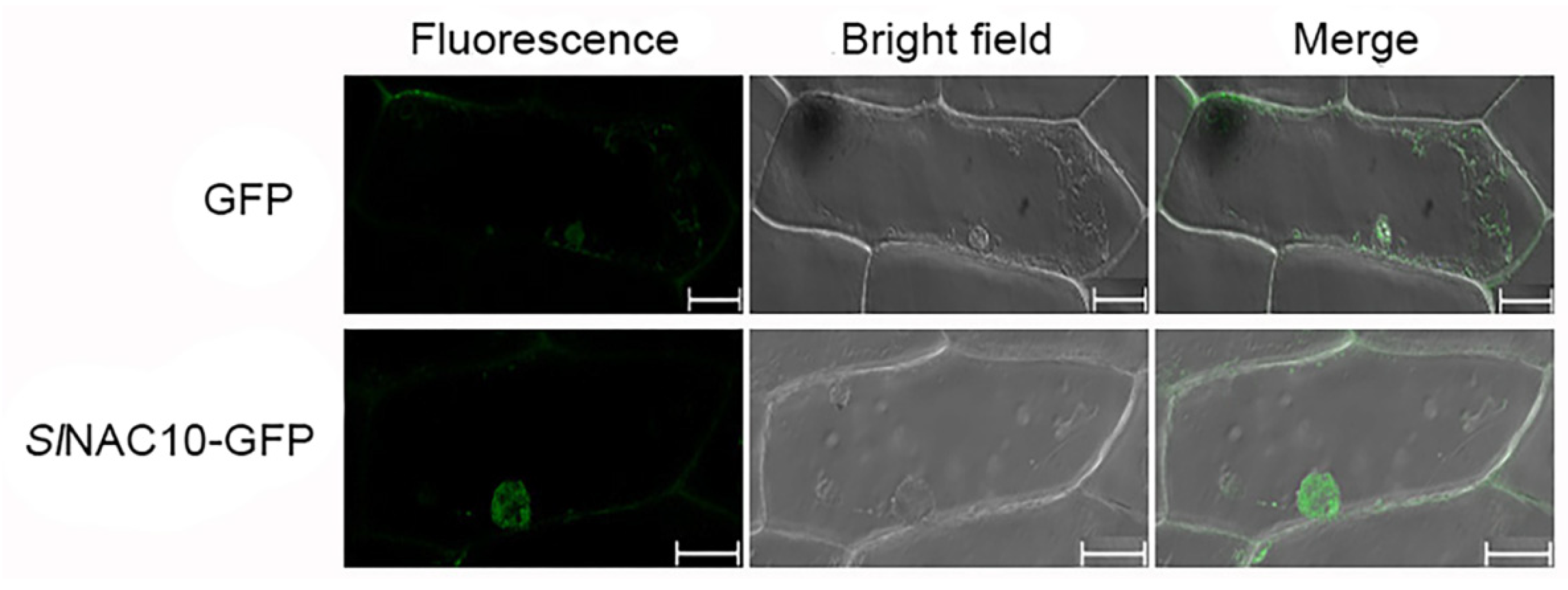
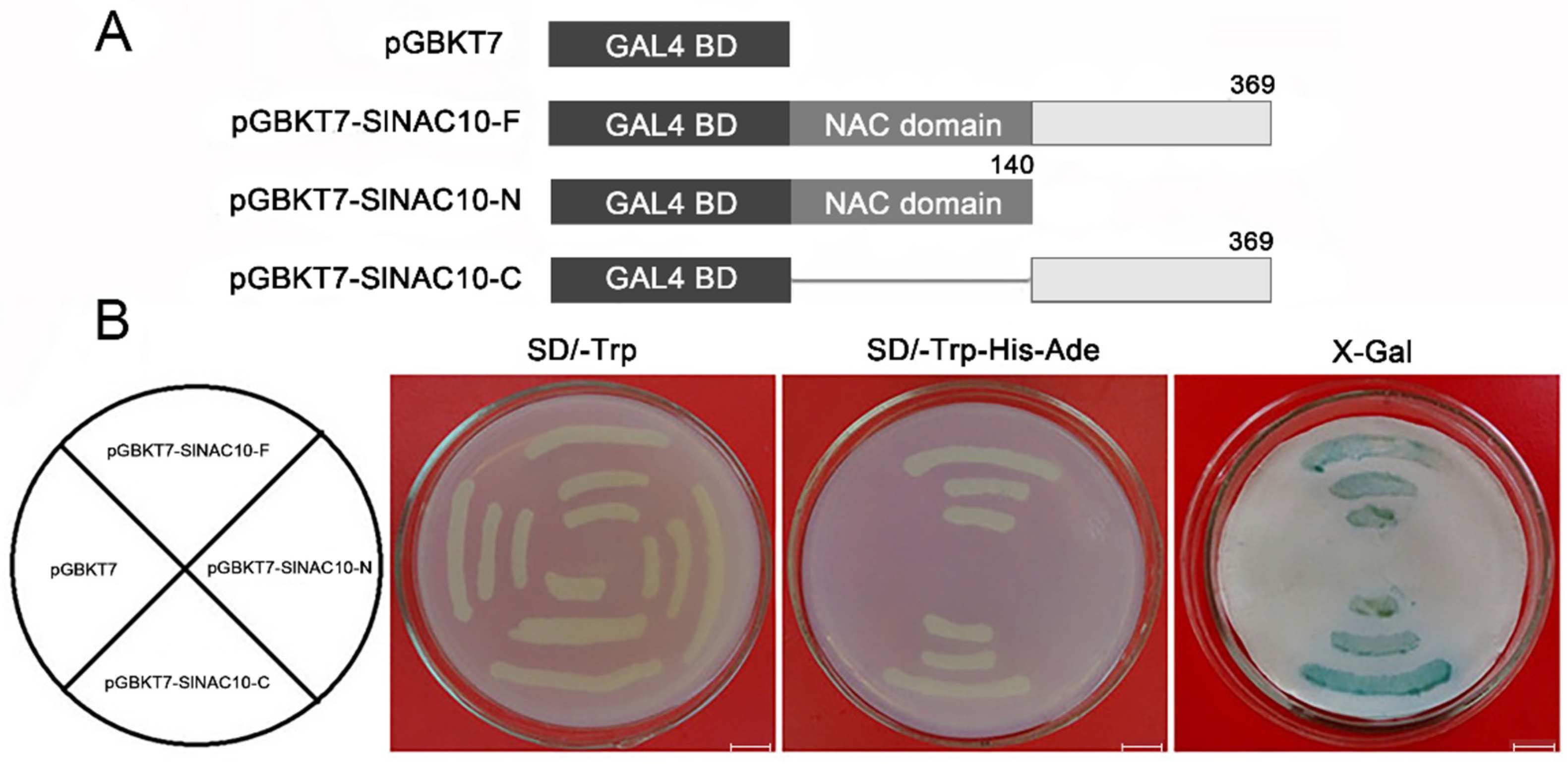
2.3. SlNAC10 Is Located in the Nucleus and Has Transcriptional Activation Activity
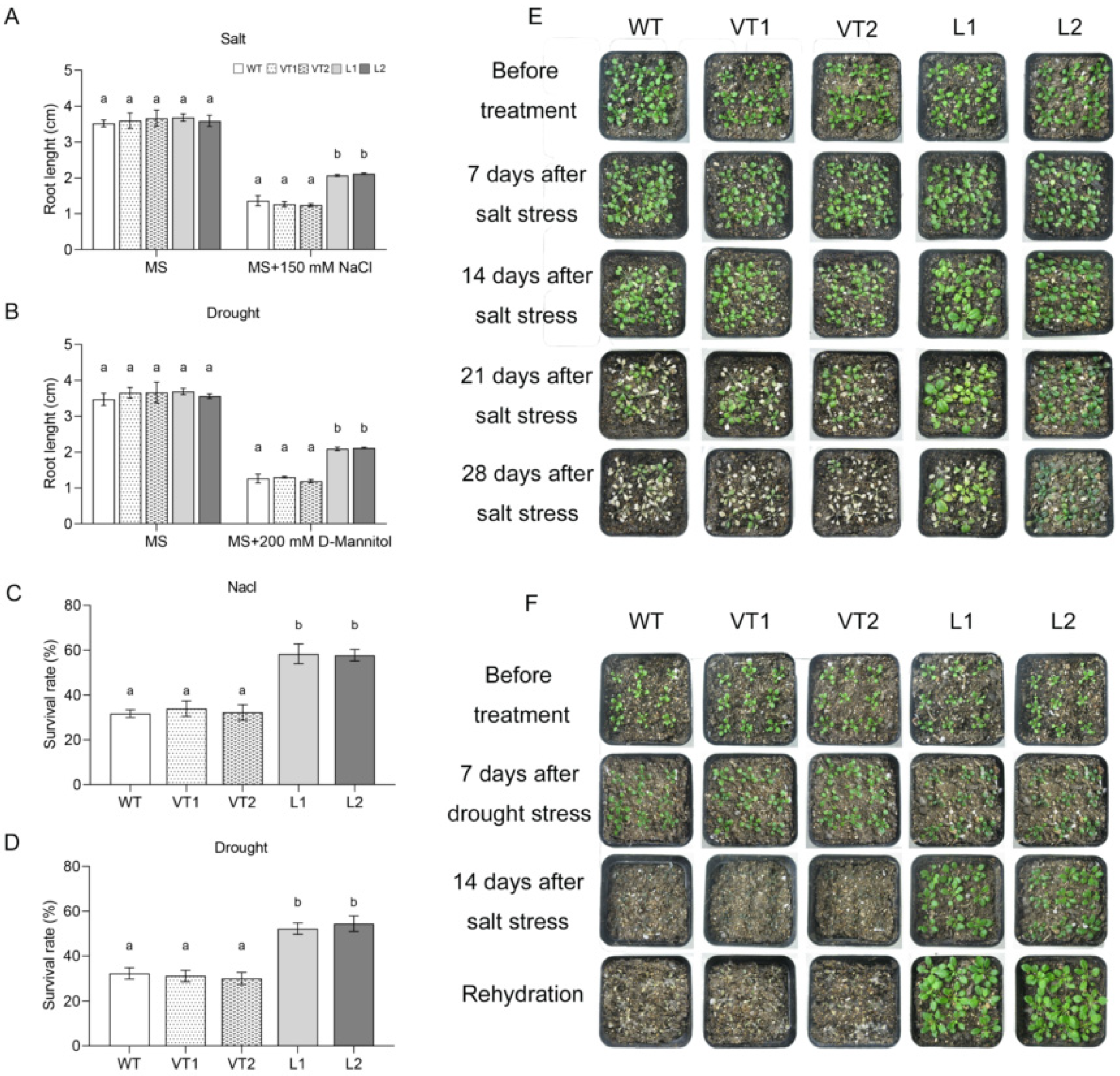
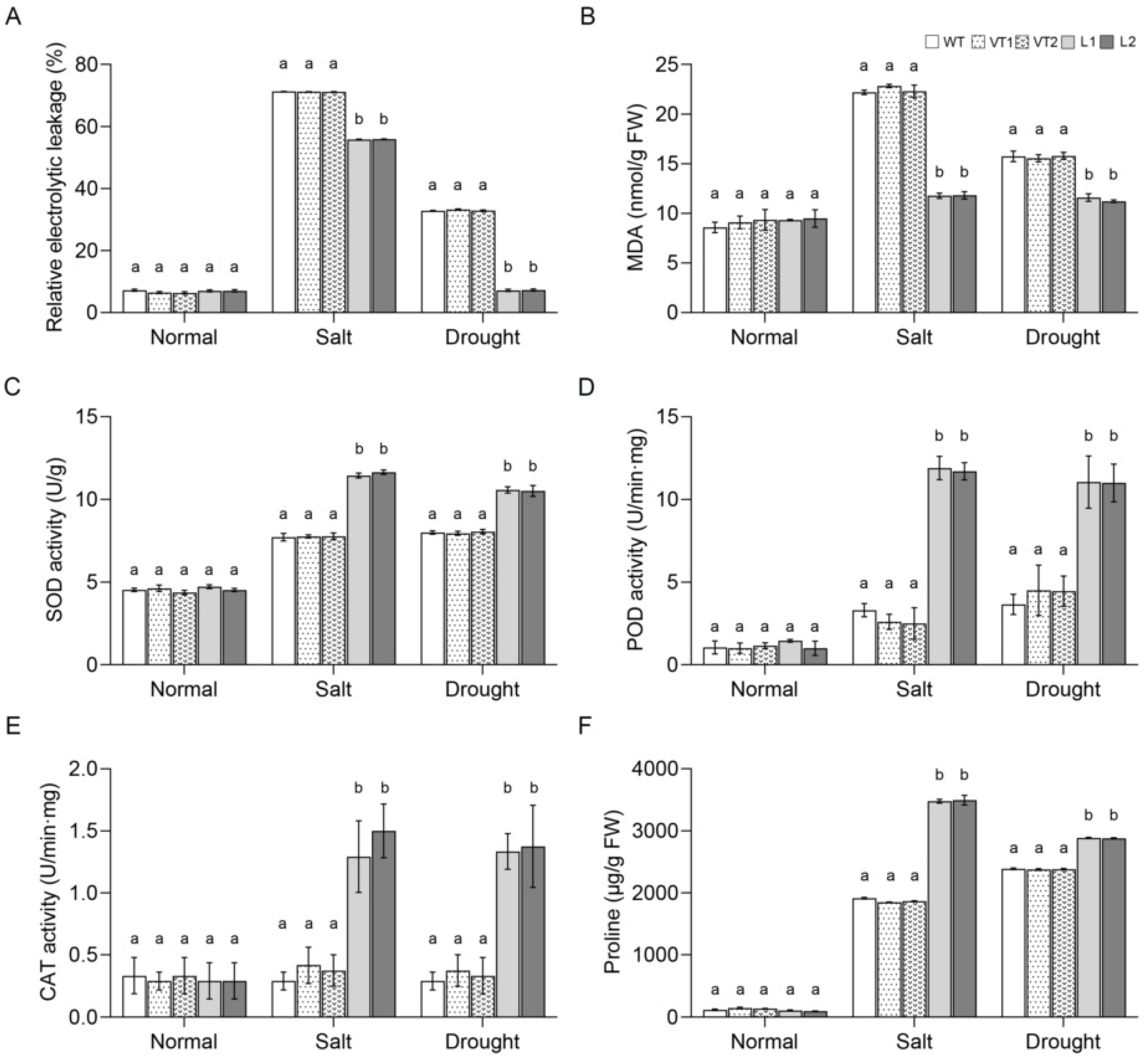
2.4. Overexpression of SlNAC10 in Transgenic Arabidopsis Thaliana Improves Salt and Drought Tolerance
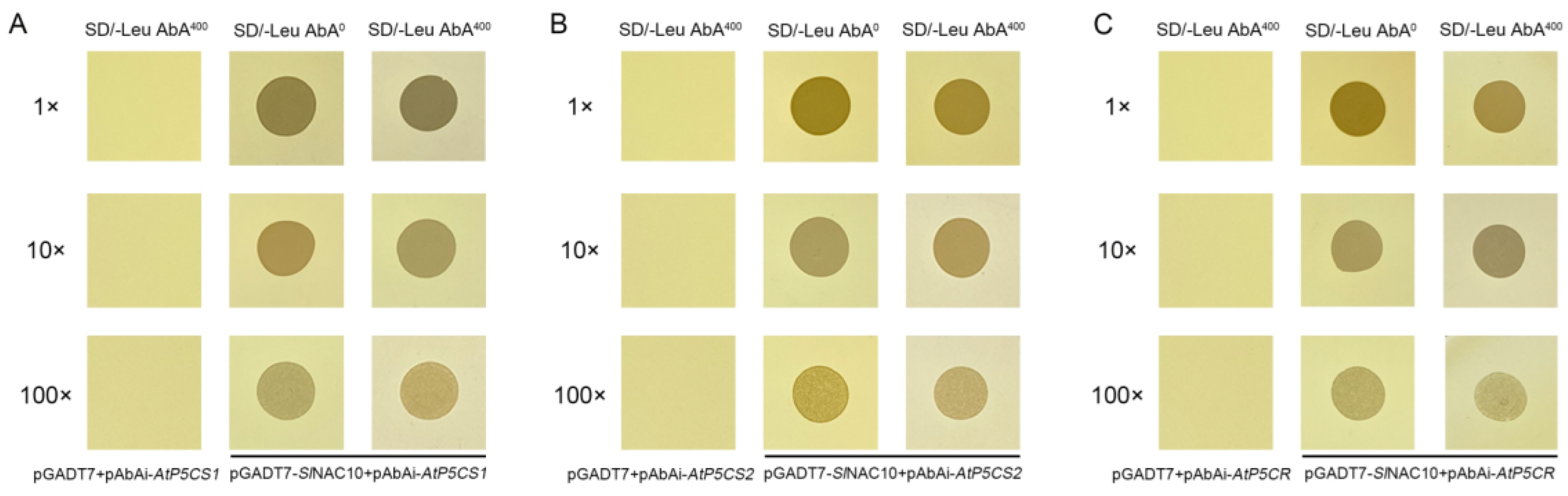
2.5. SlNAC10 Upregulates the Expression of Proline Synthesis-Related Enzyme Genes
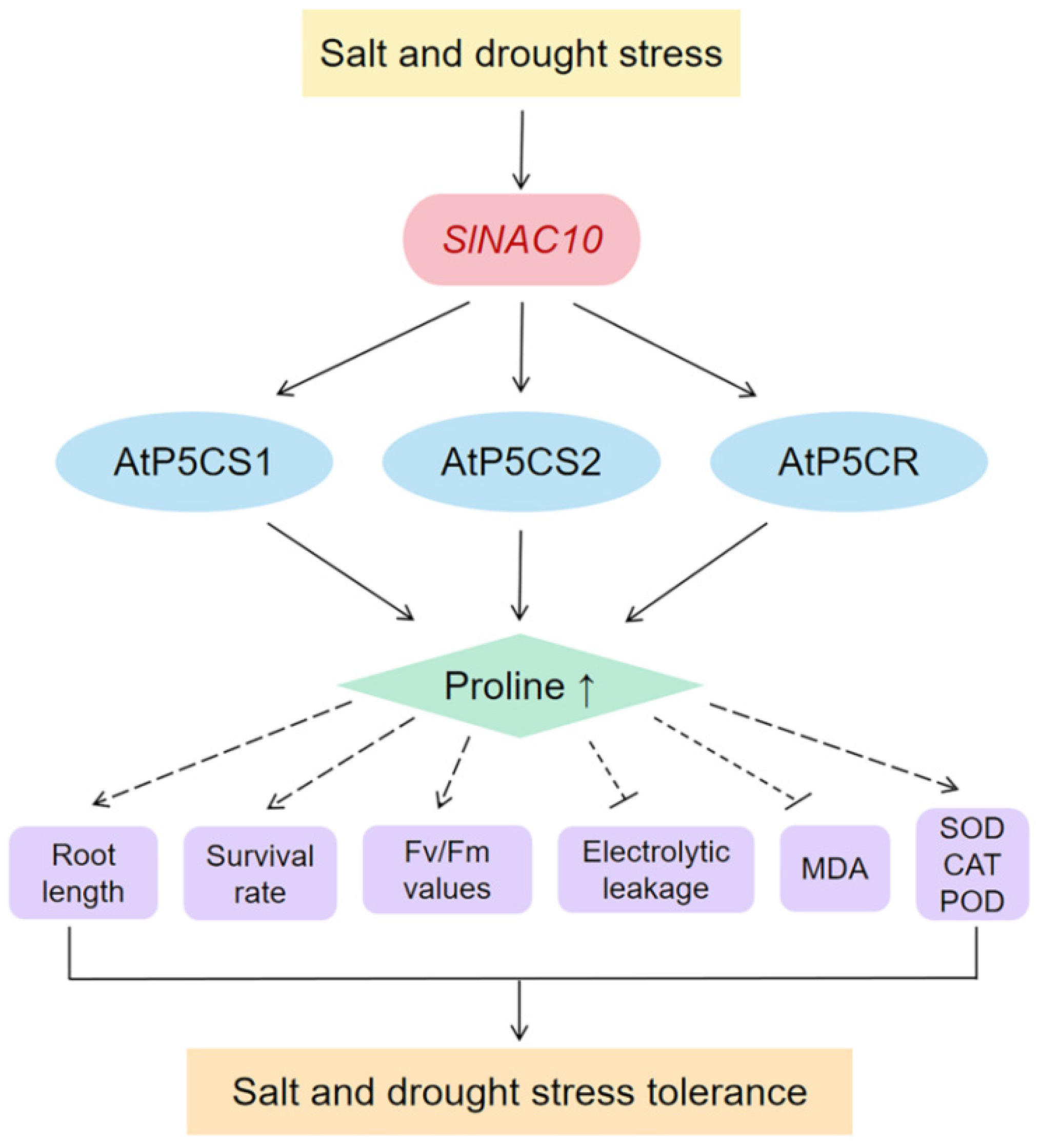
3. Discussion
4. Materials and Methods
4.1. Plant Material and Treatment
4.2. Cloning and Sequence Analysis of the SlNAC10 Gene
4.3. Analysis of SlNAC10 Gene Expression Profile
4.4. Subcellular Localization Analysis of SlNAC10
4.5. Transcriptional Activation Analysis of SlNAC10
4.6. Growth Assay of SlNAC10 Transgenic Arabidopsis under Salt and Drought Stress
4.7. Determination of Physiological Indicators of SlNAC10 Transgenic Arabidopsis Thaliana
4.8. Expression of P5CS1, P5CS2, and P5CR in Arabidopsis thaliana Overexpressing the SlNAC10 Gene
4.9. Analysis of the Interaction between SlNAC10 and the P5CS1, P5CS2, and P5CR Promoters
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shao, H.; Wang, H.; Tang, X. NAC transcription factors in plant multiple abiotic stress responses: Progress and prospects. Front. Plant Sci. 2015, 6, 902. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Koyama, H.; Bhati, K.K.; Alok, A. The biotechnological importance of the plant-specific NAC transcription factor family in crop improvement. J. Plant Res. 2021, 134, 475–495. [Google Scholar] [CrossRef] [PubMed]
- Puranik, S.; Sahu, P.P.; Srivastava, P.S.; Prasad, M. NAC proteins: Regulation and role in stress tolerance. Trends Plant Sci. 2012, 17, 369–381. [Google Scholar] [CrossRef]
- Ooka, H.; Satoh, K.; Doi, K.; Nagata, T.; Otomo, Y.; Murakami, K.; Matsubara, K.; Osato, N.; Kawai, J.; Carninci, P. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 2003, 10, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Sakamoto, S.; Kawai, T.; Kobayashi, Y.; Sato, K.; Ichinose, Y.; Yaoi, K.; Akiyoshi-Endo, M.; Sato, H.; Takamizo, T. Engineering the Oryza sativa cell wall with rice NAC transcription factors regulating secondary wall formation. Front. Plant Sci. 2013, 4, 383. [Google Scholar] [CrossRef]
