“Omic” Approaches to Bacteria and Antibiotic Resistance Identification
Abstract
1. Introduction
2. MALDI-TOF MS Technique
3. Bacteria Identification
3.1. Proteomic
3.1.1. Database
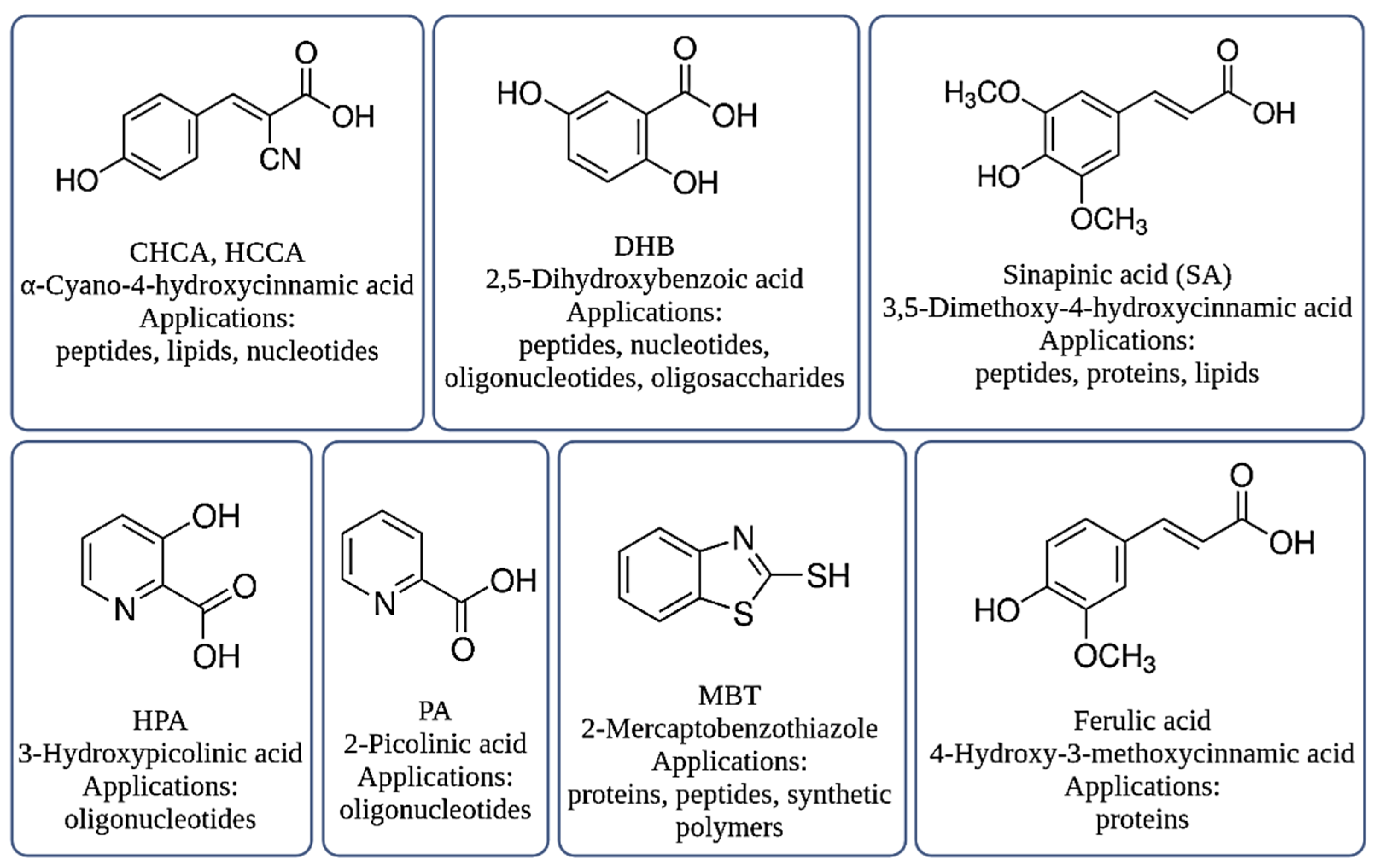
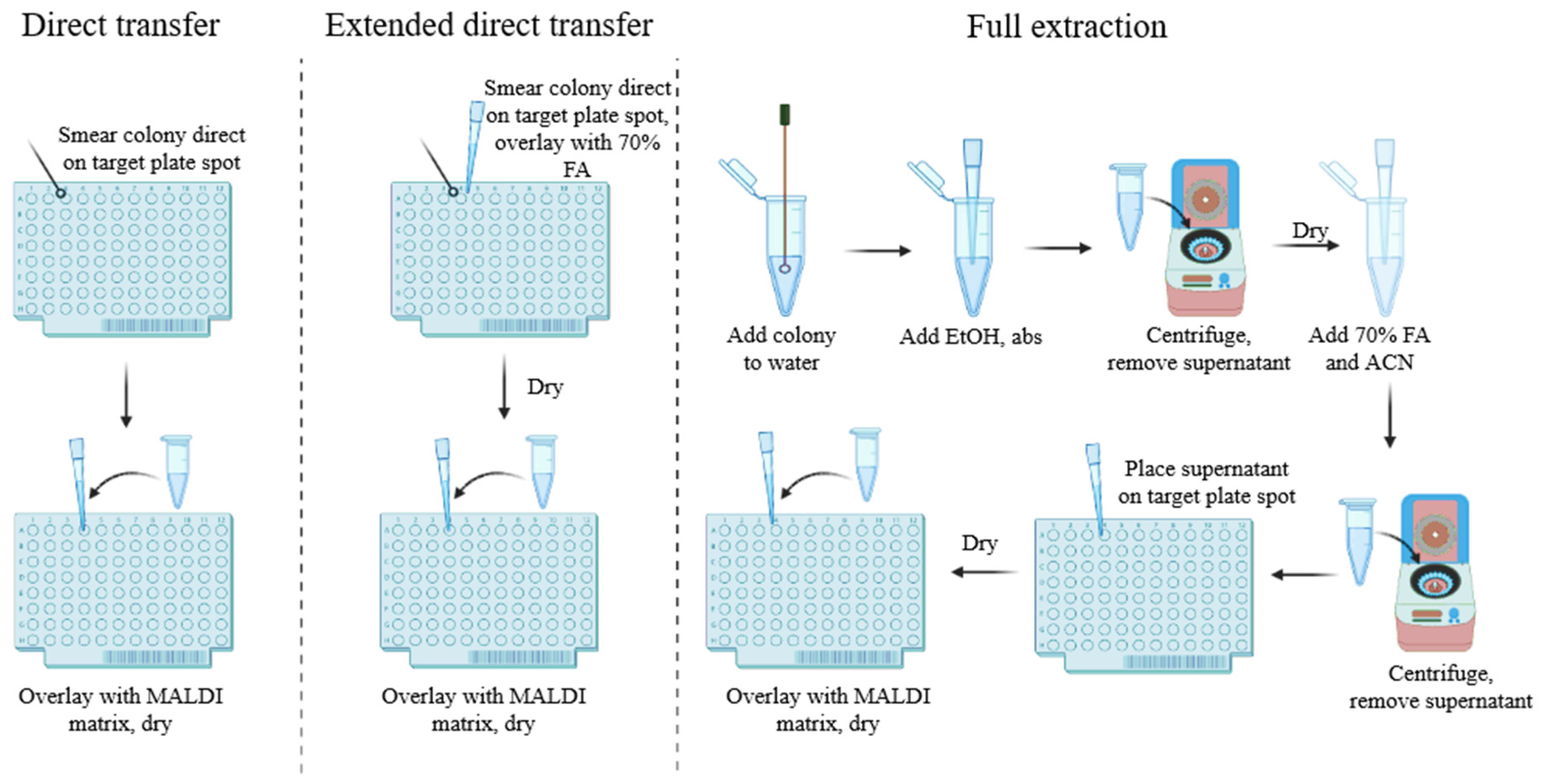
3.1.2. Sample Preparation
3.1.3. Identification Problems
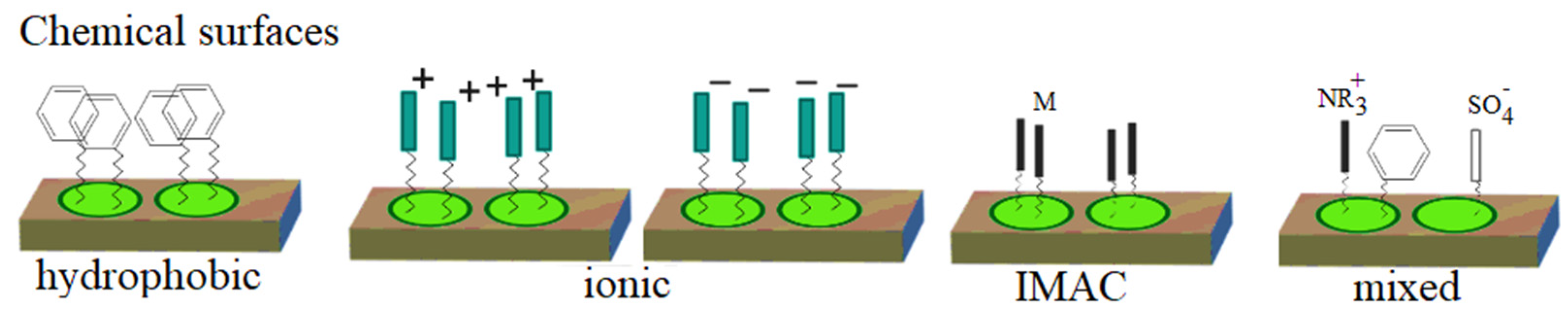
3.1.4. ProteinChip Arrays
3.2. Lipidomic
3.3. Metabolomic
3.4. Genomic
4. Antibiotic Resistance
4.1. Proteomics
4.2. Lipidomic
4.3. Metabolomic
4.4. Genomic
5. Biofilm and Development of Antibiotic Resistance
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kaplan, W.; Laing, R. Priority Medicines for Europe and the World; World Health Organization Department of Essential Drugs and Medicines Policy: Geneva, Switzerland, 2004; pp. 1–134. [Google Scholar]
- Rappuoli, R.; Bloom, D.E.; Black, S. Deploy vaccines to fight superbugs. Nature 2017, 552, 165–167. [Google Scholar] [CrossRef] [PubMed]
- Cassini, A.; Högberg, L.D.; Plachouras, D.; Quattrocchi, A.; Hoxha, A.; Simonsen, G.S.; Colobm-Cotinat, M.; Kretzschmar, M.E.; Devleesschaower, B.; Cecchini, M.; et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: A population-level modelling analysis. Lancet Infect. Dis. 2019, 19, 55–66. [Google Scholar] [CrossRef]
- Pauter, K.; Szultka-Młyńska, M.; Buszewski, B. Determination and identification of antibiotic drugs and bacterial strains in biological samples. Molecules 2020, 25, 2556. [Google Scholar] [CrossRef] [PubMed]
- Park, S.H.; Aydin, M.; Khatiwara, A.; Dolan, M.C.; Gilmore, D.F.; Bouldin, J.L.; Ahn, S.; Ricke, S.C. Current and emerging technologies for rapid detection and characterization of Salmonella in poultry and poultry products. Food Microbiol. 2013, 38, 250–262. [Google Scholar] [CrossRef]
- Omiccioli, E.; Amagliani, G.; Brandi, G.; Magnani, M. A new platform for Real-Time PCR detection of Salmonella spp.; Listeria monocytogenes and Escherichia coli O157 in milk. Food Microbiol. 2009, 26, 615–622. [Google Scholar] [CrossRef]
- Tewari, A.; Abdullah, S. Bacillus cereus food poisoning: International and Indian perspective. J. Food Sci. Technol. 2015, 52, 2500. [Google Scholar] [CrossRef]
- Zhao, X.; Lin, C.W.; Wang, J.; Oh, D.H. Advances in rapid detection methods for foodborne pathogens. J. Microbiol. Biotechnol. 2014, 24, 297–312. [Google Scholar] [CrossRef]
- Złoch, M.; Maślak, E.; Kupczyk, W.; Jackowski, M.; Pomastowski, P.; Buszewski, B. Culturomics approach to identify diabetic foot infection bacteria. Int. J. Mol. Sci. 2021, 22, 9574. [Google Scholar] [CrossRef]
- Fan, S.L.; Miller, N.S.; Lee, J.; Remick, D.G. Diagnosing sepsis—The role of laboratory medicine. Clin. Chim. Acta 2016, 460, 203–210. [Google Scholar] [CrossRef]
- Unemo, M.; Golparian, D.; Eyre, D.W. Antimicrobial resistance in Neisseria gonorrhoeae and treatment of gonorrhea. Methods Mol. Biol. 2019, 1997, 37–58. [Google Scholar]
- Karas, M.; Bachmann, D.; Hillenkamp, F. Influence of the Wavelength in High-Irradiance Ultraviolet Laser Desorption Mass Spectrometry of organic molecules. Anal. Chem. 1985, 57, 2935–2939. [Google Scholar] [CrossRef]
- Tanaka, K.; Waki, H.; Ido, Y.; Akita, S.; Yoshida, Y.; Yoshida, T. Protein and polymer analyses up to m/z 100,000 by laser ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 1988, 2, 151–153. [Google Scholar] [CrossRef]
- Griffiths, J. A Brief history of mass spectrometry. Anal. Chem. 2008, 80, 5678–5683. [Google Scholar] [CrossRef]
- Gao, J.; Cassady, C.J. Negative ion production from peptides and proteins bymatrix-assisted laser desorption/ionization time-of-flightmass spectrometry. Rapid Commun. Mass Spectrom. 2008, 22, 4066–4072. [Google Scholar] [CrossRef]
- Boesl, U. Time-of-flight mass spectrometry: Introduction to the basics. Mass Spectrom. Rev. 2016, 36, 86–109. [Google Scholar] [CrossRef]
- Fenselau, C.; Demirev, F.A. Characterization of intact microorganisms by MALDI mass spectrometry. Mass Spectrom. Rev. 2001, 20, 157–171. [Google Scholar] [CrossRef]
- Lay, J.O. MALDI-TOF mass spectrometry of bacteria. Mass Spectrom. Rev. 2001, 20, 172–194. [Google Scholar] [CrossRef]
- Faqerquist, C.K.; Garbus, B.R.; Williams, K.E.; Bares, A.H.; Harden, L.A. Covalent attachment and dissociative loss of sinapinic acid to/from cysteine-containing proteins from bacterial cell lysates analyzed by MALDI-TOF-TOF mass spectrometry. J. Am. Soc. Mass Spectrom. 2010, 21, 819–832. [Google Scholar] [CrossRef][Green Version]
- Šedo, O.; Nemec, A.; Krizova, L.; Kačalová, M.; Zdráhalab, Z. Improvement of MALDI-TOF MS profiling for the differentiation of species within the Acinetobacter calcoaceticus—Acinetobacter baumannii complex. Syst. Appl. Microbiol. 2013, 36, 572–578. [Google Scholar] [CrossRef]
- Harvey, D.J. Analysis of carbohydrates and glycoconjugates by matrix-assisted laser desorption/ionization mass spectrometry: An update for 2003–2004. Mass Spectrom. Rev. 2009, 28, 273–361. [Google Scholar] [CrossRef]
- Luxembourg, S.L.; McDonnell, L.A.; Duursma, M.C.; Guo, X.; Heeren, R.M.A. Effect of local matrix crystal variations in matrix-assisted ionization techniques for mass spectrometry. Anal. Chem. 2003, 75, 2333–2341. [Google Scholar] [CrossRef]
- Shu, X.; Liang, M.; Yang, B.; Li, Y.; Liu, C.; Wang, Y.; Shu, J. Lipid fingerprinting of Bacillus spp. using online MALDI-TOF mass spectrometry. Anal. Methods 2012, 4, 3111–3117. [Google Scholar] [CrossRef]
- Xu, N.; Huang, Z.-H.; de Jonge, B.L.M.; Cage, D.A. Structural characterization of peptidoglycan muropeptides by Matrix-Assisted Laser Desorption Ionization Mass Spectrometry and postsource decay analysis. Anal. Biochem. 1997, 248, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Altman, E.; Perry, M.B.; Li, J. Study of matrix additives for sensitive analysis of lipid a by matrix-assisted laser desorption ionization mass spectrometry. Appl. Environ. Microbiol. 2010, 76, 3437–3443. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Schilling, B.; McLendon, M.K.; Phillips, N.J.; Apicella, M.A.; Gibson, B.W. Characterization of lipid A acylation patterns in Francisella tularensis, Francisella novicida, and Francisella philomiragia using multiple-stage mass spectrometry and matrix-assisted laser desorption/ionization on an intermediate vacuum source linear ion trap. Anal. Chem. 2007, 79, 1034–1042. [Google Scholar] [PubMed]
- Liu, H.; Du, Z.; Wang, J.; Yang, R. Universal sample preparation method for characterization of bacteria by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Appl. Environ. Microbiol. 2007, 73, 1899–1907. [Google Scholar] [CrossRef] [PubMed]
- Horneffer, V.; Haverkamp, J.; Janssen, H.G.; Notz, R. MALDI-TOF-MS analysis of bacterial spores: Wet heat-treatment as a new releasing technique for biomarkers and the influence of different experimental parameters and microbiological handling. J. Am. Soc. Mass Spectrom. 2004, 15, 1444–1454. [Google Scholar] [CrossRef] [PubMed]
- Elhanany, E.; Barak, R.; Fisher, M.; Kobiler, D.; Altboum, Z. Detection of specific Bacillus anthracis spore biomarkers by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 2001, 15, 2110–2116. [Google Scholar] [CrossRef]
- Amiri-Eliasi, B.; Fenselau, C. Characterization of protein biomarkers desorbed by MALDI from whole fungal cells. Anal. Chem. 2001, 73, 5228–5231. [Google Scholar] [CrossRef]
- Valentine, N.B.; Wahl, J.H.; Kingsley, M.T.; Wahl, K.L. Direct surface analysis of fungal species by matrix-assisted laser desorption/ionization mass spectrometry. Rapid Commun. Mass Spectrom. 2002, 16, 1352–1357. [Google Scholar] [CrossRef]
- Li, T.-Y.; Liu, B.-H.; Chen, Y.-C. Characterization of Aspergillus spores by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 2000, 14, 2393–2400. [Google Scholar] [CrossRef]
- Bright, J.J.; Claydon, M.A.; Soufian, M.; Gordon, D.B. Rapid typing of bacteria using matrix-assisted laser desorption ionisation time-of-flight mass spectrometry and pattern recognition software. J. Microbiol. Methods 2002, 48, 127–138. [Google Scholar] [CrossRef]
- Evason, D.J.; Claydon, M.A.; Gordon, D.B. Effects of ion mode and matrix additives in the identification of bacteria by intact cell mass spectrometry. Rapid Commun. Mass Spectrom. 2000, 14, 669–672. [Google Scholar] [CrossRef]
- Williams, T.L.; Andrzejewski, D.; Lay, J.O.; Musser, S.M. Experimental factors affecting the quality and reproducibility of MALDI TOF mass spectra obtained from whole bacteria cells. J. Am. Soc. Mass Spectrom. 2003, 14, 342–351. [Google Scholar] [CrossRef]
- Large, R.; Knof, H. A comparison of negative and positive ion mass spectrometry. Org. Mass Spectrom. 1976, 11, 582–598. [Google Scholar] [CrossRef]
