DNA Damage Induction Alters the Expression of Ubiquitin and SUMO Regulators in Preimplantation Stage Pig Embryos
Abstract
:1. Introduction
2. Results
2.1. Effect of UV-Exposure on Embryo Development and DNA Damage
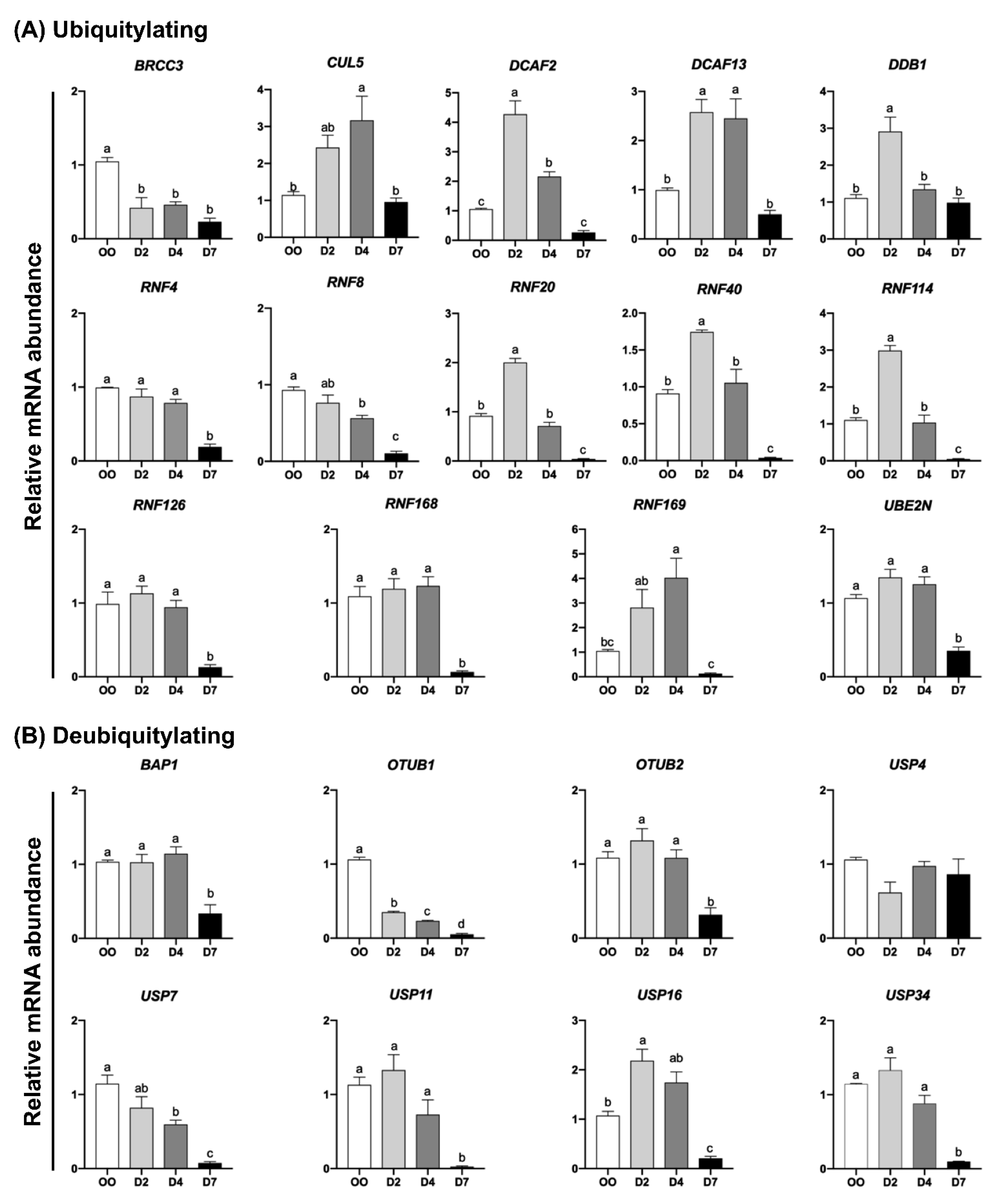
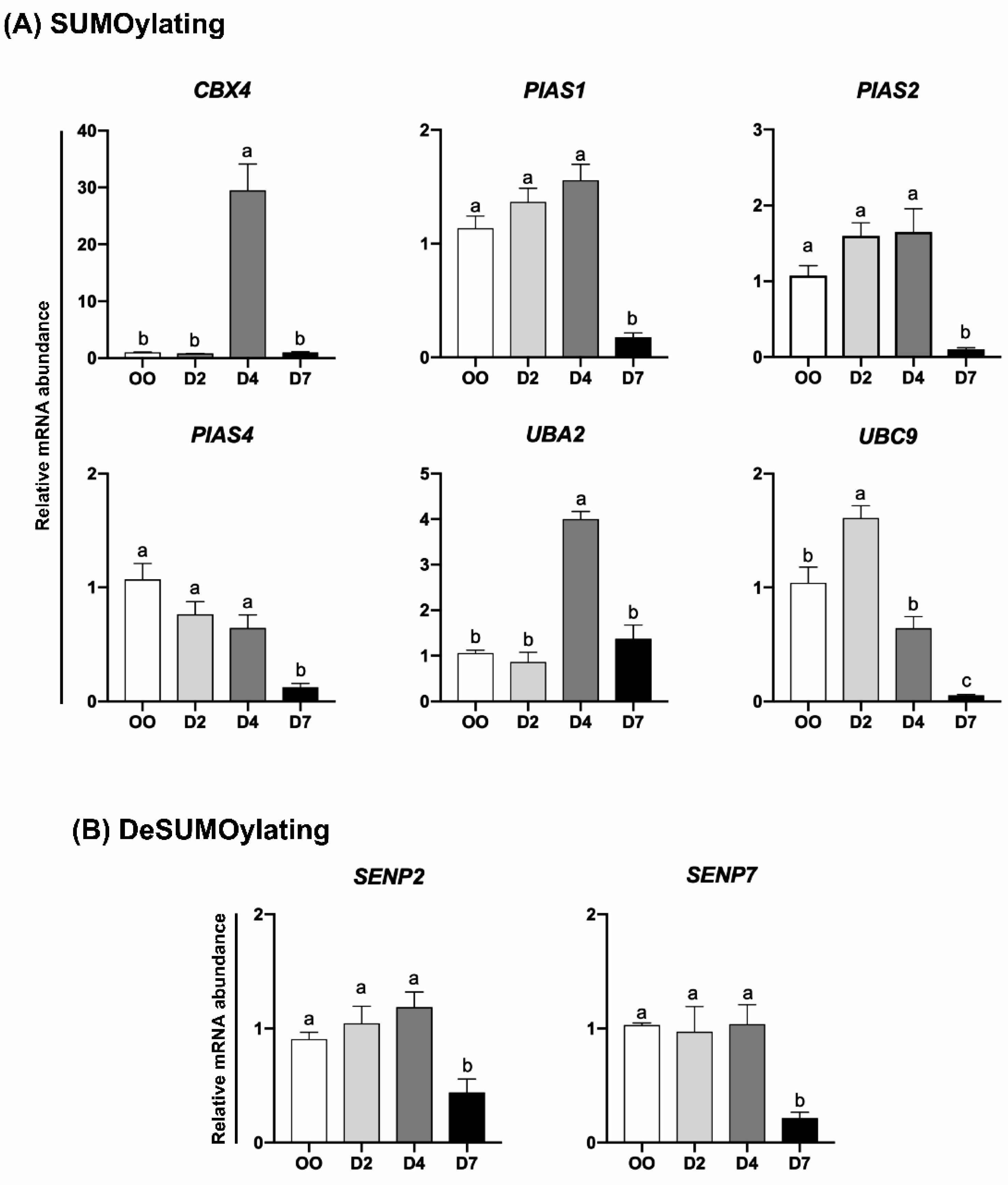
2.2. mRNA Levels of UBs, DUBs, SUMOs and deSUMOs during Embryo Development
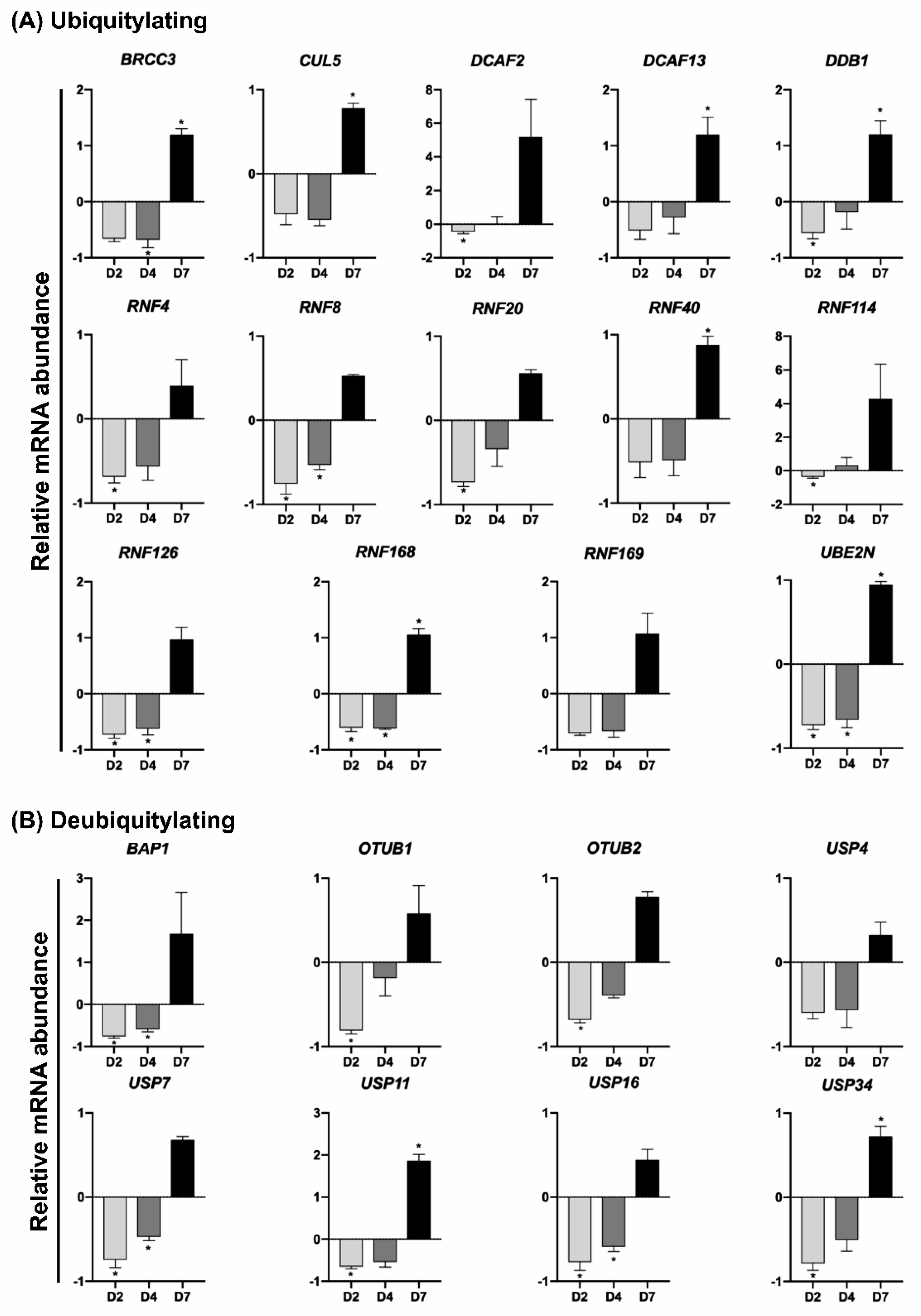
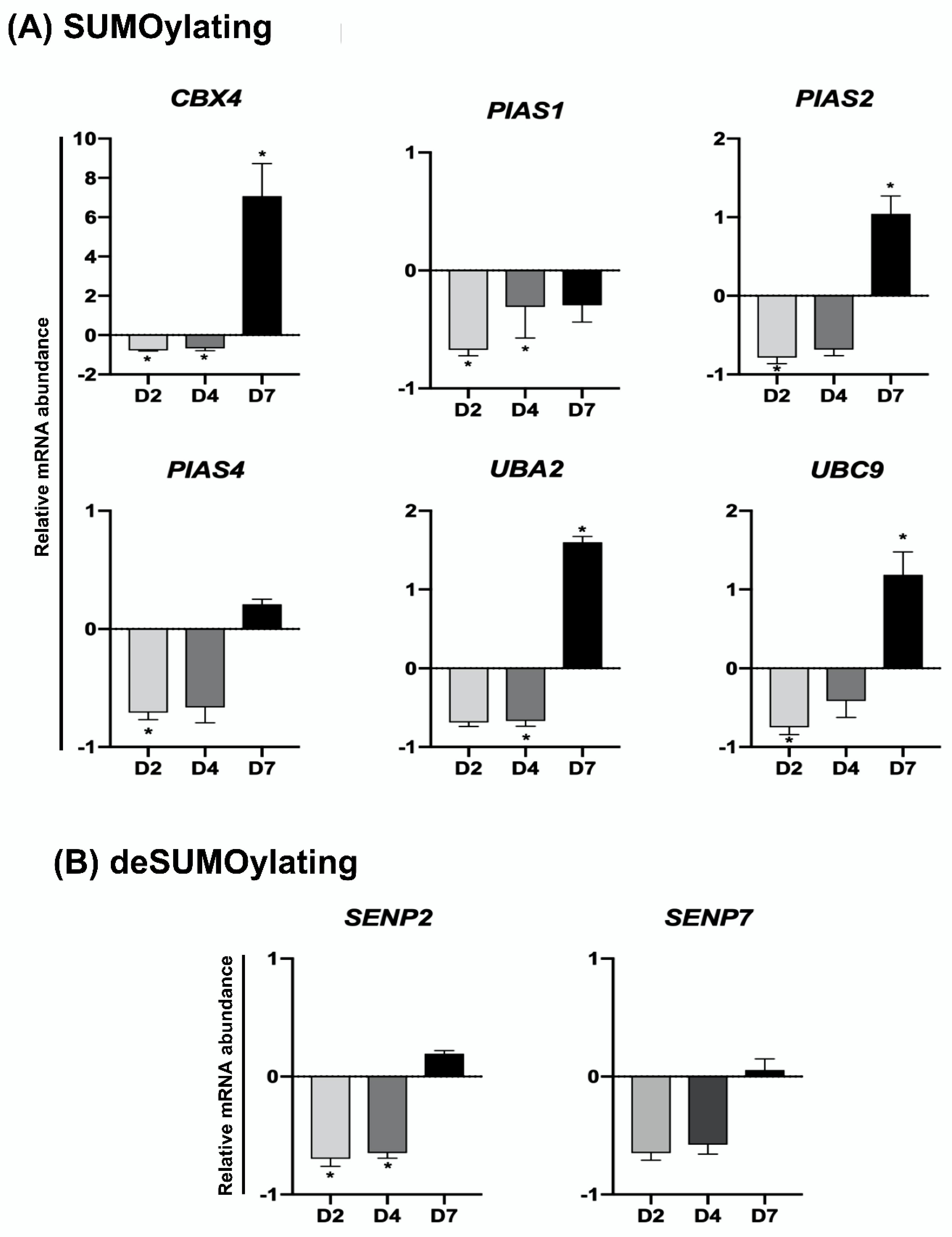
2.3. UV-Induced DNA Damage Alters mRNA Levels of UBs, DUBs, SUMOs and deSUMOs during Embryo Development
3. Discussion
4. Material and Methods
4.1. In Vitro Embryo Production
4.2. Ultraviolet Treatment
4.3. RNA Extraction and Quantitative Real-Time PCR (qPCR)
4.4. Immunofluorescence Detection of DNA Breaks
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Telford, N.A.; Watson, A.J.; Schultz, G.A. Transition from maternal to embryonic control in early mammalian development: A comparison of several species. Mol. Reprod. Dev. 1990, 26, 90–100. [Google Scholar] [CrossRef] [PubMed]
- Tadros, W.; Lipshitz, H.D. The maternal-to-zygotic transition: A play in two acts. Development 2009, 136, 3033–3042. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Lu, X.; Dean, J. The maternal to zygotic transition in mammals. Mol. Asp. Med. 2013, 34, 919–938. [Google Scholar] [CrossRef] [PubMed]
- Minami, N.; Suzuki, T.; Tsukamoto, S. Zygotic gene activation and maternal factors in mammals. J. Reprod. Dev. 2007, 53, 707–715. [Google Scholar] [CrossRef]
- Madeja, Z.E.; Pawlak, P.; Piliszek, A. Beyond the mouse: Non-rodent animal models for study of early mammalian development and biomedical research. Int. J. Dev. Biol. 2019, 63, 187–201. [Google Scholar] [CrossRef]
- Bohrer, R.C.; Dicks, N.; Gutierrez, K.; Duggavathi, R.; Bordignon, V. Double-strand DNA breaks are mainly repaired by the homologous recombination pathway in early developing swine embryos. FASEB J. 2018, 32, 1818–1829. [Google Scholar] [CrossRef] [PubMed]
- Gupta, D.; Beisel, C.L. Illuminating the path to DNA repair. Cell 2021, 184, 5503–5505. [Google Scholar] [CrossRef]
- Takatsuka, H.; Shibata, A.; Umeda, M. Genome Maintenance Mechanisms at the Chromatin Level. Int. J. Mol. Sci. 2021, 22, 10384. [Google Scholar] [CrossRef]
- Rich, T.; Allen, R.L.; Wyllie, A.H. Defying death after DNA damage. Nature 2000, 407, 777–783. [Google Scholar] [CrossRef]