- Yamaguchi, M.; Demura, T. Transcriptional regulation of secondary wall formation controlled by NAC domain proteins. Plant Biotechnol. 2010, 27, 237–242. [Google Scholar] [CrossRef]
- Zhang, L.; Yao, L.; Zhang, N.; Yang, J.; Zhu, X.; Tang, X.; Calderón-Urrea, A.; Si, H. Lateral root development in potato is mediated by stu-mi164 regulation of NAC transcription factor. Front. Plant Sci. 2018, 9, 383. [Google Scholar] [CrossRef]
- Yang, X.; Kim, M.Y.; Ha, J.; Lee, S.H. Overexpression of the soybean NAC gene GmNAC109 increases lateral root formation and abiotic stress tolerance in transgenic Arabidopsis plants. Front. Plant Sci. 2019, 10, 1036. [Google Scholar] [CrossRef]
- Peng, H.; Cheng, H.Y.; Chen, C.; Yu, X.W.; Yang, J.N.; Gao, W.R.; Shi, Q.H.; Zhang, H.; Li, J.G.; Ma, H. A NAC transcription factor gene of Chickpea (Cicer arietinum), CarNAC3, is involved in drought stress response and various developmental processes. J. Plant Physiol. 2009, 166, 1934–1945. [Google Scholar] [CrossRef]
- Kato, H.; Motomura, T.; Komeda, Y.; Saito, T.; Kato, A. Overexpression of the NAC transcription factor family gene ANAC036 results in a dwarf phenotype in Arabidopsis thaliana. J. Plant Physiol. 2010, 167, 571–577. [Google Scholar] [CrossRef]
- Takeda, S.; Hanano, K.; Kariya, A.; Shimizu, S.; Zhao, L.; Matsui, M.; Tasaka, M.; Aida, M. CUP-SHAPED COTYLEDON1 transcription factor activates the expression of LSH4 and LSH3, two members of the ALOG gene family, in shoot organ boundary cells. Plant J. 2011, 66, 1066–1077. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Qiu, K.; Kuai, B.; Ding, Y. Identification of an NAP-like transcription factor BeNAC1 regulating leaf senescence in bamboo (Bambusa emeiensis ‘Viridiflavus’). Physiol. Plant. 2011, 142, 361–371. [Google Scholar] [CrossRef] [PubMed]