- Gandhi, K.; Kumar, A.; Sarkar, P.; Aghav, A.; Lal, D. MALDI-TOF MS: Application in dairy and related sectors. Res. Rev. J. Dairy Sci. Technol. 2013, 2, 2319–3409. [Google Scholar]
- Jackson, S.N.; Wang, H.-Y.J.; Woods, A.S. Direct profiling of lipid distribution in brain tissue using MALDI-TOFMS. Anal. Chem. 2005, 77, 4523–4527. [Google Scholar] [CrossRef]
- Wang, J.; Sporns, P. MALDI-TOF MS analysis of food flavonol glycosides. J. Agric. Food. Chem. 2000, 48, 1657–1662. [Google Scholar] [CrossRef]
- Dhiman, N.; Hall, L.; Wohlfiel, S.L.; Buckwalter, S.P.; Wengenack, N.L. Notes: Performance and cost analysis of matrix-assisted laser desorption ionization–iime of flight mass spectrometry for routine identification of yeast. J. Clin. Microbiol. 2011, 49, 1614. [Google Scholar] [CrossRef]
- Rychert, J. Benefits and limitations of MALDI-TOF mass spectrometry for the identification of microorganisms. J. Infect. 2019, 2, 1–5. [Google Scholar] [CrossRef]
- Patel, T.S.; Kaakeh, R.; Nagel, J.L.; Newton, D.W.; Stevenson, J.G. Cost analysis of implementing matrix-assisted laser desorption ionization–time of flight mass spectrometry plus real-time antimicrobial stewardship intervention for bloodstream infections. J. Clin. Microbiol. 2017, 55, 60. [Google Scholar] [CrossRef] [PubMed]
- Scott, J.R.; Schürch, S.; Moore, S.; Wilkins, C.L. Evaluation of MALDI-FTMS for analysis of peptide mixtures generated by ladder sequencing. Int. J. Mass Spectrom. Ion Process. 1997, 160, 291–302. [Google Scholar] [CrossRef]
- Aichler, M.; Walch, A. MALDI Imaging mass spectrometry: Current frontiers and perspectives in pathology research and practice. Lab. Investig. 2015, 95, 422–431. [Google Scholar] [CrossRef] [PubMed]
- Buck, A.; Ly, A.; Balluff, B.; Sun, N.; Gorzolka, K.; Feuchtinger, A.; Janssen, K.-P.; Kuppen, P.J.K.; van de Velde, C.J.H.; Erlmeier, F.; et al. High-resolution MALDI-FT-ICR MS imaging for the analysis of metabolites from formalin-fixed, paraffin-embedded clinical tissue samples. J. Pathol. 2015, 237, 123–132. [Google Scholar] [CrossRef]
- Ban, N.; Beckmann, R.; Cate, J.H.D.; Dinman, J.D.; Dragon, F.; Ellis, S.R.; Lafontaine, D.L.J.; Lindahl, L.; Liljas, A.; Lipton, J.M.; et al. A new system for naming ribosomal proteins. Curr. Opin. Struct. Biol. 2014, 24, 165. [Google Scholar] [CrossRef]
- Złoch, M.; Rodzik, A.; Pauter, K.; Szultka-Młynska, M.; Rogowska, A.; Kupczyk, W.; Pomastowski, P.; Buszewski, B. Problems with identifying and distinguishing salivary streptococci: A multi-instrumental approach. Future Microbiol. 2020, 15, 1157–1171. [Google Scholar] [CrossRef]
- Singhal, N.; Kumar, M.; Kanaujia, P.K.; Virdi, J.S. MALDI-TOF mass spectrometry: An emerging technology for microbial identification and diagnosis. Front. Microbiol. 2015, 6, 791. [Google Scholar] [CrossRef]
- Ashfaq, M.Y.; Da’na, D.A.; Al-Ghouti, M.A. Application of MALDI-TOF MS for identification of environmental bacteria: A review. J. Environ. Manag. 2022, 305, 114359. [Google Scholar] [CrossRef]
- Nomura, F. Proteome-based bacterial identification using matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS): A revolutionary shift in clinical diagnostic microbiology. Biochim. Biophys. Acta-Proteins Proteom. 2015, 1854, 528–537. [Google Scholar] [CrossRef]
- Gregorich, Z.R.; Ge, Y. Top-down proteomics in health and disease: Challenges and opportunities. Proteomics 2014, 14, 1195–1210. [Google Scholar] [CrossRef]
- Han, X.; Aslanian, A.; Yates, J.R. Mass Spectrometry for Proteomics. Curr. Opin. Chem. Biol. 2008, 12, 483. [Google Scholar] [CrossRef]
- Jabbour, R.E.; Wade, M.M.; Deshpande, S.V.; Stanford, M.F.; Wick, C.H.; Zulich, A.W.; Snyder, P.J. Identification of yersinia pestis and escherichia coli strains by whole cell and outer membrane protein extracts with mass spectrometry-based proteomics. J. Proteome Res. 2010, 9, 3647–3655. [Google Scholar] [CrossRef]
- Dickinson, D.N.; La Duc, M.T.; Haskins, W.E.; Gornushkin, I.; Winefordner, J.D.; Powell, D.H.; Venkateswaran, K. Species differentiation of a diverse suite of Bacillus spores by mass spectrometry-based protein profiling. Appl. Environ. Microbiol. 2004, 70, 475–482. [Google Scholar] [CrossRef]
- Fagerquist, C.K.; Miller, W.G.; Harden, L.A.; Bates, A.H.; Vensel, W.H.; Wang, G.; Mandrell, R.E. Genomic and proteomic identification of a DNA-binding protein used in the “fingerprinting” of Campylobacter species and strains by MALDI-TOF-MS protein biomarker analysis. Anal. Chem. 2005, 77, 4897–4907. [Google Scholar] [CrossRef]
- Schaller, A.; Troller, R.; Molina, D.; Gallati, S.; Aebi, C.; Meier, P.S. Rapid typing of Moraxella catarrhalis subpopulations based on outer membrane proteins using mass spectrometry. Proteomics 2006, 6, 172–180. [Google Scholar] [CrossRef]
- Sun, L.; Teramoto, K.; Sato, H.; Torimura, M.; Tao, H.; Shintani, T. Characterization of ribosomal proteins as biomarkers for matrix-assisted laser desorption/ionization mass spectral identification of Lactobacillus plantarum. Rapid Commun. Mass Spectrom. 2006, 20, 3789–3798. [Google Scholar] [CrossRef]
- Schmidt, F.; Fiege, T.; Hustoft, H.K.; Kneist, S.; Thiede, B. Shotgun mass mapping of Lactobacillus species and subspecies from caries related isolates by MALDI-MS. Proteomics 2009, 9, 1994–2003. [Google Scholar] [CrossRef]
- Camara, J.E.; Hays, F.A. Discrimination between wild-type and ampicillin-resistant Escherichia coli by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal. Bioanal. Chem. 2007, 389, 1633–1638. [Google Scholar] [CrossRef]
- Rozanova, S.; Barkovits, K.; Nikolov, M.; Schmidt, C.; Urlaub, H.; Marcus, K. Quantitative mass spectrometry-based proteomics: An overview. Methods Mol. Biol. 2021, 2228, 85–116. [Google Scholar]
- Bilecen, K.; Yaman, G.; Ciftci, U.; Laleli, Y.R. Performances and reliability of Bruker microflex LT and VITEK MS MALDI-TOF mass spectrometry systems for the identification of clinical microorganisms. Biomed. Res. Int. 2015, 2015, 516410. [Google Scholar] [CrossRef]
- Clark, A.E.; Kaleta, E.J.; Arora, A.; Wolk, D.M. Matrix-assisted laser desorption ionization-time of flight mass spectrometry: A fundamental shift in the routine practice of clinical microbiology. Clin. Microbiol. Rev. 2013, 26, 547–603. [Google Scholar] [CrossRef]
- Carolis, E.; De Vella, A.; Vaccaro, L.; Torelli, R.; Spanu, T.; Fiori, B.; Posteraro, B.; Sanguinetti, M. Application of MALDI-TOF mass spectrometry in clinical diagnostic microbiology. J. Infect. Dev. Ctries 2014, 8, 1081–1088. [Google Scholar] [CrossRef]
- Rodríguez-Sánchez, B.; Ruiz-Serrano, M.J.; Ruiz, A.; Timke, M.; Kostrzewa, M.; Bouza, E. Evaluation of MALDI Biotyper Mycobacteria Library v3.0 for identification of nontuberculous mycobacteria. J. Clin. Microbiol. 2016, 54, 1144–1147. [Google Scholar] [CrossRef]
- Leyer, C.; Gregorowicz, G.; Mougari, F.; Raskine, L.; Cambau, E.; De Briel, D. Comparison of Saramis 4.12 and IVD 3.0 Vitek MS matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of Mycobacteria from solid and liquid culture media. J. Clin. Microbiol. 2017, 55, 2045–2054. [Google Scholar] [CrossRef]
- Lee, H.-S.; Shin, J.H.; Choi, M.J.; Won, E.J.; Kee, S.J.; Kim, S.H.; Shin, M.G.; Suh, S.P. Comparison of the Bruker Biotyper and VITEK MS matrix-assisted laser desorption/ionization time-of-flight mass spectrometry systems using a formic acid extraction method to identify common and uncommon yeast isolates. Ann. Lab. Med. 2017, 37, 223. [Google Scholar] [CrossRef]
- Lévesque, S.; Dufresne, P.J.; Soualhine, H.; Domingo, M.-C.; Bekal, S.; Lefebvre, B.; Tremblay, C. A Side by Side Comparison of Bruker Biotyper and VITEK MS: Utility of MALDI-TOF MS technology for microorganism identification in a public health feference laboratory. PLoS ONE 2015, 10, e0144878. [Google Scholar] [CrossRef] [PubMed]
- Weis, C.W.; Jutzeler, C.R.; Borgwardt, K. Machine learning for microbial identification and antimicrobial susceptibility testing on MALDI-TOF mass spectra: A systematic review. Clin. Microbiol. Infect. 2020, 26, 1310–1317. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.-Y.; Chen, C.-H.; Lee, T.-Y.; Horng, J.-T.; Liu, T.-P.; Tseng, Y.-J.; Lu, J.-J. Rapid detection of heterogeneous vancomycin-intermediate Staphylococcus aureus based on matrix-assisted laser desorption ionization time-of-flight: Using a machine learning approach and unbiased validation. Front Microbiol. 2018, 9, 2393. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, C.R.; Covington, B.C.; Derewacz, D.K.; McNees, C.R.; Wikswo, J.P.; McLean, J.A.; Bachmann, B.O. Phenotypic mapping of metabolic profiles using self-organizing maps of high-dimensional mass spectrometry data. Anal. Chem. 2014, 86, 6563–6571. [Google Scholar] [CrossRef]