- Mjelle, R.; Hegre, S.A.; Aas, P.A.; Slupphaug, G.; Drablos, F.; Saetrom, P.; Krokan, H.E. Cell cycle regulation of human DNA repair and chromatin remodeling genes. DNA Repair 2015, 30, 53–67. [Google Scholar] [CrossRef] [Green Version]
- Bétermier, M.; Borde, V.; de Villartay, J.-P. Coupling DNA Damage and Repair: An Essential Safeguard during Programmed DNA Double-Strand Breaks? Trends Cell Biol. 2020, 30, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Rogakou, E.P.; Pilch, D.R.; Orr, A.H.; Ivanova, V.S.; Bonner, W.M. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998, 273, 5858–5868. [Google Scholar] [CrossRef] [PubMed]
- Jackson, S.P.; Durocher, D. Regulation of DNA Damage Responses by Ubiquitin and SUMO. Mol. Cell 2013, 49, 795–807. [Google Scholar] [CrossRef] [PubMed]
- Scheffner, M.; Nuber, U.; Huibregtse, J.M. Protein ubiquitination involving an E1-E2-E3 enzyme ubiquitin thioester cascade. Nature 1995, 373, 81–83. [Google Scholar] [CrossRef] [PubMed]
- Eifler, K.; Vertegaal, A.C. Mapping the SUMOylated landscape. FEBS J. 2015, 282, 3669–3680. [Google Scholar] [CrossRef] [PubMed]
- Lange, S.M.; Armstrong, L.A.; Kulathu, Y. Deubiquitinases: From mechanisms to their inhibition by small molecules. Mol. Cell 2022, 82, 15–29. [Google Scholar] [CrossRef]
- Yeh, E.T.H. SUMOylation and De-SUMOylation: Wrestling with Life’s Processes. J. Biol. Chem. 2009, 284, 8223–8227. [Google Scholar] [CrossRef] [PubMed]
- Bergink, S.; Jaspers, N.G.J.; Vermeulen, W. Regulation of UV-induced DNA damage response by ubiquitylation. DNA Repair 2007, 6, 1231–1242. [Google Scholar] [CrossRef] [PubMed]
- Cruz Walma, D.A.; Chen, Z.; Bullock, A.N.; Yamada, K.M. Ubiquitin ligases: Guardians of mammalian development. Nat. Rev. Mol. Cell Biol. 2022, 23, 350–367. [Google Scholar] [CrossRef]
- Yang, Y.; Zhou, C.; Wang, Y.; Liu, W.; Liu, C.; Wang, L.; Liu, Y.; Shang, Y.; Li, M.; Zhou, S.; et al. The E3 ubiquitin ligase RNF114 and TAB1 degradation are required for maternal-to-zygotic transition. EMBO Rep. 2017, 18, 205–216. [Google Scholar] [CrossRef] [Green Version]