- de Zélicourt, A.; Diet, A.; Marion, J.; Laffont, C.; Ariel, F.; Moison, M.; Zahaf, O.; Crespi, M.; Gruber, V.; Frugier, F. Dual involvement of a Medicago truncatula NAC transcription factor in root abiotic stress response and symbiotic nodule senescence. Plant J. 2012, 70, 220–230. [Google Scholar] [CrossRef]
- Yang, J.; Duan, G.; Li, C.; Liu, L.; Han, G.; Zhang, Y.; Wang, C. The crosstalks between JA and other plant hormone signaling highlight the involvement of JA as a core component in plant response to biotic and abiotic stresses. Front. Plant Sci. 2019, 10, 1349. [Google Scholar] [CrossRef] [PubMed]
- Johnsson, C.; Jin, X.; Xue, W.; Dubreuil, C.; Lezhneva, L.; Fischer, U. The plant hormone auxin directs timing of xylem development by inhibition of secondary cell wall deposition through repression of secondary wall NAC-domain transcription factors. Physiol. Plant. 2019, 165, 673–689. [Google Scholar] [CrossRef]
- Yuan, X.; Wang, H.; Cai, J.; Li, D.; Song, F. NAC transcription factors in plant immunity. Phytopathol. Res. 2019, 1, 3. [Google Scholar] [CrossRef]
- Yang, R.; Deng, C.; Ouyang, B.; Ye, Z. Molecular analysis of two salt-responsive NAC-family genes and their expression analysis in tomato. Mol. Biol. Rep. 2011, 38, 857–863. [Google Scholar] [CrossRef]
- Alshareef, N.O.; Wang, J.Y.; Ali, S.; Al-Babili, S.; Tester, M.; Schmöckel, S.M. Overexpression of the NAC transcription factor JUNGBRUNNEN1 (JUB1) increases salinity tolerance in tomato. Plant Physiol. Biochem. 2019, 140, 113–121. [Google Scholar] [CrossRef]
- Wang, J.; Lian, W.; Cao, Y.; Wang, X.; Wang, G.; Qi, C.; Liu, L.; Qin, S.; Yuan, X.; Li, X. Overexpression of BoNAC019, a NAC transcription factor from Brassica oleracea, negatively regulates the dehydration response and anthocyanin biosynthesis in Arabidopsis. Sci. Rep. 2018, 8, 13349. [Google Scholar] [CrossRef]
- Yuan, X.; Wang, H.; Cai, J.; Bi, Y.; Li, D.; Song, F. Rice NAC transcription factor ONAC066 functions as a positive regulator of drought and oxidative stress response. BMC Plant Biol. 2019, 19, 278. [Google Scholar] [CrossRef]