- Mortier, T.; Wieme, A.D.; Vandamme, P.; Waegeman, W. Bacterial species identification using MALDI-TOF mass spectrometry and machine learning techniques: A large-scale benchmarking study. Comput. Struct. Biotechnol. J. 2021, 19, 6157–6168. [Google Scholar] [CrossRef]
- Park, J.H.; Jang, Y.; Yoon, I.; Kim, T.S.; Park, H. Comparison of Autof ms1000 and Bruker Biotyper MALDI-TOF MS platforms for routine identification of clinical microorganisms. Hindawi 2021, 2021, 6667623. [Google Scholar]
- Buchan, B.W.; Riebe, K.M.; Timke, M.; Kostrzewa, M.; Ledeboer, N.A. Comparison of MALDI-TOF MS with HPLC and nucleic acid sequencing for the identification of Mycobacterium species in cultures using solid medium and broth. Am. J. Clin. Pathol. 2014, 141, 25. [Google Scholar] [CrossRef]
- Farfour, E.; Leto, J.; Barritault, M.; Barberis, C.; Meyer, J.; Dauphin, B.; Le Guern, A.-S.; Leflèche, A.; Badell, E.; Guiso, N.; et al. Evaluation of the andromas matrix-assisted laser desorption ionization-time of flight mass spectrometry system for identification of aerobically growing gram-positive bacilli. J. Clin. Microbiol. 2012, 50, 2702–2707. [Google Scholar] [CrossRef]
- Carbonnelle, E.; Beretti, J.-L.; Cottyn, S.; Quesne, G.; Berche, P.; Nassif, X.; Ferroni, A. Rapid identification of Staphylococci isolated in clinical microbiology laboratories by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 2007, 45, 2156–2161. [Google Scholar] [CrossRef] [PubMed]
- Degand, N.; Carbonnelle, E.; Dauphin, B.; Beretti, J.-L.; Le Bourgeois, M.; Sermet-Gaudelus, I.; Segonds, C.; Verche, P.; Nassif, X.; Ferrorni, A. Matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of nonfermenting gram-negative bacilli isolated from cystic fibrosis patients. J. Clin. Microbiol. 2008, 46, 3361–3367. [Google Scholar] [CrossRef]
- Regoui, S.; Hennebique, A.; Girard, T.; Boisset, S.; Caspar, Y.; Maurin, M. Optimized MALDI TOF mass spectrometry identification of Francisella tularensis subsp. holarctica. Microorganisms 2020, 8, 1143. [Google Scholar] [CrossRef]
- Legaria, M.C.; Nastro, M.; Camporro, J.; Heger, F.; Barberis, C.; Stecher, D.; Rodriguez, C.H.; Vay, C.A. Peptostreptococcus anaerobius: Pathogenicity, identification, and antimicrobial susceptibility. Review of monobacterial infections and addition of a case of urinary tract infection directly identified from a urine sample by MALDI-TOF MS. Anaerobe 2021, 72, 102461. [Google Scholar] [CrossRef]
- Kim, D.; Ji, S.; Kim, J.R.; Kim, M.; Byun, J.H.; Yum, J.H.; Yong, D.; Lee, K. Performance evaluation of a new matrix-assisted laser desorption/ionization time-of-flight mass spectrometry, ASTA MicroIDSys system, in bacterial identification against clinical isolates of anaerobic bacteria. Anaerobe 2020, 61, 102131. [Google Scholar] [CrossRef]
- Yoo, I.Y.; Shim, H.J.; Yun, S.A.; Kang, O.K.; Chung, Y.N.; Kim, T.Y.; Lee, H.; Park, Y.-J.; Huh, H.J.; Lee, N.Y. Evaluation of the ASTA MicroIDSys matrix-assisted laser desorption ionization time-of-flight mass spectrometry system for identification of mycobacteria directly from positive MGIT liquid cultures. Int. J. Infect. Dis. 2021, 102, 172–177. [Google Scholar] [CrossRef]
- Yi, Q.; Xiao, M.; Fan, X.; Zhang, G.; Yang, F.; Zhang, J.-J.; Duan, S.-M.; Cheng, J.-W.; Li, Y.; Zhou, M.-L.; et al. Evaluation of Autof MS 1000 and Vitek MS MALDI-TOF MS system in identification of closely-related yeasts causing invasive fungal diseases. Front. Cell. Infect. Microbiol. 2021, 11, 3. [Google Scholar] [CrossRef]
- Lee, Y.; Sung, J.Y.; Kim, H.; Yong, D.; Lee, K. Comparison of a new matrix-assisted laser desorption/ionization time-of-flight mass spectrometry platform, ASTA MicroIDSys, with Bruker Biotyper for species Identification. Ann. Lab. Med. 2017, 37, 531–535. [Google Scholar] [CrossRef][Green Version]
- Ziegler, D.; Pothier, J.F.; Ardley, J.; Fossou, R.K.; Pflüger, V.; de Meyer, S.; Vogel, G.; Tonolla, M.; Howieson, J.; Reeve, W.; et al. Ribosomal protein biomarkers provide root nodule bacterial identification by MALDI-TOF MS. Appl. Microbiol. Biotechnol. 2015, 99, 5547–5562. [Google Scholar] [CrossRef] [PubMed]
- Suarez, S.; Ferroni, A.; Lotz, A.; Jolley, K.A.; Guérin, P.; Leto, J.; Dauphin, B.; Jamet, A.; Maiden, M.C.J.; Nassif, X.; et al. Ribosomal proteins as biomarkers for bacterial identification by mass spectrometry in the clinical microbiology laboratory. J. Microbiol. Methods. 2013, 94, 390–396. [Google Scholar] [CrossRef] [PubMed]
- Rothen, J.; Pothier, J.F.; Foucault, F.; Blom, J.; Nanayakkara, D.; Li, C.; Ip, M.; Tanner, M.; Vpgel, G.; Pflüger, V.; et al. Subspecies typing of Streptococcus agalactiae based on ribosomal subunit protein mass variation by MALDI-TOF MS. Front. Microbiol. 2019, 10, 471. [Google Scholar] [CrossRef]
- Matsumura, Y.; Yamamoto, M.; Nagao, M.; Tanaka, M.; Machida, K.; Ito, Y.; Takakura, S.; Ichiyama, S. Detection of extended-spectrum-β-lactamase-producing Escherichia coli ST131 and ST405 clonal groups by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 2014, 52, 1034–1040. [Google Scholar] [CrossRef]
- Toh, B.E.W.; Zowawi, H.M.; Krizova, L.; Paterson, D.L.; Kamolvit, W.; Peleg, A.Y.; Sidjabat, H.; Nemec, A.; Pflüger, V.; Huber, C.A. Differentiation of Acinetobacter genomic species 13BJ/14TU from Acinetobacter haemolyticus by use of matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS). J. Clin. Microbiol. 2015, 53, 3384. [Google Scholar] [CrossRef]
- Drevinek, M.; Dresler, J.; Klimentova, J.; Pisa, L.; Hubalek, M. Evaluation of sample preparation methods for MALDI-TOF MS identification of highly dangerous bacteria. Lett. Appl. Microbiol. 2012, 55, 40–46. [Google Scholar] [CrossRef]
- Schulthess, B.; Bloemberg, G.V.; Zbinden, R.; Böttger, E.C.; Hombach, M. Evaluation of the Bruker MALDI Biotyper for identification of Gram-positive rods: Development of a diagnostic algorithm for the clinical labolatory. J. Clin. Microbiol. 2014, 52, 1089–1097. [Google Scholar] [CrossRef]
- Freiwald, A.; Sauer, S. Phylogenetic classification and identification of bacteria by mass spectrometry. Nat. Protoc. 2009, 4, 732–742. [Google Scholar] [CrossRef]
- Pascale, M.R.; Mazzotta, M.; Salaris, S.; Girolamini, L.; Grottola, A.; Simone, M.L.; Cordovana, M.; Bisognin, F.; Dal Monte, P.; Bucci Sabattini, M.A.; et al. Evaluation of MALDI-TOF mass spectrometry in diagnostic and environmental surveillance of Legionella species: A comparison with culture and Mip-Gene Sequencing technique. Front. Microbiol. 2020, 11, 589369. [Google Scholar] [CrossRef]
- Tsuchida, S.; Umemura, H.; Nakayama, T.; Mauri, P.L.; Marchetti-Deschmann, M.; Canetti, D. Current Status of matrix-assisted laser desorption/ionization–time-of-flight mass spectrometry (MALDI-TOF MS) in clinical diagnostic microbiology. Molecules 2020, 25, 4775. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, H.; Cai, K.; Yu, P.; Liu, Y.; Zhao, G.; Chen, R.; Xu, R.; Yu, M. Evaluation of three sample preparation methods for the identification of clinical strains by using two MALDI-TOF MS systems. J. Mass Spectrom. 2021, 56, e4696. [Google Scholar] [CrossRef]
- Huang, C.H.; Huang, L.; Chang, M.T.; Chen, K.L. Establishment and application of an analytical in-house database (IHDB) for rapid discrimination of Bacillus subtilis group (BSG) using whole-cell MALDI-TOF MS technology. Mol. Cell Probes. 2016, 30, 312–319. [Google Scholar] [CrossRef]
- Veloo, A.C.M.; Elgersma, P.E.; Friedrich, A.W.; Nagy, E.; van Winkelhoff, A.J. The influence of incubation time, sample preparation and exposure to oxygen on the quality of the MALDI-TOF MS spectrum of anaerobic bacteria. Clin. Microbiol. Infect. 2014, 20, 1091–1097. [Google Scholar] [CrossRef]
- Rotcheewaphan, S.; Lemon, J.K.; Desai, U.U.; Henderson, C.M.; Zelazny, A.M. Rapid one-step protein extraction method for the identification of mycobacteria using MALDI-TOF MS. Diagn. Microbiol. Infect. Dis. 2019, 94, 355–360. [Google Scholar] [CrossRef]
- Bizzini, A.; Durussel, C.; Bille, J.; Greub, G.; Prod’hom, G. Performance of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of bacterial strains routinely isolated in a clinical microbiology laboratory. J. Clin. Microbiol. 2010, 48, 1549–1554. [Google Scholar] [CrossRef]
- La Scola, B.; Raoult, D. Direct identification of bacteria in positive blood culture bottles by Matrix-Assisted Laser Desorption Ionisation Time-of-Flight Mass Spectrometry. PLoS ONE 2009, 4, e8041. [Google Scholar] [CrossRef]