- Higuchi, C.; Shimizu, N.; Shin, S.W.; Morita, K.; Nagai, K.; Anzai, M.; Kato, H.; Mitani, T.; Yamagata, K.; Hosoi, Y.; et al. Ubiquitin-proteasome system modulates zygotic genome activation in early mouse embryos and influences full-term development. J. Reprod. Dev. 2018, 64, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Zhou, J.; Liu, P.; Hu, J.; Jin, H.; Shimono, Y.; Takahashi, M.; Xu, G. Polycomb protein Cbx4 promotes SUMO modification of de novo DNA methyltransferase Dnmt3a. Biochem. J. 2007, 405, 369–378. [Google Scholar] [CrossRef] [PubMed]
- Baek, K.-H.; Choi, J.; Pei, C.-Z. Cellular Functions of OCT-3/4 Regulated by Ubiquitination in Proliferating Cells. Cancers 2020, 12, 663. [Google Scholar] [CrossRef] [PubMed]
- Glanzner, W.G.; Gutierrez, K.; Rissi, V.B.; de Macedo, M.P.; Lopez, R.; Currin, L.; Dicks, N.; Baldassarre, H.; Agellon, L.B.; Bordignon, V. Histone Lysine Demethylases KDM5B and KDM5C Modulate Genome Activation and Stability in Porcine Embryos. Front. Cell Dev. Biol. 2020, 8, 151. [Google Scholar] [CrossRef]
- Glanzner, W.G.; Rissi, V.B.; de Macedo, M.P.; Mujica, L.K.S.; Gutierrez, K.; Bridi, A.; de Souza, J.R.M.; Gonçalves, P.B.D.; Bordignon, V. Histone 3 lysine 4, 9, and 27 demethylases expression profile in fertilized and cloned bovine and porcine embryos. Biol. Reprod. 2018, 98, 742–751. [Google Scholar] [CrossRef]
- Hirvonen-Santti, S.J.; Sriraman, V.; Anttonen, M.; Savolainen, S.; Palvimo, J.J.; Heikinheimo, M.; Richards, J.S.; Jänne, O.A. Small Nuclear RING Finger Protein Expression during Gonad Development: Regulation by Gonadotropins and Estrogen in the Postnatal Ovary. Endocrinology 2004, 145, 2433–2444. [Google Scholar] [CrossRef]
- Yamaguchi, L.; Nishiyama, A.; Misaki, T.; Johmura, Y.; Ueda, J.; Arita, K.; Nagao, K.; Obuse, C.; Nakanishi, M. Usp7-dependent histone H3 deubiquitylation regulates maintenance of DNA methylation. Sci. Rep. 2017, 7, 55. [Google Scholar] [CrossRef]
- Yan, Y.-L.; Zhang, C.; Hao, J.; Wang, X.-L.; Ming, J.; Mi, L.; Na, J.; Hu, X.; Wang, Y. DPPA2/4 and SUMO E3 ligase PIAS4 opposingly regulate zygotic transcriptional program. PLoS Biol. 2019, 17, e3000324. [Google Scholar] [CrossRef]
- Wang, Z.-B.; Ou, X.-H.; Tong, J.-S.; Li, S.; Wei, L.; Ouyang, Y.-C.; Hou, Y.; Schatten, H.; Sun, Q.-Y. The SUMO pathway functions in mouse oocyte maturation. Cell Cycle 2010, 9, 2640–2646. [Google Scholar] [CrossRef]
- Chiu, S.-Y.; Asai, N.; Costantini, F.; Hsu, W. SUMO-Specific Protease 2 Is Essential for Modulating p53-Mdm2 in Development of Trophoblast Stem Cell Niches and Lineages. PLoS Biol. 2008, 6, e310. [Google Scholar] [CrossRef]