- Chen, X.; Wang, Y.; Lv, B.; Li, J.; Luo, L.; Lu, S.; Zhang, X.; Ma, H.; Ming, F. The NAC family transcription factor OsNAP confers abiotic stress response through the ABA pathway. Plant Cell Physiol. 2014, 55, 604–619. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Huang, F.; Cheng, H.; Song, H.; Yu, D. Overexpression of the GmNAC2 gene, an NAC transcription factor, reduces abiotic stress tolerance in tobacco. Plant Mol. Biol. Rep. 2013, 31, 435–442. [Google Scholar] [CrossRef]
- Hao, Y.J.; Wei, W.; Song, Q.X.; Chen, H.W.; Zhang, Y.Q.; Wang, F.; Zou, H.F.; Lei, G.; Tian, A.G.; Zhang, W.K. Soybean NAC transcription factors promote abiotic stress tolerance and lateral root formation in transgenic plants. Plant J. 2011, 68, 302–313. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, K.H.; Mostofa, M.G.; Li, W.; Van Ha, C.; Watanabe, Y.; Le, D.T.; Thao, N.P.; Tran, L.-S.P. The soybean transcription factor GmNAC085 enhances drought tolerance in Arabidopsis. Environ. Exp. Bot. 2018, 151, 12–20. [Google Scholar] [CrossRef]
- Ju, Y.L.; Yue, X.F.; Min, Z.; Wang, X.H.; Fang, Y.L.; Zhang, J.X. VvNAC17, a novel stress-responsive grapevine (Vitis vinifera L.) NAC transcription factor, increases sensitivity to abscisic acid and enhances salinity, freezing, and drought tolerance in transgenic Arabidopsis. Plant Physiol. Biochem. 2020, 146, 98–111. [Google Scholar] [CrossRef]
- An, J.P.; Yao, J.F.; Xu, R.R.; You, C.X.; Wang, X.F.; Hao, Y.J. An apple NAC transcription factor enhances salt stress tolerance by modulating the ethylene response. Physiol. Plant. 2018, 164, 279–289. [Google Scholar] [CrossRef]
- Hou, X.M.; Zhang, H.F.; Liu, S.Y.; Wang, X.K.; Zhang, Y.M.; Meng, Y.C.; Luo, D.; Chen, R.G. The NAC transcription factor CaNAC064 is a regulator of cold stress tolerance in peppers. Plant Sci. 2020, 291, 110346. [Google Scholar] [CrossRef]
- Zhang, H.; Ma, F.; Wang, X.; Liu, S.; Saeed, U.H.; Hou, X.; Zhang, Y.; Luo, D.; Meng, Y.; Zhang, W. Molecular and functional characterization of CaNAC035, an NAC transcription factor from pepper (Capsicum annuum L.). Front. Plant Sci. 2020, 11, 14. [Google Scholar] [CrossRef]