- Dai, Y.; Xu, X.; Yan, X.; Li, D.; Cao, W.; Tang, L.; Hu, M.; Jiang, C. Evaluation of a rapid and simplified protocol for direct identification of microorganisms from positive blood cultures by using Matrix Assisted Laser Desportion Ionization Time-of-Flight Mass spectrometry (MALDI-TOF MS). Front. Cell Infect. Microbiol. 2021, 11, 632679. [Google Scholar] [CrossRef]
- Oviaño, M.; de la Luna Ramírez, C.; Barbeyto, L.P.; Bou, G. Rapid direct detection of carbapenemase-producing Enterobacteriaceae in clinical urine samples by MALDI-TOF MS analysis. J. Antimicrob. Chemother. 2017, 72, 1350–1354. [Google Scholar] [CrossRef]
- Mohan, B.; Gautam, N.; Sethuraman, N.; Kaur, H.; Taneja, N. Evaluation of matrix assisted laser desorption ionisation-time of flight mass spectrometry in direct identification of bacteriuria from urine samples. Indian J. Med. Microbiol. 2020, 38, 293–298. [Google Scholar] [CrossRef]
- Sun, C.; Zhang, X.; Wang, J.; Cheng, C.; Kang, H.; Gu, B.; Ma, P. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry combined with UF-5000i urine flow cytometry to directly identify pathogens in clinical urine specimens within 1 hour. Ann. Transl. Med. 2020, 8, 602. [Google Scholar] [CrossRef] [PubMed]
- Ying, J.; Gao, W.; Huang, D.; Ding, C.; Ling, L.; Pan, T.; Yu, S. Application of MALDI-TOF MS Profiling Coupled With Functionalized Magnetic Enrichment for Rapid Identification of Pathogens in a Patient With Open Fracture. Front. Chem. 2021, 9, 672744. [Google Scholar] [CrossRef] [PubMed]
- Birmingham, J.; Demirev, P.; Ho, Y.-P.; Thomas, J.; Bryden, W.; Fenselau, C. Corona Plasma Discharge for Rapid Analysis of Microorganisms by Mass Spectrometry. Rapid. Commun. Mass Spectrom. 1999, 13, 604–606. [Google Scholar] [CrossRef]
- Ryzhov, V.; Hathout, Y.; Fenselau, C. Rapid Characterization of Spores of Bacillus cereus Group Bacteria by Matrix-Assisted Laser Desorption-Ionization Time-of-Flight Mass Spectrometry. Appl. Environ. Microbiol. 2000, 66, 3828. [Google Scholar] [CrossRef]
- Afonso, C.; Fenselau, C. Use of Bioactive Glass Slides for Matrix-Assisted Laser Desorption/Ionization Analysis: Application to Microorganisms. Anal. Chem. 2003, 75, 694–697. [Google Scholar] [CrossRef]
- Saleen, P.G.; Drake, S.K.; Murray, P.R.; Zelazny, A.M. Identification of mycobacteria in solid-culture media by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 2011, 49, 1790–1794. [Google Scholar]
- Noumi, E.; Merghni, A.; Alreshidi, M.; Del Campo, R.; Adnan, M.; Haddad, O.; De Feo, V.; Snoussi, M. Phenotypic and Genotypic Characterization with MALDI-TOF-MS Based Identification of Staphylococcus spp. Isolated from Mobile Phones with their Antibiotic Susceptibility, Biofilm Formation, and Adhesion Properties. Int. J. Environ. Res. Public Health 2020, 17, 3761. [Google Scholar] [CrossRef]
- Pierce, C.Y.; Barr, J.R.; Woolfitt, A.R.; Moura, H.; Shaw, E.I.; Thompson, H.A.; Massung, R.F.; Fernandez, F.M. Strain and phase identification of the U.S. category B agent Coxiella burnetii by matrix assisted laser desorption/ionization time-of-flight mass spectrometry and multivariate pattern recognition. Anal. Chim. Acta 2007, 583, 23–31. [Google Scholar] [CrossRef]
- Jones, J.J.; Stump, M.J.; Fleming, R.C.; Lay, J.O.; Wilkins, C.L. Investigation of MALDI-TOF and FT-MS techniques for analysis of Escherichia coli whole cells. Anal. Chem. 2003, 75, 1340–1347. [Google Scholar] [CrossRef]
- Shah, H.N.; Chilton, C.; Rajakaruna, L.; Gaulton, T.; Hallas, G.; Atanassov, H.; Khoder, G.; Rakowska, P.D.; Cerasoli, E.; Gharbia, S.E. Changing Concepts in the Characterisation of Microbes and the Influence of Mass Spectrometry. In Mass Spectrometry for Microbial Proteomics; Shah, H.N., Gharbia, S.E., Eds.; Wiley Online Library: Hoboken, NJ, USA, 2010; pp. 1–34. [Google Scholar]
- Reddy, G.; Dalmasso, E.A. SELDI ProteinChip® Array Technology: Protein-Based Predictive Medicine and Drug Discovery Applications. J. Biomed. Biotechnol. 2003, 2003, 237. [Google Scholar] [CrossRef]
- Rajakaruna, L.K. Proteomics as a Tool for the Characterisation of Nosocomial Pathogens; ProQuest LLC.: London, UK, 2010; pp. 1–235. [Google Scholar]
- Shah, H.N.; Rajakaruna, L.; Ball, G.; Misra, R.; Al-Shahib, A.; Fang, M.; Gharbia, S.E. Tracing the transition of methicillin resistance in sub-populations of Staphylococcus aureus, using SELDI-TOF Mass Spectrometry and Artificial Neural Network Analysis. Syst. Appl. Microbiol. 2011, 34, 81–86. [Google Scholar] [CrossRef]
- Schmid, O.; Ball, G.; Lancashire, L.; Culak, R.; Shah, H. New approaches to identification of bacterial pathogens by surface enhanced laser desorption/ionization time of flight mass spectrometry in concert with artificial neural networks, with special reference to Neisseria gonorrhoeae. J. Med. Microbiol. 2005, 54, 1205–1211. [Google Scholar] [CrossRef]
- Nakamura, M.T.; Yudell, B.E.; Loor, J.J. Regulation of energy metabolism by long-chain fatty acids. Prog. Lipid. Res. 2014, 53, 124–144. [Google Scholar] [CrossRef]
- Van Meer, G.; Voelker, D.R.; Feigenson, G.W. Membrane lipids: Where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008, 9, 112–124. [Google Scholar] [CrossRef]
- Fahy, E.; Subramaniam, S.; Brown, H.A.; Glass, C.K.; Merrill, A.H.; Murphy, R.C.; Reatz, C.R.H.; Russel, D.W.; Seyama, Y.; Shaw, W.; et al. A comprehensive classification system for lipids. J. Lipid Res. 2005, 46, 839–861. [Google Scholar] [CrossRef]
- Sohlenkamp, C.; Geiger, O. Bacterial membrane lipids: Diversity in structures and pathways. FEMS Microbiol. Rev. 2015, 40, 133–159. [Google Scholar] [CrossRef]
- Liu, B.; Knirel, Y.A.; Leng, L.; Perepelov, A.V.; Senchenkova, S.N.; Reeves, P.R.; Wang, L. Structural diversity in Salmonella O antigens and its genetic basis. FEMS Microbiol. Rev. 2014, 38, 56–89. [Google Scholar] [CrossRef]
- Froning, M.; Helmer, P.O.; Hayen, H. Identification and structural characterization of lipid A from Escherichia coli, Pseudomonas putida and Pseudomonas taiwanensis using liquid chromatography coupled to high-resolution tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2020, 34, e8897. [Google Scholar] [CrossRef]
- Casabuono, A.C.; van der Ploeg, C.A.; Roge, A.D.; Bruno, S.B.; Couto, A.S. Characterization of lipid A profiles from Shigella flexneri variant Xlipopolysaccharide. Rapid Commun. Mass Spectrom. 2012, 26, 2011–2020. [Google Scholar] [CrossRef]
- Fischer, W. Lipoteichoic acids and lipoglycans. In New Comprehensive Biochemistry; Ghuysen, J., Hakenbeck, R., Eds.; Elsevier Science: Amsterdam, The Netherlands, 1994; pp. 199–215. [Google Scholar]
- Schneewind, O.; Missiakas, D. Lipoteichoic Acids, Phosphate-Containing Polymers in the Envelope of Gram-Positive Bacteria. J. Bacteriol. 2014, 196, 1133–1142. [Google Scholar] [CrossRef]
- Koch, H.U.; Fischer, W. Acyldiglucosyldiacylglycerol-containing lipoteichoic acid with a poly(3-O-galabiosyl-2-O-galactosyl-sn-glycero-1-phosphate) chain from Streptococcus lactis Kiel 42172. Biochemistry 1978, 17, 5275–5281. [Google Scholar] [CrossRef]
- Fischer, W. Pneumococcal lipoteichoic and teichoic acid. Microb. Drug Resist. 1997, 3, 309–325. [Google Scholar] [CrossRef]
- Fischer, W. One-step purification of bacterial lipid macroamphiphiles by hydrophobic interaction chromatography. Anal. Biochem. 1991, 194, 353–358. [Google Scholar] [CrossRef]
- Jackson, M. The mycobacterial cell envelope-lipids. Cold Spring Harb. Perspect. Med. 2014, 4, a021105. [Google Scholar] [CrossRef]
- Ripoll, F.; Deshayes, C.; Pasek, S.; Laval, F.; Beretti, J.-L.; Biet, F.; Risler, J.-L.; Daffè, M.; Etienne, G.; Gillard, J.-L.; et al. Genomics of glycopeptidolipid biosynthesis in Mycobacterium abscessus and M. chelonae. BMC Genom. 2007, 8, 114. [Google Scholar] [CrossRef] [PubMed]
- Batt, S.M.; Minnikin, D.E.; Besra, G.S. The thick waxy coat of mycobacteria, a protective layer against antibiotics and the host’s immune system. Biochem. J. 2020, 477, 1983–2006. [Google Scholar] [CrossRef]
- Marrakchi, H.; Lanéelle, M.-A.; Daffé, M. Mycolic acids: Structures, biosynthesis, and beyond. Chem. Biol. 2014, 21, 67–85. [Google Scholar] [CrossRef]
- Burbaund, S.; Laval, F.; Lemassu, A.; Daffé, M.; Guilhot, C.; Chalut, C. Trehalose Polyphleates Are Produced by a Glycolipid Biosynthetic Pathway Conserved across Phylogenetically Distant Mycobacteria. Cell Chem. Biol. 2016, 23, 278–289. [Google Scholar] [CrossRef] [PubMed]
- Jarrad, A.M.; Karoli, T.; Blaskovich, M.A.T.; Lyras, D.; Cooper, M.A. Clostridium difficile drug pipeline: Challenges in discovery and development of new agents. J. Med. Chem. 2015, 58, 5164–5185. [Google Scholar] [CrossRef]
- Abel, K.; Deschmerting, H.; Peterson, J.I. Classification of microorganisms by analysis of chemical composition. J. Bacteriol. 1963, 85, 1039–1044. [Google Scholar] [CrossRef]