- Huang, C.J.; Wu, D.; Jiao, X.F.; Khan, F.A.; Xiong, C.L.; Liu, X.M.; Yang, J.; Yin, T.L.; Huo, L.J. Maternal SENP7 programs meiosis architecture and embryo survival in mouse. Biochim. Biophys. Acta-Mol. Cell Res. 2017, 1864, 1195–1206. [Google Scholar] [CrossRef]
- Romeo, K.; Louault, Y.; Cantaloube, S.; Loiodice, I.; Almouzni, G.; Quivy, J.-P. The SENP7 SUMO-Protease Presents a Module of Two HP1 Interaction Motifs that Locks HP1 Protein at Pericentric Heterochromatin. Cell Rep. 2015, 10, 771–782. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Guo, Y.; Sun, H.; Liu, L.; Yao, L.; Liu, C.; He, Y.; Cao, S.; Zhou, C.; Li, M.; et al. Maternal RNF114-mediated target substrate degradation regulates zygotic genome activation in mouse embryos. Development 2021, 148, dev199426. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.L.; Zhao, L.W.; Zhang, J.; Le, R.; Ji, S.Y.; Chen, C.; Gao, Y.; Li, D.; Gao, S.; Fan, H.Y. DCAF13 promotes pluripotency by negatively regulating SUV39H1 stability during early embryonic development. EMBO J. 2018, 37, e98981. [Google Scholar] [CrossRef]
- Rong, Y.; Zhu, Y.-Z.; Yu, J.-l.; Wu, Y.-W.; Ji, S.-Y.; Zhou, Y.; Jiang, Y.; Jin, J.; Fan, H.-Y.; Shen, L.; et al. USP16-mediated histone H2A lysine-119 deubiquitination during oocyte maturation is a prerequisite for zygotic genome activation. Nucleic Acids Res. 2022, 50, 5599–5616. [Google Scholar] [CrossRef]
- Portney, B.A.; Khatri, R.; Meltzer, W.A.; Mariano, J.M.; Zalzman, M. ZSCAN4 is negatively regulated by the ubiquitin-proteasome system and the E3 ubiquitin ligase RNF20. Biochem. Biophys. Res. Commun. 2018, 498, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Falco, G.; Lee, S.L.; Stanghellini, I.; Bassey, U.C.; Hamatani, T.; Ko, M.S. Zscan4: A novel gene expressed exclusively in late 2-cell embryos and embryonic stem cells. Dev. Biol. 2007, 307, 539–550. [Google Scholar] [CrossRef]
- Hino, K.; Simó, S.; Cooper, J.A. Comparative Analysis of cul5 and rbx2 Expression in the Developing and Adult Murine Brain and Their Essentiality during Mouse Embryogenesis. Dev. Dyn. 2018, 247, 1227–1236. [Google Scholar] [CrossRef]