- Han, X.; Feng, Z.; Xing, D.; Yang, Q.; Wang, R.; Qi, L.; Li, G. Two NAC transcription factors from Caragana intermedia altered salt tolerance of the transgenic Arabidopsis. BMC Plant Biol. 2015, 15, 208. [Google Scholar] [CrossRef]
- Borgohain, P.; Saha, B.; Agrahari, R.; Chowardhara, B.; Sahoo, S.; van der Vyver, C.; Panda, S.K. SlNAC2 overexpression in Arabidopsis results in enhanced abiotic stress tolerance with alteration in glutathione metabolism. Protoplasma 2019, 256, 1065–1077. [Google Scholar] [CrossRef]
- Lu, M.; Ying, S.; Zhang, D.F.; Shi, Y.S.; Song, Y.C.; Wang, T.Y.; Li, Y. A maize stress-responsive NAC transcription factor, ZmSNAC1, confers enhanced tolerance to dehydration in transgenic Arabidopsis. Plant Cell Rep. 2012, 31, 1701–1711. [Google Scholar] [CrossRef] [PubMed]
- Mao, H.; Yu, L.; Han, R.; Li, Z.; Liu, H. ZmNAC55, a maize stress-responsive NAC transcription factor, confers drought resistance in transgenic Arabidopsis. Plant Physiol. Biochem. 2016, 105, 55–66. [Google Scholar] [CrossRef]
- Li, X.L.; Yang, X.; Hu, Y.X.; Yu, X.D.; Li, Q.L. A novel NAC transcription factor from Suaeda liaotungensis K. enhanced transgenic Arabidopsis drought, salt, and cold stress tolerance. Plant Cell Rep. 2014, 33, 767–778. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Hu, Y.X.; Li, X.L.; Yu, X.D.; Li, Q.L. Molecular characterization and function analysis of SlNAC2 in Suaeda liaotungensis K. Gene 2014, 543, 190–197. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.F.; Shan, H.Y.; Shi, H.; Wu, D.D.; Li, T.T.; Li, Q.L. Characterization of a transcription factor SlNAC7 gene from Suaeda liaotungensis and its role in stress tolerance. J. Plant Res. 2021, 134, 1105–1120. [Google Scholar] [CrossRef]
- Wu, D.; Sun, Y.; Wang, H.; Shi, H.; Su, M.; Shan, H.; Li, T.; Li, Q. The SlNAC8 gene of the halophyte Suaeda liaotungensis enhances drought and salt stress tolerance in transgenic Arabidopsis thaliana. Gene 2018, 662, 10–20. [Google Scholar] [CrossRef]