- Hu, T.; Zhang, J.L. Mass-spectrometry-based lipidomics. J. Sep. Sci. 2018, 41, 351–372. [Google Scholar] [CrossRef] [PubMed]
- Solntceva, V.; Kostrzewa, M.; Larrouy-Maumus, G. Detection of species-specific lipids by routine MALDI TOF Mass Spectrometry to unlock the challenges of microbial identification and antimicrobial susceptibility testing. Front. Cell Infect. Microbiol. 2020, 10, 621452. [Google Scholar] [CrossRef] [PubMed]
- Pomastowski, P.; Złoch, M.; Rodzik, A.; Ligor, M.; Kostrzewa, M.; Buszewski, B. Analysis of bacteria associated with honeys of different geographical and botanical origin using two different identification approaches: MALDI-TOF MS and 16S rDNA PCR technique. PLoS ONE 2019, 14, e0217078. [Google Scholar] [CrossRef] [PubMed]
- Ratiu, I.A.; Railean Plugaru, V.; Pomastowski, P.; Milanowski, M.; Mametov, R.; Bocos-Bintintan, V.; Buszewski, B. Temporal influence of different antibiotics onto the inhibition of Escherichia coli bacterium grown in different media. Anal. Biochem. 2019, 585, 113407. [Google Scholar] [CrossRef]
- Walczak-Skierska, J.; Złoch, M.; Pauter, K.; Pomastowski, P.; Buszewski, B. Lipidomic analysis of lactic acid bacteria strains by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J. Dairy Sci. 2020, 103, 11062–11078. [Google Scholar] [CrossRef]
- Leung, L.M.; Fondrie, W.E.; Doi, Y.; Johnson, J.K.; Strickland, D.K.; Ernst, R.K.; Goodlett, D.R. Identification of the ESKAPE pathogens by mass spectrometric analysis of microbial membrane glycolipids. Sci. Rep. 2017, 7, 6403. [Google Scholar] [CrossRef]
- Ryu, S.Y.; Wendt, G.A.; Chandler, C.E.; Ernst, R.K.; Goodlett, D.R. Model-based spectral library approach for bacterial identification via membrane glycolipids. Anal. Chem. 2019, 91, 11482–11487. [Google Scholar] [CrossRef]
- Liang, T.; Leung, L.M.; Opene, B.; Fondrie, W.E.; Lee, Y.I.; Chandler, C.E.; Yoon, S.H.; Ernst, R.K.; Goodlett, D.R. Rapid microbial identification and antibiotic resistance detection by mass spectrometric analysis of membrane lipids. Anal. Chem. 2019, 91, 1286–1294. [Google Scholar] [CrossRef]
- Patel, A.; Mikes, F.; Matsakas, L. An overview of current pretreatment methods used to improve lipid extraction from oleaginous microorganisms. Molecules 2018, 23, 1562. [Google Scholar] [CrossRef]
- Matyash, V.; Liebisch, G.; Kurzchalia, T.V.; Shevchenko, A.; Schwudke, D. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J. Lipid Res. 2008, 49, 1137–1146. [Google Scholar] [CrossRef]
- El Hamidi, A.; Tirsoaga, A.; Novikov, A.; Hussein, A.; Caroff, M. Microextraction of bacterial lipid A: Easy and rapid method for mass spectrometric characterization. J. Lipid Res. 2005, 46, 1773–1778. [Google Scholar] [CrossRef]
- Angelini, R.; Babudri, F.; Lobasso, S.; Corcelli, A. MALDI-TOF/MS analysis of archaebacterial lipids in lyophilized membranes dry-mixed with 9-aminoacridine. J. Lipid Res. 2010, 51, 2818–2825. [Google Scholar] [CrossRef]
- Calvano, C.D.; Zambonin, C.G.; Palmisano, F. Lipid fingerprinting of gram-positive lactobacilli by intact—Matrix-assisted laser desorption/ionization mass spectrometry using a proton sponge based matrix. Rapid Commun. Mass Spectrom. 2011, 25, 1757–1764. [Google Scholar] [CrossRef]
- Larrouy-Maumus, G.; Puzo, G. Mycobacterial envelope lipids fingerprint from direct MALDI-TOF MS analysis of intact bacilli. Tuberculosis 2015, 95, 75–85. [Google Scholar] [CrossRef]
- Larrouy-Maumus, G.; Clements, A.; Filloux, A.; McCarthy, R.R.; Mostowy, S. Direct detection of lipid A on intact Gram-negative bacteria by MALDI-TOF mass spectrometry. J. Microbiol. Methods 2016, 120, 68–71. [Google Scholar] [CrossRef]
- Gonzalo, X.; Broda, A.; Drobniewski, F.; Larrouy-Maumus, G. Performance of lipid fingerprint-based MALDI-ToF for the diagnosis of mycobacterial infections. Clin. Microbiol. Infect. 2021, 27, 912.e1–912.e5. [Google Scholar] [CrossRef]
- Dortet, L.; Potron, A.; Bonnin, R.A.; Plesiat, P.; Naas, T.; Filloux, A.; Larrouy-Maumus, G. Rapid detection of colistin resistance in Acinetobacter baumannii using MALDI-TOF-based lipidomics on intact bacteria. Sci. Rep. 2018, 8, 16910. [Google Scholar] [CrossRef]
- Furniss, C.R.D.; Kostrzewa, M.; Mavridou, D.A.I.; Larrouy-Maumus, G. The clue is in the lipid A: Rapid detection of colistin resistance. PLoS Pathog. 2020, 16, e1008331. [Google Scholar] [CrossRef]
- Dortet, L.; Broda, A.; Bernabeu, S.; Glupczynski, Y.; Bogaerts, P.; Bonnin, R.; Naas, T.; Filloux, A.; Larrouy-Maumus, G. Optimization of the MALDIxin test for the rapid identification of colistin resistance in Klebsiella pneumoniae using MALDI-TOF MS. J. Antimicrob. Chemother. 2020, 75, 110–116. [Google Scholar] [CrossRef]
- Khor, M.J.; Broda, A.; Kostrzewa, M.; Drobniewski, F.; Larrouy-Maumus, G. An improved method for rapid detection of Mycobacterium abscessus complex based on species-specific lipid fingerprint by routine MALDI-TOF. Front. Chem. 2021, 9, 715890. [Google Scholar] [CrossRef]
- Cox, G.; Wright, G.D. Intrinsic antibiotic resistance: Mechanisms, origins, challenges and solutions. Int. J. Med. Microbiol. 2013, 303, 287–292. [Google Scholar] [CrossRef] [PubMed]
- Saichek, N.R.; Cox, C.R.; Kim, S.; Harrington, P.B.; Stambach, N.R.; Voorhees, K.J. Strain-level Staphylococcus differentiation by CeO2-metal oxide laser ionization mass spectrometry fatty acid profiling. BMC Microbiol. 2016, 16, 72. [Google Scholar] [CrossRef] [PubMed]
- Cox, C.R.; Jensen, K.R.; Saichek, N.R.; Voorhees, K.J. Strain-level bacterial identification by CeO2-catalyzed MALDI-TOF MS fatty acid analysis and comparison to commercial protein-based methods. Sci. Rep. 2015, 5, 10470. [Google Scholar] [CrossRef] [PubMed]
- Meitei, N.S.; Shulaev, V. Bioinformatics in Lipidomics: Automating Large-Scale LC-MS-Based Untargeted Lipidomics Profiling with SimLipid Software. Plant Metab. Eng. 2022, 2396, 197–214. [Google Scholar]
- Verma, A.; Meitei, N.S.; Gajbhiye, P.U.; Raftery, M.J.; Ambatipudi, K. Comparative Analysis of Milk Triglycerides Profile between Jaffarabadi Buffalo and Holstein Friesian Cow. Metabolites 2021, 10, 507. [Google Scholar] [CrossRef] [PubMed]
- Jinno, K.; Sawada, H. Recent trends in open-tubular capillary electrochromatography. TrAC Trends Anal. Chem. 2000, 19, 664–675. [Google Scholar] [CrossRef]
- Jurowski, K.; Kochan, K.; Walczak, J.; Barańska, M.; Piekoszewski, W.; Buszewski, B. Analytical techniques in lipidomics: State of the art. Crit. Rev. Anal. Chem. 2017, 47, 418–437. [Google Scholar] [CrossRef]
- Szultka-Młyńska, M.; Buszewski, B. Study of in-vitro metabolism of selected antibiotic drugs in human liver microsomes by liquid chromatography coupled with tandem mass spectrometry. Anal. Bioanal. Chem. 2016, 408, 8273–8287. [Google Scholar] [CrossRef]
- Szultka-Młyńska, M.; Buszewski, B. Electrochemical oxidation of selected immunosuppressants and identification of their oxidation products by means of liquid chromatography and tandem mass spectrometry (EC-HPLC-MS/MS). J. Pharm. Biomed. Anal. 2019, 176, 112799. [Google Scholar] [CrossRef]
- Szultka-Młyńska, M.; Pauter, K.; Buszewski, B. Identification of in vitro and in vivo potential metabolites of novel cardiovascular and adrenolytic drugs by liquid chromatography-mass spectrometry with the aid of experimental design. Nov. Biotechnol. Chim. 2019, 18, 179–194. [Google Scholar] [CrossRef][Green Version]
- Weckwerth, W. Metabolomics in systems biology. Annu. Rev. Plant Biol. 2003, 54, 669–689. [Google Scholar] [CrossRef]
- Megha, P.; Prasad, T. Metabolomics: A Promising Tool to Study Disease Biomarkers and Host-Pathogen Interactions. Integr. Omics Approaches Infect. Dis. 2021, 403–423. [Google Scholar] [CrossRef]
- Mashego, M.R.; Van Gulik, W.M.; Heijnen, J.J. Metabolome dynamic responses of Saccharomyces cerevisiae to simultaneous rapid perturbations in external electron acceptor and electron donor. FEMS Yeast Res. 2007, 7, 48–66. [Google Scholar] [CrossRef]
- Pinu, F.R.; Villas-Boas, S.G. Extracellular microbial metabolomics: The state of the art. Metabolites 2017, 7, 43. [Google Scholar] [CrossRef]
- Shi, Y.; Yang, H.; Chu, M.; Niu, X.; Huo, X.; Gao, Y.; Zeng, J.; Zhang, T.; Li, Y.G.; Outi, K.E.; et al. Klebsiella. In Beneficial Microbes in Agro-Ecology; Academic Press: Cambridge, MA, USA, 2020; pp. 233–257. [Google Scholar] [CrossRef]
- Grim, C.M.; Luu, G.T.; Sanchez, L.M. Staring into the void: Demystifying microbial metabolomics. FEMS Microbiol. Lett. 2019, 366, 135. [Google Scholar] [CrossRef]
- Bundy, J.G.; Willey, T.L.; Castell, R.S.; Ellar, D.J.; Brindle, K.M. Discrimination of pathogenic clinical isolates and laboratory strains of Bacillus cereus by NMR-based metabolomic profiling. FEMS Microbiol. Lett. 2005, 242, 127–136. [Google Scholar] [CrossRef]