- Xu, Y.W.; Cao, L.R.; Wang, M.; Xu, Y.; Wu, X.; Liu, J.; Tong, C.; Fan, H.Y. Maternal DCAF2 is crucial for maintenance of genome stability during the first cell cycle in mice. J. Cell Sci. 2017, 130, 3297–3307. [Google Scholar] [CrossRef]
- Yu, C.; Zhang, Y.L.; Pan, W.W.; Li, X.M.; Wang, Z.W.; Ge, Z.J.; Zhou, J.J.; Cang, Y.; Tong, C.; Sun, Q.Y.; et al. CRL4 complex regulates mammalian oocyte survival and reprogramming by activation of TET proteins. Science 2013, 342, 1518–1521. [Google Scholar] [CrossRef]
- Yu, C.; Ji, S.-Y.; Sha, Q.-Q.; Sun, Q.-Y.; Fan, H.-Y. CRL4–DCAF1 ubiquitin E3 ligase directs protein phosphatase 2A degradation to control oocyte meiotic maturation. Nat. Commun. 2015, 6, 8017. [Google Scholar] [CrossRef] [PubMed]
- Nacerddine, K.; Lehembre, F.; Bhaumik, M.; Artus, J.; Cohen-Tannoudji, M.; Babinet, C.; Pandolfi, P.P.; Dejean, A. The SUMO pathway is essential for nuclear integrity and chromosome segregation in mice. Dev. Cell 2005, 9, 769–779. [Google Scholar] [CrossRef]
- Xu, D.; Sun, F.; Bi, J.; Guan, Y.; Luo, X.; Chen, X.; Lv, Y.; Jin, Y. Effects of E2 binding enzyme UBC9 on porcine oocyte maturation, apoptosis and embryo development. Reprod. Domest. Anim. 2020, 55, 1526–1534. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Bi, J.; Guan, Y.; Luo, X.; Chen, X.; Lv, Y.; Jin, Y. Effects of the E1 activating enzyme UBA2 on porcine oocyte maturation, apoptosis, and embryonic development in vitro. Anim. Sci. J. 2021, 92, e13548. [Google Scholar] [CrossRef]
- Bartocci, C.; Lazzerini Denchi, E. Put a RING on it: Regulation and inhibition of RNF8 and RNF168 RING finger E3 ligases at DNA damage sites. Front. Genet. 2013, 4, 128. [Google Scholar] [CrossRef] [PubMed]
- Walser, F.; Mulder, M.P.C.; Bragantini, B.; Burger, S.; Gubser, T.; Gatti, M.; Botuyan, M.V.; Villa, A.; Altmeyer, M.; Neri, D.; et al. Ubiquitin Phosphorylation at Thr12 Modulates the DNA Damage Response. Mol. Cell 2020, 80, 423–436.e9. [Google Scholar] [CrossRef] [PubMed]
- Mattiroli, F.; Vissers, J.H.A.; van Dijk, W.J.; Ikpa, P.; Citterio, E.; Vermeulen, W.; Marteijn, J.A.; Sixma, T.K. RNF168 Ubiquitinates K13-15 on H2A/H2AX to Drive DNA Damage Signaling. Cell 2012, 150, 1182–1195. [Google Scholar] [CrossRef] [PubMed]
- Barreta, M.H.; Gasperin, B.G.; Rissi, V.B.; de Cesaro, M.P.; Ferreira, R.; de Oliveira, J.F.; Gonçalves, P.B.D.; Bordignon, V. Homologous recombination and non-homologous end-joining repair pathways in bovine embryos with different developmental competence. Exp. Cell Res. 2012, 318, 2049–2058. [Google Scholar] [CrossRef]