- Duval, M.; Hsieh, T.-F.; Kim, S.Y.; Thomas, T.L. Molecular characterization of AtNAM: A member of theArabidopsis NAC domain superfamily. Plant Mol. Biol. 2002, 50, 237–248. [Google Scholar] [CrossRef]
- Shan, W.; Kuang, J.F.; Chen, L.; Xie, H.; Peng, H.H.; Xiao, Y.Y.; Li, X.P.; Chen, W.X.; He, Q.G.; Chen, J.Y. Molecular characterization of banana NAC transcription factors and their interactions with ethylene signalling component EIL during fruit ripening. Environ. Exp. Bot. 2012, 63, 5171–5187. [Google Scholar] [CrossRef]
- Wang, F.; Wang, J.W.; Sun, L.J.; Song, X.S. The molecular cloning and functional characterization of ChNAC1, a NAC transcription factor in Cerasus humilis. Plant Growth Regul. 2019, 89, 331–343. [Google Scholar] [CrossRef]
- Zhong, H.; Guo, Q.Q.; Chen, L.; Ren, F.; Wang, Q.Q.; Zheng, Y.; Li, X.B. Two Brassica napus genes encoding NAC transcription factors are involved in response to high-salinity stress. Plant Cell Rep. 2012, 31, 1991–2003. [Google Scholar] [CrossRef]
- Yang, X.; He, K.; Chi, X.; Chai, G.; Wang, Y.; Jia, C.; Zhang, H.; Zhou, G.; Hu, R. Miscanthus NAC transcription factor MlNAC12 positively mediates abiotic stress tolerance in transgenic Arabidopsis. Plant Sci. 2018, 277, 229–241. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Chen, R.; Jiang, Q.; Sun, X.; Zhang, H.; Hu, Z. GmNAC06, a NAC domain transcription factor enhances salt stress tolerance in soybean. Plant Mol. Biol. 2021, 105, 333–345. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Wang, Y.; Li, B.; Chang, J.; Chen, M.; Li, K.; Yang, G.; He, G. TaNAC29, a NAC transcription factor from wheat, enhances salt and drought tolerance in transgenic Arabidopsis. BMC Plant Biol. 2015, 15, 268. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.K.; Chung, P.J.; Jeong, J.S.; Jang, G.; Bang, S.W.; Jung, H.; Kim, Y.S.; Ha, S.H.; Choi, Y.D.; Kim, J.K. The rice Os NAC 6 transcription factor orchestrates multiple molecular mechanisms involving root structural adaptions and nicotianamine biosynthesis for drought tolerance. Plant Biotechnol. J. 2017, 15, 754–764. [Google Scholar] [CrossRef] [PubMed]