- T’Kindt, R.; Scheltema, R.A.; Jankevics, A.; Brunker, K.; Rijal, S.; Dujardin, J.-C.; Breitling, R.; Watson, D.G.; Coombs, G.H.; Decuypere, S. Metabolomics to Unveil and Understand Phenotypic Diversity between Pathogen Populations. PLoS Negl. Trop. Dis. 2010, 4, e904. [Google Scholar] [CrossRef]
- Bean, H.D.; Rees, C.A.; Hill, J.E. Comparative analysis of the volatile metabolomes of Pseudomonas aeruginosa clinical isolates. J. Breath Res. 2016, 10, 047102. [Google Scholar] [CrossRef]
- Jørgensen, K.M.; Wassermann, T.; Johansen, H.K.; Christiansen, L.E.; Molin, S.; Høiby, N.; Ciofu, O. Diversity of metabolic profiles of cystic fibrosis pseudomonas aeruginosa during the early stages of lung infection. Microbiology 2015, 161, 1447–1462. [Google Scholar] [CrossRef]
- Karami, N.; Karimi, A.; Aliahmadi, A.; Mirzajan, F.; Rezadoost, H.; Ghassempour, A.; Fallah, F. Identification of bacteria using volatile organic compounds. Cell Mol. Biol. 2017, 63, 112–121. [Google Scholar] [CrossRef]
- Allegretta, G.; Maurer, C.K.; Eberhard, J.; Maura, D.; Hartmann, R.W.; Rahme, L.; Empting, M. In-depth profiling of MvfR-regulated small molecules in Pseudomonas aeruginosa after Quorum Sensing inhibitor treatment. Front. Microbiol. 2017, 8, 924. [Google Scholar] [CrossRef]
- Moyne, O.; Castelli, F.; Bicout, D.J.; Boccard, J.; Camara, B.; Cournoyer, B.; Faudry, E.; Terrier, S.; Hannani, D.; Huot-Marchand, S.; et al. Metabotypes of Pseudomonas aeruginosa correlate with antibiotic resistance, virulence and clinical outcome in cystic fibrosis chronic infections. Metabolites 2021, 11, 63. [Google Scholar] [CrossRef]
- Morrison, D.J.; Preston, T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes 2016, 7, 189–200. [Google Scholar] [CrossRef]
- Sharon, G.; Garg, N.; Debelius, J.; Knight, R.; Dorrestein, P.C.; Mazmanian, S.K. Specialized metabolites from the microbiome in health and disease. Cell Metab. 2014, 20, 719–730. [Google Scholar] [CrossRef]
- Sun, M.; Wu, W.; Liu, Z.; Cong, Y. Microbiota metabolite short chain fatty acids, GPCR, and inflammatory bowel diseases. J. Gastroenterol. 2017, 52, 1–8. [Google Scholar] [CrossRef]
- Yang, Y.L.; Xu, Y.; Straight, P.; Dorrestein, P.C. Translating metabolic exchange with imaging mass spectrometry. Nat. Chem. Biol. 2009, 5, 885–887. [Google Scholar] [CrossRef]
- Yang, J.Y.; Phelan, V.V.; Simkovsky, R.; Watrous, J.D.; Trial, R.M.; Fleming, T.C.; Wenter, R.; Moore, B.S.; Golden, S.S.; Pogliano, K.; et al. Primer on agar-based microbial imaging mass spectrometry. J. Bacteriol. 2012, 194, 6023–6028. [Google Scholar] [CrossRef]
- Bleich, R.; Watrous, J.D.; Dorrestein, P.C.; Bowers, A.A.; Shank, E.A. Thiopeptide antibiotics stimulate biofilm formation in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 2015, 112, 3086–3091. [Google Scholar] [CrossRef]
- Moree, W.J.; Phelan, V.V.; Wu, C.H.; Bandeira, N.; Cornett, D.S.; Duggan, B.M.; Dorrestein, P.C. Interkingdom metabolic transformations captured by microbial imaging mass spectrometry. Proc. Natl. Acad. Sci. USA 2012, 109, 13811–13816. [Google Scholar] [CrossRef]
- De Bruijn, I.; Cheng, X.; de Jager, V.; Expósito, R.G.; Watrous, J.; Patel, N.; Postma, J.; Dorrestein, P.C.; Kobayashi, D.; Raaijmakers, J.M. Comparative genomics and metabolic profiling of the genus Lysobacter. BMC Genom. 2015, 16, 991. [Google Scholar] [CrossRef]
- Prideaux, B.; Dartois, V.; Staab, D.; Weiner, D.M.; Goh, A.; Via, L.E.; Barry, C.E., 3rd; Stoeckli, M. High-Sensitivity MALDI-MRM-MS Imaging of Moxifloxacin Distribution in Tuberculosis-Infected Rabbit Lungs and Granulomatous Lesions. Anal. Chem. 2011, 83, 2112. [Google Scholar] [CrossRef] [PubMed]
- Hensel, M.; Shea, J.E.; Gleeson, C.; Jones, M.D.; Dalton, E.; Holden, D.W. Simultaneous identification of bacterial virulence genes by negative selection. Science 1995, 269, 400–403. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.J.; Stump, M.J.; Fleming, R.C.; Lay, J.O.; Wilkins, C.L. Strategies and data analysis techniques for lipid and phospholipid chemistry elucidation by intact cell MALDI-FTMS. J. Am. Soc. Mass Spectrom. 2004, 15, 1665–1674. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Clark, C.M.; Costa, M.S.; Sanchez, L.M.; Murphy, B.T. Coupling MALDI-TOF mass spectrometry protein and specialized metabolite analyses to rapidly discriminate bacterial function. Proc. Natl. Acad. Sci. USA 2018, 115, 4981–4986. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.D.; Saharuka, V.; Kovalev, V.; Stuart, L.; Del Prete, M.; Lubowiecka, K.; De Mot, R.; Venturi, V.; Alexandrov, T. Facilitating imaging mass spectrometry of microbial specialized metabolites with METASPACE. Metabolites 2021, 11, 477. [Google Scholar] [CrossRef] [PubMed]
- Lewis, L.; Onsongo, M.; Njapau, H.; Schurz-Rogers, H.; Luber, G.; Kieszak, S.; Nyamongo, J.; Backer, L.; Mohamud Dahiye, A.; Misore, A.; et al. Aflatoxin contamination of commercial maize products during an outbreak of acute aflatoxicosis in eastern and central Kenya. Environ. Health Perspect. 2005, 113, 1763–1767. [Google Scholar] [CrossRef]
- Richard, J.L. Some major mycotoxins and their mycotoxicoses—An overview. Int. J. Food Microbiol. 2007, 119, 3–10. [Google Scholar] [CrossRef]
- Hleba, L.; Císarová, M.; Shariati, M.A.; Tančinová, D. Detection of mycotoxins using maldi-tof mass spectrometry. J. Microbiol. Biotech. Food Sci. 2017, 7, 181–185. [Google Scholar] [CrossRef]
- Sivagnanam, K.; Komatsu, E.; Rampitsch, C.; Perreault, H.; Gräfenhan, T. Rapid screening of Alternaria mycotoxins using MALDI-TOF mass spectrometry. J. Sci. Food Agric. 2017, 97, 357–361. [Google Scholar] [CrossRef]
- Jerome Jeyakumar, J.M.; Zhang, M.; Thiruvengadam, M. Determination of mycotoxins by HPLC, LC-ESI-MS/MS, and MALDI-TOF MS in Fusarium species-infected sugarcane. Microb. Pathog. 2018, 123, 98–110. [Google Scholar] [CrossRef]
- Cleary, J.L.; Luu, G.T.; Pierce, E.C.; Dutton, R.J.; Sanchez, L.M. BLANKA: An Algorithm for blank subtraction in mass spectrometry of complex biological samples. J. Am. Soc. Mass Spectrom. 2019, 30, 1426–1434. [Google Scholar] [CrossRef]
- Sévin, D.C.; Kuehne, A.; Zamboni, N.; Sauer, U. Biological insights through nontargeted metabolomics. Curr. Opin. Biotechnol. 2015, 34, 1–8. [Google Scholar] [CrossRef]
- Hursf, G.B.; Doktycz, M.J.; Vass, A.A.; Buchanan, M.V. Detection of bacterial DNA polymerase chain reaction products by Matrix-assisted Laser Desorption/Ionization Mass Spectrometry. RAPID Commun. Mass Spectrom. 1996, 10, 377–382. [Google Scholar]
- Honisch, C.; Chen, Y.; Mortimer, C.; Arnold, C.; Schmidt, O.; Van Den Boom, D.; Cantor, C.R.; Shah, H.N.; Sharbia, S.E. Automated comparative sequence analysis by base-specific cleavage and mass spectrometry for nucleic acid-based microbial typing. Proc. Natl. Acad. Sci. USA 2007, 104, 10649–10654. [Google Scholar] [CrossRef]
- Von Wintzingerode, F.; Böcker, S.; Schlötelburg, C.; Chiu, N.H.L.; Storm, N.; Jurinke, C.; Cantor, C.R.; Göbel, U.B.; van den Boom, D. Base-specific fragmentation of amplified 16S rRNA genes analyzed by mass spectrometry: A tool for rapid bacterial identification. Proc. Natl. Acad. Sci. USA 2002, 99, 7039–7044. [Google Scholar] [CrossRef]
- Lefmann, M.; Honisch, C.; Böcker, S.; Storm, N.; Von Wintzingerode, F.; Schlötelburg, C.; Moter, A.; van den Boom, D.; Göbel, U.B. Novel mass spectrometry-based tool for genotypic identification of Mycobacteria. J. Clin. Microbiol. 2004, 42, 339–346. [Google Scholar] [CrossRef]
- Cuénod, A.; Wüthrich, D.; Seth-Smith, H.M.B.; Ott, C.; Gehringer, C.; Foucault, F.; Mouchet, R.; Kassim, A.; Revathi, G.; Vogt, D.R.; et al. Whole-genome sequence-informed MALDI-TOF MS diagnostics reveal importance of Klebsiella oxytoca group in invasive infections: A retrospective clinical study. Genome Med. 2021, 13, 150. [Google Scholar] [CrossRef]
- Dunne, E.M.; Kok Ong, E.; Moser, R.J.; Siba, P.M.; Phuanukoonnon, S.; Greenhill, A.R.; Robins-Browne, R.M.; Mulholland, E.K.; Satzke, C. Multiocus sequence typing of Streptococcus pneumoniae by use mass spectrometry. J. Clin. Microbiol. 2011, 49, 3756–3760. [Google Scholar] [CrossRef]
- Ha, S.M.; Kim, C.K.; Roh, J.; Byun, J.H.; Yang, S.J.; Choi, S.B.; Chun, J.; Yong, D. Application of the whole genome-based bacterial identification system, TRUEBAC ID, using clinical isolates that were not identified with three matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) systems. Ann. Lab. Med. 2019, 39, 530–536. [Google Scholar] [CrossRef]