- Foglizzo, M.; Middleton, A.J.; Day, C.L. Structure and Function of the RING Domains of RNF20 and RNF40, Dimeric E3 Ligases that Monoubiquitylate Histone H2B. J. Mol. Biol. 2016, 428, 4073–4086. [Google Scholar] [CrossRef]
- Yu, M.; Liu, K.; Mao, Z.; Luo, J.; Gu, W.; Zhao, W. USP11 Is a Negative Regulator to γH2AX Ubiquitylation by RNF8/RNF168. J. Biol. Chem. 2016, 291, 959–967. [Google Scholar] [CrossRef] [Green Version]
- Han, M.M.; Hirakawa, M.; Yamauchi, M.; Matsuda, N. Roles of the SUMO-related enzymes, PIAS1, PIAS4, and RNF4, in DNA double-strand break repair by homologous recombination. Biochem. Biophys. Res. Commun. 2022, 591, 95–101. [Google Scholar] [CrossRef] [PubMed]
- Sy, S.M.; Jiang, J.; O, W.S.; Deng, Y.; Huen, M.S. The ubiquitin specific protease USP34 promotes ubiquitin signaling at DNA double-strand breaks. Nucleic Acids Res. 2013, 41, 8572–8580. [Google Scholar] [CrossRef] [PubMed]
- Garvin, A.J.; Walker, A.K.; Densham, R.M.; Chauhan, A.S.; Stone, H.R.; Mackay, H.L.; Jamshad, M.; Starowicz, K.; Daza-Martin, M.; Ronson, G.E.; et al. The deSUMOylase SENP2 coordinates homologous recombination and nonhomologous end joining by independent mechanisms. Genes Dev. 2019, 33, 333–347. [Google Scholar] [CrossRef] [PubMed]
- Galanty, Y.; Belotserkovskaya, R.; Coates, J.; Jackson, S.P. RNF4, a SUMO-targeted ubiquitin E3 ligase, promotes DNA double-strand break repair. Genes Dev. 2012, 26, 1179–1195. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Deng, O.; Feng, Z.; Du, Z.; Xiong, X.; Lai, J.; Yang, X.; Xu, M.; Wang, H.; Taylor, D.; et al. RNF126 promotes homologous recombination via regulation of E2F1-mediated BRCA1 expression. Oncogene 2016, 35, 1363–1372. [Google Scholar] [CrossRef] [PubMed]
- Wan, Q.; Chen, Q.; Cai, D.; Zhao, Y.; Wu, X. OTUB2 Promotes Homologous Recombination Repair Through Stimulating Rad51 Expression in Endometrial Cancer. Cell Transpl. 2020, 29, 963689720931433. [Google Scholar] [CrossRef]
- Swift, M.L.; Azizkhan-Clifford, J. DNA damage-induced sumoylation of Sp1 induces its interaction with RNF4 and degradation in S phase to remove 53BP1 from DSBs and permit HR. DNA Repair 2022, 111, 103289. [Google Scholar] [CrossRef]