- Ding, A.; Li, S.; Li, W.; Hao, Q.; Wan, X.; Wang, K.; Liu, Q.; Liu, Q.; Jiang, X. RhNAC31, a novel rose NAC transcription factor, enhances tolerance to multiple abiotic stresses in Arabidopsis. Acta Physiol. Plant. 2019, 41, 75. [Google Scholar] [CrossRef]
- Tak, H.; Negi, S.; Ganapathi, T.R. Banana NAC transcription factor MusaNAC042 is positively associated with drought and salinity tolerance. Protoplasma 2017, 254, 803–816. [Google Scholar] [CrossRef]
- Cao, H.; Wang, L.; Nawaz, M.A.; Niu, M.; Sun, J.; Xie, J.; Kong, Q.; Huang, Y.; Cheng, F.; Bie, Z. Ectopic expression of pumpkin NAC transcription factor CmNAC1 improves multiple abiotic stress tolerance in Arabidopsis. Front. Plant Sci. 2017, 8, 2052. [Google Scholar] [CrossRef]
- Mergby, D.; Hanin, M.; Saidi, M.N. The durum wheat NAC transcription factor TtNAC2A enhances drought stress tolerance in Arabidopsis. Environ. Exp. Bot. 2021, 186, 104439. [Google Scholar] [CrossRef]
- Wang, K.; Zhong, M.; Wu, Y.H.; Bai, Z.Y.; Liang, Q.Y.; Liu, Q.L.; Pan, Y.Z.; Zhang, L.; Jiang, B.B.; Jia, Y. Overexpression of a chrysanthemum transcription factor gene DgNAC1 improves the salinity tolerance in chrysanthemum. Plant Cell Rep. 2017, 36, 571–581. [Google Scholar] [CrossRef]
- Wang, Y.; Cao, S.; Guan, C.; Kong, X.; Wang, Y.; Cui, Y.; Liu, B.; Zhou, Y.; Zhang, Y. Overexpressing the NAC transcription factor LpNAC13 from Lilium pumilum in tobacco negatively regulates the drought response and positively regulates the salt response. Plant Physiol. Biochem. 2020, 149, 96–110. [Google Scholar] [CrossRef]
- Tang, H.; Yu, Q.; Li, Z.; Liu, F.; Su, W.; Zhang, C.; Ling, H.; Luo, J.; Su, Y.; Que, Y. A PIP-mediated osmotic stress signaling cascade plays a positive role in the salt tolerance of sugarcane. BMC Plant Biol. 2021, 21, 589. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Li, Y.; Hu, L.; Ye, Y.; Chen, J.; Li, J.; Pei, B.; Wang, G.; Chen, S.; Cheng, Y. The cotton GhMYB4 gene enhances salt and drought tolerance in transgenic Arabidopsis. Agron. J. 2021, 113, 4762–4776. [Google Scholar] [CrossRef]