- Zhang, C.; Xiu, L.; Xiao, Y.; Xie, Z.; Ren, L.; Peng, J. Simultaneous detection of key bacterial pathogens related to pneumonia and meningitis using multiplex PCR coupled with mass spectrometry. Front. Cell Infect. Microbiol. 2018, 8, 107. [Google Scholar] [CrossRef]
- Peng, L.; Li, K.; Zhang, C.; Jin, Q. MW polyomavirus and STL polyomavirus present in tonsillar tissues from children with chronic tonsillar disease. Clin. Microbiol. Infect. 2016, 22, 97. [Google Scholar] [CrossRef]
- Nyasinga, J.; Kyany’a, C.; Okoth, R.; Oundo, C.; Matano, D.; Wacira, S.; Sang, W.; Musembi, S.; Musila, L. A six-member SNP assay on the iPlex MassARRAY platform provides a rapid and affordable alternative for typing major African Staphylococcus aureus types. Access Microbiol. 2019, 1, e000018. [Google Scholar] [CrossRef] [PubMed]
- Syrmis, M.W.; Moser, R.J.; Whileu, D.M.; Vaska, V.; Coombs, G.W.; Nissen, M.D.; Sloots, T.P.; Nimmo, G.R. Comparison of a multiplexed MassARRAY system with real-time allele-specific PCR technology for genotyping of methicillin-resistant Staphylococcus aureus. Clin. Microbiol. Infect. 2011, 17, 1804–1810. [Google Scholar] [CrossRef]
- Read, A.F.; Woods, R.J. Antibiotic resistance management. Evol. Med. Public Health 2014, 2014, 147. [Google Scholar] [CrossRef]
- Levy, S.B.; Bonnie, M. Antibacterial resistance worldwide: Causes, challenges and responses. Nat. Med. 2004, 10, S122–S129. [Google Scholar] [CrossRef]
- Spellberg, B.; Gilbert, D.N. The future of antibiotics and resistance: A tribute to a career of leadership by John Bartlett. Clin. Infect. Dis. 2014, 59, S71–S75. [Google Scholar] [CrossRef]
- Sengupta, S.; Chattopadhyay, M.K.; Grossart, H.P. The multifaceted roles of antibiotics and antibiotic resistance in nature. Front. Microbiol. 2013, 4, 47. [Google Scholar] [CrossRef]
- Alekshun, M.N.; Levy, S.B. Molecular mechanisms of antibacterial multidrug resistance. Cell 2007, 128, 1037–1050. [Google Scholar] [CrossRef]
- Davies, J.; Davies, D. Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 2010, 74, 417–433. [Google Scholar] [CrossRef]
- Żabicka, D.; Literacka, E. MDR, XDR, PDR-jednolite, międzynarodowe definicje nabytej oporności drobnoustrojów na antybiotyki. Aktual. Nar. Programu Ochr. Antybiot. 2012, 3, 1–7. [Google Scholar]
- Sotgiu, G.; Ferrara, G.; Matteelli, A.; Richardson, M.D.; Centis, R.; Ruesch-Gerdes, S.; Toungoussova, O.; Zellweger, J.-P.; Spanevello, A.; Cirillo, D.; et al. Epidemiology and clinical management of XDR-TB: A systematic review by TBNET. Eur. Respir. J. 2009, 33, 871–881. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention (CDC). Vital signs: Carbapenem-resistant Enterobacteriaceae. MMWR Morb. Mortal. Wkly. Rep. 2013, 62, 165–170. [Google Scholar]
- Patel, G.; Huprikar, S.; Factor, S.H.; Jenkins, S.G.; Calfee, D.P. Outcomes of carbapenem-resistant Klebsiella pneumoniae infection and the impact of antimicrobial and adjunctive therapies. Infect. Control. Hosp. Epidemiol. 2008, 29, 1099–1106. [Google Scholar] [CrossRef]
- Bauer, A.W.; Kirby, W.M.; Sherris, J.C.; Truck, M. Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 1966, 45, 493–496. [Google Scholar] [CrossRef]
- Picard, J. Applied Veterinary Bacteriology and Mycology: Bacteriological Techniques; University of Pretoria, Afrivip: Pretoria, South Africa, 1990. [Google Scholar]
- Liu, B.; Pop, M. ARDB—Antibiotic Resistance Genes Database. Nucleic. Acids Res. 2009, 37, D443–D447. [Google Scholar] [CrossRef]
- Yin, X.; Jiang, X.-T.; Chai, B.; Li, L.; Yang, Y.; Cole, J.R.; Tiedje, J.M.; Zhang, T. ARGs-OAP v2.0 with an expanded SARG database and Hidden Markov Models for enhancement characterization and quantification of antibiotic resistance genes in environmental metagenomes. Bioinformatics 2018, 34, 2263–2270. [Google Scholar] [CrossRef]
- McArthur, A.G.; Weglechner, N.; Nizam, F.; Yan, A.; Azad, M.A.; Baylay, A.J.; Bhullar, K.; Canova, M.J.; De Pascale, G.; Ejim, L.; et al. The comprehensive antibiotic resistance database. Antimicrob. Agents Chemother. 2013, 57, 3348–3357. [Google Scholar] [CrossRef]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Voldby Larsen, M. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef]
- Wang, Y.R.; Chen, Q.; Cui, S.H.; Li, F.Q. Characterization of Staphylococcus aureus isolated from clinical specimens by matrix assisted laser desorption/ionization time-of-flight mass spectrometry. Biomed. Environ. Sci. 2013, 26, 430–436. [Google Scholar]
- Rhoads, D.D.; Wang, H.; Karichu, J.; Richter, S.S. The presence of a single MALDI-TOF mass spectral peak predicts methicillin resistance in staphylococci. Diagn. Microbiol. Infect. Dis. 2016, 86, 257–261. [Google Scholar] [CrossRef]
- Schuster, D.; Josten, M.; Janssen, K.; Bodenstein, I.; Albert, C.; Schallenberg, A.; Gajdiss, M.; Sib, E.; Szekat, C.; Kehl, K.; et al. Detection of methicillin-resistant coagulase-negative staphylococci harboring the class A mec complex by MALDI-TOF mass spectrometry. Int. J. Med. Microbiol. 2018, 308, 522–526. [Google Scholar] [CrossRef] [PubMed]
- Ho, P.-L.; Yau, C.-Y.; Ho, L.-Y.; Chen, J.H.K.; Lai, E.L.Y.; Lo, S.W.U.; Tse, C.W.S.; Chow, K.-H. Rapid detection of cfiA metallo-β-lactamase-producing Bacteroides fragilis by the combination of MALDI-TOF MS and CarbaNP. J. Clin. Pathol. 2017, 70, 868–873. [Google Scholar] [CrossRef] [PubMed]
- Nix, I.D.; Idelevich, E.A.; Strock, L.M.; Sparbier, K.; Drews, O.; Kostrzewa, M.; Becker, K. Detection of Methicillin Resistance in Staphylococcus aureus From Agar Cultures and Directly From Positive Blood Cultures Using MALDI-TOF Mass Spectrometry-Based Direct-on-Target Microdroplet Growth Assay. Front. Microbiol. 2020, 11, 232. [Google Scholar] [CrossRef] [PubMed]
- Idelevich, E.A.; Storck, L.M.; Sparbier, K.; Drews, O.; Kostrzewa, M.; Becker, K. Rapid Direct Susceptibility Testing from Positive Blood Cultures by the Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry-Based Direct-on-Target Microdroplet Growth Assay. J. Clin. Microbiol. 2018, 56, e00913–e00918. [Google Scholar] [CrossRef]
- Paskova, V.; Chudejova, K.; Sramkova, A.; Kraftova, L.; Jakubu, V.; Petinaki, E.A.; Zamlickova, H.; Neradova, K.; Papagiannitsis, C.C.; Hrabak, J. Insufficient repeatability and reproducibility of MALDI-TOF MS-based identification of MRSA. Folia Microbiol. 2020, 65, 895–900. [Google Scholar] [CrossRef]
- Li, M.; Liu, M.; Song, Q.; Xiong, L.; Chen, Z.; Kang, M.; Xie, Y. Rapid antimicrobial susceptibility testing by matrix-assisted laser desorption ionization–time of flight mass spectrometry using a qualitative method in Acinetobacter baumannii complex. J. Microbiol. Methods 2018, 153, 60–65. [Google Scholar] [CrossRef]
- Watkins, R.R.; David, M.Z.; Salata, R.A. Current concepts on the virulence mechanisms of meticillin-resistant Staphylococcus aureus. J. Med. Microbiol. 2012, 61, 1179–1193. [Google Scholar] [CrossRef]
- Liu, Y.-Y.; Chandler, C.E.; Leung, L.M.; McElheny, C.L.; Mettus, R.T.; Shanks, R.M.Q.; Liu, J.-H.; Goodlett, D.R.; Ernst, R.K.; Doi, Y. Structural modification of lipopolysaccharide conferred by mcr-1 in gram-negative ESKAPE pathogens. Antimicrob. Agents Chemother. 2017, 61, e00580-17. [Google Scholar] [CrossRef]
- Lopalco, P.; Stahl, J.; Annese, C.; Averhoff, B.; Corcelli, A. Identification of unique cardiolipin and monolysocardiolipin species in Acinetobacter baumannii. Sci. Rep. 2017, 7, 2972. [Google Scholar] [CrossRef]
- Bisignano, C.; Ginestra, G.; Smeriglio, A.; La Camera, E.; Crisafi, G.; Franchina, F.A.; Tranchida, P.Q.; Alibrandi, A.; Trombetta, D.; Mondello, L.; et al. Study of the lipid profile of ATCC and clinical strains of staphylococcus aureus in relation to their antibiotic resistance. Molecules 2019, 24, 1276. [Google Scholar] [CrossRef]
- Sparbier, K.; Schubert, S.; Kostrzewa, M. MBT-ASTRA: A suitable tool for fast antibiotic susceptibility testing? Methods 2016, 104, 48–54. [Google Scholar] [CrossRef]
- Hrabák, J.; Walková, R.; Študentová, V.; Chudáčková, E.; Bergerová, T. Carbapenemase activity detection by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 2011, 49, 3222–3227. [Google Scholar] [CrossRef]
- Burckhardt, I.; Zimmermann, S. Using matrix-assisted laser desorption ionization-time of flight mass spectrometry to detect carbapenem resistance within 1 to 2.5 hours. J. Clin. Microbiol. 2011, 49, 3321–3324. [Google Scholar] [CrossRef]
- Ota, Y.; Furuhashi, K.; Hirai, N.; Ishikawa, J.; Nagura, O.; Yamanaka, K.; Maekawa, M. Evaluation of MBT STAR-Cepha and MBT STAR-Carba kits for the detection of extended-spectrum β-lactamases and carbapenemase producing microorganisms using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J. Microbiol. Methods 2021, 183, 106166. [Google Scholar] [CrossRef]