- Ng, H.M.; Wei, L.; Lan, L.; Huen, M.S. The Lys63-deubiquitylating Enzyme BRCC36 Limits DNA Break Processing and Repair. J. Biol. Chem. 2016, 291, 16197–16207. [Google Scholar] [CrossRef]
- Kato, K.; Nakajima, K.; Ui, A.; Muto-Terao, Y.; Ogiwara, H.; Nakada, S. Fine-tuning of DNA damage-dependent ubiquitination by OTUB2 supports the DNA repair pathway choice. Mol. Cell 2014, 53, 617–630. [Google Scholar] [CrossRef] [Green Version]

| Stage of Development | UV Exposure | Cultured Embryos | Blastocyst n (%) |
|---|---|---|---|
| Day 2 | − | 103 | 22 (21.36) a |
| + | 126 | 6 (4.76) b | |
| Day 4 | − | 150 | 33 (22) a |
| + | 156 | 7 (4.49) b | |
| Day 7 (blastocysts) | − | 39 | 39 (100) |
| + | 39 | 39 (100) |
| Gene Name | Forward (5′ to 3′) | Reverse (5′ to 3′) | Reference or Accession Number |
|---|---|---|---|
| BRCC3 | TTGAGCTGCCCAAGATCCTC | CGGACATCTGACTGCACAGA | a |
| CUL5 | CCCAAAGCTCAAACGGCAAG | AGTGAGCTGTAAGCGTCCAA | b |
| DCAF2 | GCTGCCTATATCTGGAAGGTCT | ACCTCTTGCGAATGACCCAG | XM_013979970.2 |
| DCAF13 | TTCCTTGCTTCCCTGGATGG | CGTACAAAACCTTCATGCGC | XM_005662912.3 |
| DDB1 | GAACGAAAGACTGAACCGGC | GCCATCGTCGTACTGTAGGT | XM_003122651.6 |
| RNF4 | GTGGACCTGACGCACAATGA | CGTCATCACTGCTCACCACA | c |
| RNF8 | AGAGGACCTGAAGCAACAGC | GTCCTTCCTGCTGCGGTTTA | d |
| RNF20 | ACATTGGCACAGGGGAGAAG | TGCCTCCATTGCCTTACGTT | XM_001926594 |
| RNF40 | CCTTCAGGAGAAGCATCGCA | ACAGACACTCTTGACTCCGC | e |
| RNF114 | GTCCATAGCACGGACACCAA | CCGACGCTGGATGTGTTCTA | NM_001001869.1 |
| RNF126 | CTTGGATGCCATCATCACGC | GGAGGGCCTGGATTTTCTCTT | f |
| RNF168 | GTGTCTTGGCATTGCCCTTG | CAGACTGTGATGCTGACCCA | g |
| RNF169 | GTTGTCCTGCACGTCTCTCA | CCGACCGGTGTGTCTGTATC | h |
| UBE2N | GCCGTACTCGGGATTTGACA | GAGCGTTGCTCTCATCTGGT | XM_005664262.3 |
| USP4 | TAAATACGATGCCGGGGAGC | GCTGTCGAAGCCCACATACT | i |
| USP7 | TAAATACGATGCCGGGGAGC | GTTTTGGTCCGTCTGAGGGT | j |
| USP11 | ATTCGCTCGACCTCGTCTTA | TCTGCGTTCTTGATCCACAG | XM_003484105.2 |
| USP16 | GCCAAGATTGTAAGACGGACA | AGCCCTGATGGCCACATTTA | k |
| USP34 | GCAAGTTTGTTGCTGCTGGA | TGCCACACAGTCCAGGATTC | l |
| OTUB1 | AGTATGCCGAGGACGACAAC | TGGTCTTGCGGATGTACGAG | NM_001162403.1 |
| OTUB2 | TCAGCCTTCATCAGGAACCG | GGCTGTGATCTGGATGTGGT | XM_001925368.4 |
| BAP1 | TTCAGCCCTGAGAGCAAAGG | GGGCCTGGCATGGCTATTAT | m |
| PIAS1 | TCAGCTCTTCACCCAGTCCA | AGCGCTGACTGTTGTCTGAT | XM_003121749.6 |