- Yuan, C.; Huang, D.; Yang, Y.; Sun, M.; Cheng, T.; Wang, J.; Pan, H.; Zhang, Q. CmCYC2-like transcription factors may interact with each other or bind to the promoter to regulate floral symmetry development in Chrysanthemum morifolium. Plant Mol. Biol. 2020, 103, 159–171. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, L.; Cao, Y.; Qi, C.; Li, S.; Liu, L.; Wang, G.; Mao, A.; Ren, S.; Guo, Y.D. CsATAF1 positively regulates drought stress tolerance by an ABA-dependent pathway and by promoting ROS scavenging in cucumber. Plant Cell Physiol. 2018, 59, 930–945. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Cao, W.H.; Liu, J.; He, X.J.; Mu, R.L.; Zhou, H.L.; Chen, S.Y.; Zhang, J.S. Modulation of ethylene responses affects plant salt-stress responses. Plant Physiol. 2007, 143, 707–719. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Du, X.; Su, M.; Jiao, Y.; Xu, S.; Song, J.; Wang, H.; Li, Q. A Transcription Factor SlNAC10 Gene of Suaeda liaotungensis Regulates Proline Synthesis and Enhances Salt and Drought Tolerance. Int. J. Mol. Sci. 2022, 23, 9625. https://doi.org/10.3390/ijms23179625
Du X, Su M, Jiao Y, Xu S, Song J, Wang H, Li Q. A Transcription Factor SlNAC10 Gene of Suaeda liaotungensis Regulates Proline Synthesis and Enhances Salt and Drought Tolerance. International Journal of Molecular Sciences. 2022; 23(17):9625. https://doi.org/10.3390/ijms23179625
Chicago/Turabian StyleDu, Xinran, Mingxing Su, Yang Jiao, Suxiang Xu, Jieqiong Song, Hongfei Wang, and Qiuli Li. 2022. "A Transcription Factor SlNAC10 Gene of Suaeda liaotungensis Regulates Proline Synthesis and Enhances Salt and Drought Tolerance" International Journal of Molecular Sciences 23, no. 17: 9625. https://doi.org/10.3390/ijms23179625
APA StyleDu, X., Su, M., Jiao, Y., Xu, S., Song, J., Wang, H., & Li, Q. (2022). A Transcription Factor SlNAC10 Gene of Suaeda liaotungensis Regulates Proline Synthesis and Enhances Salt and Drought Tolerance. International Journal of Molecular Sciences, 23(17), 9625. https://doi.org/10.3390/ijms23179625