- Anantharajah, A.; Tossens, B.; Olive, N.; Kabamba-Mukadi, B.; Rodriguez-Villalobos, H.; Verroken, A. Performance Evaluation of the MBT STAR®-Carba IVD Assay for the Detection of Carbapenemases With MALDI-TOF MS. Front. Microbiol. 2019, 10, 1413. [Google Scholar] [CrossRef]
- Vogel, N.; Schiebel, K.; Humeny, A. Technologies in the Whole-Genome Age: MALDI-TOF-Based Genotyping. Transfus. Med. Hemotherapy 2009, 36, 253. [Google Scholar] [CrossRef]
- Sauer, S. Typing of single nucleotide polymorphisms by MALDI mass spectrometry: Principles and diagnostic applications. Clin. Chim. Acta 2006, 363, 95–105. [Google Scholar] [CrossRef][Green Version]
- Griffin, T.J.; Smith, L.M. Single-nucleotide polymorphism analysis by MALDI–TOF mass spectrometry. Trends Biotechnol. 2000, 18, 77–84. [Google Scholar] [CrossRef]
- Lau, A.F.; Wang, H.; Weingarten, R.A.; Drake, S.K.; Suffredini, A.F.; Garfield, M.K.; Chen, Y.; Gucek, M.; Youn, J.-H.; Stock, F.; et al. A rapid Matrix-Assisted Laser Desorption Ionization–Time of Flight Mass Spectrometry-Based method for single-plasmid tracking in an outbreak of carbapenem-resistant Enterobacteriaceae. J. Clin. Microbiol. 2014, 52, 2804. [Google Scholar] [CrossRef]
- Cordovana, M.; Kostrzewa, M.; Glandorf, J.; Bienia, M.; Ambretti, S.; Pranada, A.B. A full MALDI-based approach to detect plasmid-encoded KPC-producing klebsiella pneumoniae. Front. Microbiol. 2018, 9, 2854. [Google Scholar] [CrossRef]
- Rybicka, M.; Miłosz, E.; Bielawski, K.P. Superiority of MALDI-TOF Mass Spectrometry over Real-Time PCR for SARS-CoV-2 RNA Detection. Viruses 2021, 13, 730. [Google Scholar] [CrossRef]
- Ellis, J.A.; Ong, B. The MassARRAY® system for targeted SNP genotyping. Methods Mol. Biol. 2017, 1492, 77–94. [Google Scholar]
- Shi, J.; He, G.; Ning, H.; Wu, L.; Wu, Z.; Ye, X.; Qiu, C.; Jiang, X. Application of matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) in the detection of drug resistance of Mycobacterium tuberculosis in re-treated patients. Tuberculosis 2022, 135, 102209. [Google Scholar] [CrossRef]
- Pu, L.; Jian, Z.; Pan, F.; Geng, Y.; He, M.; Liao, P. Comparative genomic analysis and multi-drug resistance differences of Acinetobacter baumannii in Chongqing, China. Infect. Drug Resist. 2019, 12, 2827. [Google Scholar] [CrossRef]
- Suzuki, S.; Horinouchi, T.; Furusawa, C. Prediction of antibiotic resistance by gene expression profiles. Nat. Commun. 2014, 5, 5792. [Google Scholar] [CrossRef]
- Ikryannikova, L.N.; Shitikov, E.A.; Zhivankova, D.G.; Il’ina, E.N.; Edelstein, M.V.; Govorun, V.M. A MALDI TOF MS-based minisequencing method for rapid detection of TEM-type extended-spectrum beta-lactamases in clinical strains of Enterobacteriaceae. J. Microbiol. Methods 2008, 75, 385–391. [Google Scholar] [CrossRef]
- Stewart, P.S. Antimicrobial Tolerance in Biofilms. Microbiol. Spectr. 2015, 3, 1–30. [Google Scholar]
- Vu, B.; Chen, M.; Crawford, R.J.; Ivanova, E.P. Bacterial extracellular polysaccharides involved in biofilm formation. Molecules 2009, 14, 2535. [Google Scholar] [CrossRef]
- Houari, A.; Picard, J.; Habarou, H.; Galas, L.; Vaudry, H.; Heim, V.; Di Martino, P. Rheology of biofilms formed at the surface of NF membranes in a drinking water production unit. Biofouling 2008, 24, 235–240. [Google Scholar] [CrossRef]
- Chen, M.; Yu, Q.; Sun, H. Novel strategies for the prevention and treatment of biofilm related infections. Int. J. Mol. Sci. 2013, 14, 18488–18501. [Google Scholar] [CrossRef]
- Darouiche, R.O. Treatment of infections associated with surgical implants. N. Engl. J. Med. 2009, 350, 1422–1429. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, M.; Monteiro, F.J.; Ferraz, M.P. Infection of orthopedic implants with emphasis on bacterial adhesion process and techniques used in studying bacterial-material interactions. Biomatter 2012, 2, 176–194. [Google Scholar] [CrossRef] [PubMed]
- Kaya, E.; Grassi, L.; Benedetti, A.; Maisetta, G.; Pileggi, C.; Di Luca, M.; Batoni, G.; Esin, S. In vitro interaction of Pseudomonas aeruginosa biofilms with human peripheral blood mononuclear cells. Front. Cell Infect. Microbiol. 2020, 10, 187. [Google Scholar] [CrossRef] [PubMed]
- Høiby, N.; Bjarnsholt, T.; Givskov, M.; Molin, S.; Ciofu, O. Antibiotic resistance of bacterial biofilms. Int. J. Antimicrob. Agents 2010, 35, 322–332. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Singh, S.K.; Chowdhury, I.; Singh, R. Understanding the mechanism of bacterial biofilms resistance to antimicrobial agents. Open Microbiol. J. 2017, 11, 53. [Google Scholar] [CrossRef] [PubMed]
- Zambrano, M.M.; Kolter, R. Mycobacterial biofilms: A greasy way to hold it together. Cell 2005, 123, 762–764. [Google Scholar] [CrossRef]
- Abidi, S.H.; Sherwani, S.K.; Siddiqui, T.R.; Bashir, A.; Kazmi, S.U. Drug resistance profile and biofilm forming potential of Pseudomonas aeruginosa isolated from contact lenses in Karachi-Pakistan. BMC Ophthalmol. 2013, 13, 57. [Google Scholar] [CrossRef]
- Amin, M.; Pai, V.; Qayoom, S.; Arshi, S.; Khurshid, S. Biofilm formation and multidrug resistance in nosocomial isolates of Acinetobacter. Indian J. Microbiol. Res. 2018, 5, 425. [Google Scholar]
- Manandhar, S.; Singh, A.; Varma, A.; Pandey, S.; Shrivastava, N. Biofilm producing clinical Staphylococcus aureus isolates augmented prevalence of antibiotic resistant cases in tertiary care hospitals of Nepal. Front. Microbiol. 2018, 9, 2749. [Google Scholar] [CrossRef]
- Cepas, V.; López, Y.; Muñoz, E.; Rolo, D.; Ardanuy, C.; Martí, S.; Xervavins, M.; Horcajada, J.P.; Bosch, J.; Soto, S.M. Relationship between biofilm formation and antimicrobial resistance in Gram-negative bacteria. Microb. Drug Resist. 2019, 25, 72–79. [Google Scholar] [CrossRef]
- Avila-Novoa, M.G.; Solís-Velázquez, O.A.; Rangel-López, D.E.; González-Gómez, J.P.; Guerrero-Medina, P.J.; Gutiérrez-Lomelí, M. Biofilm formation and detection of fluoroquinolone-and carbapenem-resistant genes in multidrug-resistant Acinetobacter baumannii. Can. J. Infect. Dis. Med. Microbiol. 2019, 2019, 3454907. [Google Scholar] [CrossRef]
- Caputo, P.; Di Martino, M.C.; Perfetto, B.; Iovino, F.; Donnarumma, G. Use of MALDI-TOF MS to discriminate between biofilm-producer and non-producer strains of Staphylococcus epidermidis. Int. J. Environ. Res. Public Health 2018, 15, 1695. [Google Scholar] [CrossRef]
- Li, B.; Comi, T.J.; Si, T.; Dunham, S.J.B.; Sweedler, J.V. A one-step matrix application method for MALDI mass spectrometry imaging of bacterial colony biofilms. J. Mass Spectrom. 2016, 51, 1030–1035. [Google Scholar] [CrossRef]
- Pauter, K.; Railean-Plugaru, V.; Złoch, M.; Pomastowski, P.; Szultka-Młyńska, M.; Buszewski, B. Identification, structure and characterization of Bacillus tequilensis biofilm with the use of electrophoresis and complementary approaches. J. Clin. Med. 2022, 11, 722. [Google Scholar] [CrossRef]
- Si, T.; Li, B.; Zhang, K.; Xu, Y.; Zhao, H.; Sweedler, J.V. Characterization of Bacillus subtilis colony biofilms via mass spectrometry and fluorescence imaging. J. Proteome Res. 2016, 15, 1955–1962. [Google Scholar] [CrossRef]
- Pereira, F.D.E.S.; Bonatto, C.C.; Lopes, C.A.P.; Pereira, A.L.; Silva, L.P. Use of MALDI-TOF mass spectrometry to analyze the molecular profile of Pseudomonas aeruginosa biofilms grown on glass and plastic surfaces. Microb. Pathog. 2015, 86, 32–37. [Google Scholar] [CrossRef]
- De Carolis, E.; Soldini, S.; La Rosa, M.; Nucci, F.; Posteraro, B.; Sanguinetti, M. BIOF–HILO assay: A new MALDI–TOF mass spectrometry based method for discriminating between high- and low-biofilm-producing Candida parapsilosis isolates. Front. Microbiol. 2019, 10, 2046. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Janiszewska, D.; Szultka-Młyńska, M.; Pomastowski, P.; Buszewski, B. “Omic” Approaches to Bacteria and Antibiotic Resistance Identification. Int. J. Mol. Sci. 2022, 23, 9601. https://doi.org/10.3390/ijms23179601
Janiszewska D, Szultka-Młyńska M, Pomastowski P, Buszewski B. “Omic” Approaches to Bacteria and Antibiotic Resistance Identification. International Journal of Molecular Sciences. 2022; 23(17):9601. https://doi.org/10.3390/ijms23179601
Chicago/Turabian StyleJaniszewska, Daria, Małgorzata Szultka-Młyńska, Paweł Pomastowski, and Bogusław Buszewski. 2022. "“Omic” Approaches to Bacteria and Antibiotic Resistance Identification" International Journal of Molecular Sciences 23, no. 17: 9601. https://doi.org/10.3390/ijms23179601
APA StyleJaniszewska, D., Szultka-Młyńska, M., Pomastowski, P., & Buszewski, B. (2022). “Omic” Approaches to Bacteria and Antibiotic Resistance Identification. International Journal of Molecular Sciences, 23(17), 9601. https://doi.org/10.3390/ijms23179601