| PIAS2 | CACAAAGCAGCCCAACCAAA | ATGAAGGCGGAATAGCAGCA | n |
| PIAS4 | AGAGCGGACTCAAACACGAA | GGTGTGGGTTCTGAGCTCTT | XM_003354007.4 |
| CBX4 | ATCGCCTTCCAGAACAGGGAA | ATTGGAACGACGCGCAAAG | XM_003131145.4 |
| UBA2 | GGAGCCGACTTCAAGCAGAT | TGGGGCATCACCAACAACTT | XM_021097539.1 |
| UBC9 | CAAGCAGAGGCCTACACGAT | AAGGTCGCTGCTTATGAGGG | o |
| SENP2 | TTCTGAAGAGGGTGGCAAGG | GGAGGAACGGAGTTTCCATGA | p |
| SENP7 | CCCATTTCAAGTGTCCCTGC | AGGCAACCCAAAGAAAGAGGA | q |
| RAD51 | CGGTGGAAGAGGAGAGCTTTG | TTTAGCTGCCTCGGTCAGAAT | [24] |
| BRCA1 | CTTCTGTGGTGAAGGACCCC | TCACATGGAAGCCACTGTCC | [24] |
| KU70 | TTCAAGCCCTTGGGAATGCT | CTTGGTGAGCAGAGCAGTGA | [24] |
| KU80 | TTCCTGAGAGCCCTTCGAGA | TTTGGGCTTCCTCGACTGTG | [24] |
| EIF1AX | ACACCTCCCCGATAGGAGTC | TTGAGCACACTCTTGCCCAT | [24] |
| KDM5B | GACGTGTGCCAGTTTTGGAC | TCGAGGACACAGCACCTCTA | [24] |
| H2A | GGTGCTGGAGTATCTGACCG | GTTGAGCTCTTCGTCGTTGC | [24] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
da Silva, Z.; Glanzner, W.G.; Currin, L.; de Macedo, M.P.; Gutierrez, K.; Guay, V.; Gonçalves, P.B.D.; Bordignon, V. DNA Damage Induction Alters the Expression of Ubiquitin and SUMO Regulators in Preimplantation Stage Pig Embryos. Int. J. Mol. Sci. 2022, 23, 9610. https://doi.org/10.3390/ijms23179610
da Silva Z, Glanzner WG, Currin L, de Macedo MP, Gutierrez K, Guay V, Gonçalves PBD, Bordignon V. DNA Damage Induction Alters the Expression of Ubiquitin and SUMO Regulators in Preimplantation Stage Pig Embryos. International Journal of Molecular Sciences. 2022; 23(17):9610. https://doi.org/10.3390/ijms23179610
Chicago/Turabian Styleda Silva, Zigomar, Werner Giehl Glanzner, Luke Currin, Mariana Priotto de Macedo, Karina Gutierrez, Vanessa Guay, Paulo Bayard Dias Gonçalves, and Vilceu Bordignon. 2022. "DNA Damage Induction Alters the Expression of Ubiquitin and SUMO Regulators in Preimplantation Stage Pig Embryos" International Journal of Molecular Sciences 23, no. 17: 9610. https://doi.org/10.3390/ijms23179610
APA Styleda Silva, Z., Glanzner, W. G., Currin, L., de Macedo, M. P., Gutierrez, K., Guay, V., Gonçalves, P. B. D., & Bordignon, V. (2022). DNA Damage Induction Alters the Expression of Ubiquitin and SUMO Regulators in Preimplantation Stage Pig Embryos. International Journal of Molecular Sciences, 23(17), 9610. https://doi.org/10.3390/ijms23179610






